Abstract
Full activation of T cells and differentiation into effector T cells are essential for many immune responses and require co-stimulatory signaling via the CD28 receptor. Extracellular ligand binding to CD28 recruits protein-tyrosine kinases to its cytoplasmic tail, which contains a YMNM motif. Following phosphorylation of the tyrosine, the proteins growth factor receptor-bound protein 2 (Grb2), Grb2-related adaptor downstream of Shc (Gads), and p85 subunit of phosphoinositide 3-kinase may bind to pYMNM (where pY is phosphotyrosine) via their Src homology 2 (SH2) domains, leading to downstream signaling to distinct immune pathways. These three adaptor proteins bind to the same site on CD28 with variable affinity, and all are important for CD28-mediated co-stimulatory function. However, the mechanism of how these proteins recognize and compete for CD28 is unclear. To visualize their interactions with CD28, we have determined the crystal structures of Gads SH2 and two p85 SH2 domains in complex with a CD28-derived phosphopeptide. The high resolution structures obtained revealed that, whereas the CD28 phosphopeptide bound to Gads SH2 is in a bent conformation similar to that when bound to Grb2 SH2, it adopts a more extended conformation when bound to the N- and C-terminal SH2 domains of p85. These differences observed in the peptide-protein interactions correlated well with the affinity and other thermodynamic parameters for each interaction determined by isothermal titration calorimetry. The detailed insight into these interactions reported here may inform the development of compounds that specifically inhibit the association of CD28 with these adaptor proteins to suppress excessive T cell responses, such as in allergies and autoimmune diseases.
Keywords: crystal structure, isothermal titration calorimetry (ITC), protein-protein interaction, signal transduction, Src homology 2 domain (SH2 domain)
Introduction
For complete activation of T cells and differentiation into effector T cells, co-stimulatory signals via CD28 receptors are required in addition to signals via T cell receptors (1, 2). Binding of ligands such as CD80 (B7-1) and CD86 (B7-2) to CD28 on its extracellular surface recruits protein-tyrosine kinases to the CD28 cytoplasmic tail, which consists of 41 amino acid residues and a phosphorylated Tyr189 within a YMNM motif (3). Following phosphorylation, CD28 recruits growth factor receptor-bound protein 2 (Grb2),4 Grb2-related adaptor downstream of Shc (Gads), and p85 subunit of phosphoinositide 3-kinase (PI3K) (4, 5). These adaptor molecules primarily bind to the sequence pYMNM (where pY is phosphotyrosine) in CD28 via their SH2 domains. As one of the downstream signals of CD28-mediated co-stimulatory activity, Grb2 stimulates the CD28-mediated IL-2 promoter activation through the Vav1-NFAT/AP-1 signaling pathway (6). The binding of Gads to CD28 is essential for the CD28-mediated NF-κB activation (7–9). The regulatory subunit of PI3K, p85, has both positive and negative effects on CD28-mediated co-stimulation (10).
Both Grb2 and Gads contain a central SH2 domain flanked by two SH3 domains with Gads having an extra Pro/Glu-rich sequence between the SH2 domain and the C-terminal SH3 domain (11). Grb2 is expressed ubiquitously in mammalian cells, whereas Gads is expressed predominantly in lymphoid tissue and hematopoietic cells, particularly T cells (12–14). The p85 subunit of PI3K consists of an SH3 domain, a RhoGAP domain, and two SH2 domains, which are designated as nSH2 and cSH2 for the N- and C-terminal SH2 domains, respectively. The consensus SH2-binding sequence of Grb2 and Gads is pYXNX, whereas that of p85, either nSH2 or cSH2, is pYXXM. Although these three adaptor molecules activate different signaling pathways downstream of CD28 as mentioned above, the SH2 domains of all the molecules can bind to CD28 at the same site, YMNM, after the phosphorylation of its first Tyr. Because our previous report showed that both nSH2 and cSH2 of p85 have a higher affinity to pYMNM than Grb2 and Gads (15), it is likely that the high affinity of p85 SH2 to CD28 results in a sequestration of CD28 from other SH2 domains, although we need to know the relative concentration of the proteins to draw such a conclusion. However, both Grb2-CD28 and Gads-CD28 interactions are also important for CD28-mediated co-stimulation. It remains unclear how these three adaptor molecules structurally bind to CD28 and elicit the proper co-stimulatory function of CD28.
We have recently reported the crystal structure of Grb2 SH2 complexed with a phosphotyrosine-containing peptide derived from CD28 (16) and that the peptide adopted the twisted U-type conformation. Here, we report the crystal structures and the thermodynamic analyses of Gads SH2, p85 nSH2, and cSH2 in complex with the phosphotyrosine-containing peptide derived from CD28. The structures reveal the bent conformation of CD28-derived peptide in Gads SH2, whereas the extended conformation is observed in both p85 nSH2 and cSH2. The conformational variations of the peptide together with the detailed interaction manner between the peptide and SH2s confirm the results of the thermodynamic analyses. High resolution structures of 1.20, 0.90, and 1.10 Å for Gads SH2, p85 nSH2, and cSH2, respectively, reported here should also be good templates for further computational analysis, such as molecular dynamics simulations and rational design of new ligands.
Results
Protein Expression and Crystallization
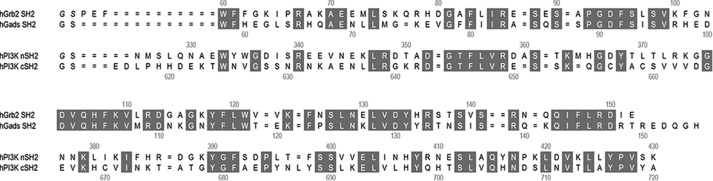
All the SH2 domains analyzed in this study were expressed as glutathione S-transferase (GST) fusion proteins but purified in GST-free form after the thrombin digestion. The sequence homology between Gads SH2 and Grb2 SH2 is 55.9%, and that between p85 nSH2 and cSH2 is 35.2% (Fig. 1). Our first construct for the Gads SH2 (Trp58–His155) expression has an additional 5 residues, GSPEF, at the N terminus that is a cloning artifact and remained attached even after digestion by thrombin, similar to the construct for Grb2 SH2 whose crystal structure was previously determined at 1.35-Å resolution (16). Although the protein was successfully crystallized in complex with CD28-derived 8-residue phosphopeptide, SDpYMNMTP, designated as OctP, its diffractions were limited to a relatively low resolution (∼3 Å). To improve the resolution, we expressed Gads SH2 with the C-terminal 6 residues (Arg150–His155) deleted, but only the free form of Gads SH2 was crystallized (data not shown). Our final construct has shorter N-terminal additional residues, Gly-Ser, and the original C terminus, His155, which produced high quality crystals in complex with OctP. The additional sequence, Gly-Ser, was necessary because of thrombin digestion. The modification of N and C termini of Gads SH2 described above had little effect on its CD28 binding (data not shown). Consequently, a similar approach was taken for both p85 nSH2 (Asn325–Lys430) and cSH2 (Glu614–Ala720) where the proteins were expressed with two additional residues, Gly-Ser, at the N terminus and co-crystallized with Oct-P.
FIGURE 1.
The amino acid sequences of SH2 domains and comparison of the backbone structures. The sequence alignments of Grb2 SH2, Gads SH2, p85 nSH2, and p85 cSH2 are shown. The gray boxes indicate the sequence similarity between Grb2 and Gads or nSH2 and cSH2, respectively. The sequence homology and the root mean square deviations of the main chain are given in the text.
For crystallization experiments, all the SH2 domains were concentrated to about 1 mm in 10 mm phosphate buffer (pH 7.5). Freeze-dried OctP was suspended in the same buffer, and the suspended solution was added to SH2 solutions in apparent molar ratios of ∼2:1. Crystals of Gads SH2, p85 nSH2, and cSH2 in complex with OctP were obtained from the following screening buffers: 100 mm HEPES-NaOH (pH 7.5), 10% (w/v) polyethylene glycol (PEG) 6000, and 20% (v/v) (±)-2-methyl-2,4-pentanediol for Gads SH2-OctP complex; 100 mm sodium acetate trihydrate (pH 4.6), 30% (w/v) PEG monomethyl ether 2000, and 200 mm ammonium sulfate for p85 nSH2-OctP complex; and 100 mm sodium acetate trihydrate (pH 4.6), 30% (w/v) PEG 4000, and 200 mm ammonium acetate for p85 cSH2-OctP complex. The crystals appeared within 2 weeks and grew with maximum dimensions as follows: 0.1 × 0.2 × 0.4 mm for Gads SH2-OctP, 0.1 × 0.1 × 0.1 mm for p85 nSH2-OctP, and 0.1 × 0.2 × 1 mm for p85 cSH2-OctP.
Crystal Structures of CD28-SH2 Complexes
The crystal structure of Gads SH2 complexed with OctP was determined at 1.20-Å resolution (Fig. 2A and Table 1). The overall structure of Gads SH2 consists of a central antiparallel β-sheet flanked by two α-helices, which is similar to that of Grb2 and other SH2 domains (Fig. 2B) reported previously (17). The crystal structures of p85 nSH2 and cSH2 complexed with OctP were also determined at 0.90- and 1.10-Å resolution, respectively (Fig. 2, C and D, and Table 1). These complexes also show similar structures to that of Gads SH2 (Fig. 2, E and F). The root mean square deviations of main-chain structures between Grb2 SH2 and Gads SH2, Gads SH2 and p85 nSH2, and p85 nSH2 and cSH2 are 0.428, 1.320, and 0.875 Å, respectively. Despite the high similarity of the overall structures, the recognition mechanism of the phosphopeptide is quite different between Gads SH2 and p85 SH2 domains, probably reflecting the differences in consensus amino acid sequences, as described below.
FIGURE 2.
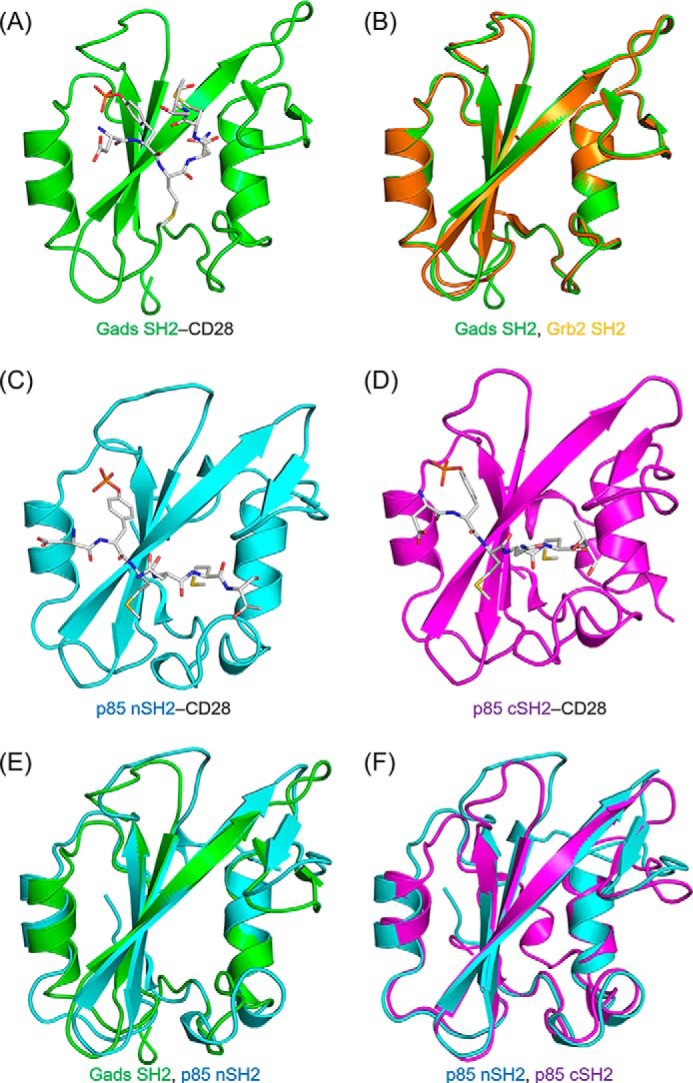
Overall structures and superposition of SH2 domain complexes with phosphopeptides. The structures of the complexes of phosphopeptide with Gads SH2 (A), p85 nSH2 (C), and p85 cSH2 (D), respectively, are shown. The SH2 domains are shown as ribbon representations, whereas the CD28-derived phosphopeptides (sequence SDpYMNMTP; OctP) are shown as gray stick representations. The superpositions of Grb2 SH2 and Gads SH2 (B), Gads SH2 and p85 nSH2 (E), and p85 nSH2 and p85 cSH2 (F) are also shown. The structure of Grb2 SH2 complexed with CD28-derived phosphopeptide was determined previously (Ref. 16; Protein Data Bank code 3WA4). The backbone structures of Grb2 SH2, Gads SH2, p85 nSH2, and p85 cSH2 are colored in orange, green, cyan, and pink, respectively.
TABLE 1.
Data collection and refinement statistics
Values in parentheses are for the highest resolution shell. r.m.s., root mean square.
| Gads-OctP | nSH2-OctP | cSH2-OctP | |
|---|---|---|---|
| Data collection | |||
| Wavelength (Å) | 0.7000 | 0.9800 | 0.9800 |
| Space group | P212121 | P1 | P212121 |
| Unit cell parameters | |||
| a/b/c (Å) | 33.60/53.34/108.30 | 27.59/31.24/32.25 | 40.60/41.44/64.85 |
| α/β/γ (°) | 73.28/85.38/71.73 | ||
| Resolution (Å) | 50-1.20 (1.27-1.20) | 30.9-0.90 (0.95-0.90) | 50-1.10 (1.13-1.10) |
| No. of observations | 403,340 | 359,798 | 385,500 |
| No. of unique reflections | 61,484 | 65,076 | 45,133 |
| Completeness (%) | 99.3 (99.3) | 89.5 (81.1) | 100 (100) |
| Average I/σ(I) | 14.5 (3.7) | 18.1 (4.0) | 17.7 (6.3) |
| Redundancy | 6.6 (6.7) | 5.5 (3.9) | 8.5 (6.9) |
| Rsyma (%) | 5.9 (38.9) | 5.5 (34.2) | 6.8 (31.9) |
| Refinement | |||
| Rb (%) | 14.8 | 12.6 | 13.8 |
| Rfreec (%) | 17.0 | 14.5 | 16.0 |
| Number of atoms | |||
| Protein | 1,833 | 1,066 | 1,105 |
| Sulfate | 15 | ||
| Glycerol | 6 | 6 | |
| Water | 247 | 193 | 184 |
| Average B factor (Å2) | 18.0 | 10.0 | 14.0 |
| r.m.s. deviation from ideal | |||
| Bonds (Å) | 0.013 | 0.007 | 0.017 |
| Angles (°) | 1.6 | 1.4 | 1.9 |
| Ramachandran plot | |||
| Favored region (%) | 97.64 | 96.92 | 98.55 |
| Allowed region (%) | 2.36 | 3.08 | 1.45 |
| Outlier region (%) | 0 | 0 | 0 |
a Rsym = ΣΣi|I(h) − I(h)i|/ΣΣiI(h) where I(h) is the mean intensity after rejection.
b R = Σ|Fo − Fc|/Σ|Fo| where Fo is the observed structure factor amplitude and Fc is the calculated structure factor amplitude.
c Rfree is the same as R but was calculated using a random set containing 5% of the data that were excluded during refinement.
The recognition mode of Gads SH2 for CD28 phosphopeptide is very similar to that of Grb2 SH2. The side chains of Arg65, Arg83, Ser87, and Ser93 together with the main chain of Gln86 are involved in a salt bridge and hydrogen bonding with the phosphate group (Fig. 3A). The main chains of Lys106 and Leu117 interact with Asn at pY+2 via hydrogen bonding (Fig. 3B). The bound OctP adopts a bent conformation (Fig. 3C), similar to a type I β-turn but different from the canonical conformation, as reported previously in Grb2 SH2-peptide complex (16). The bulky side chain of Trp121 in Grb2 SH2 is considered to force the bound peptide into the bent conformation. Because this Trp residue is conserved as Trp118 in Gads SH2 and its side chain orientation is stabilized by the cation-π interaction with another conserved residue, Arg139, OctP is also forced to bend in the complex with Gads SH2 to avoid steric hindrance.
FIGURE 3.
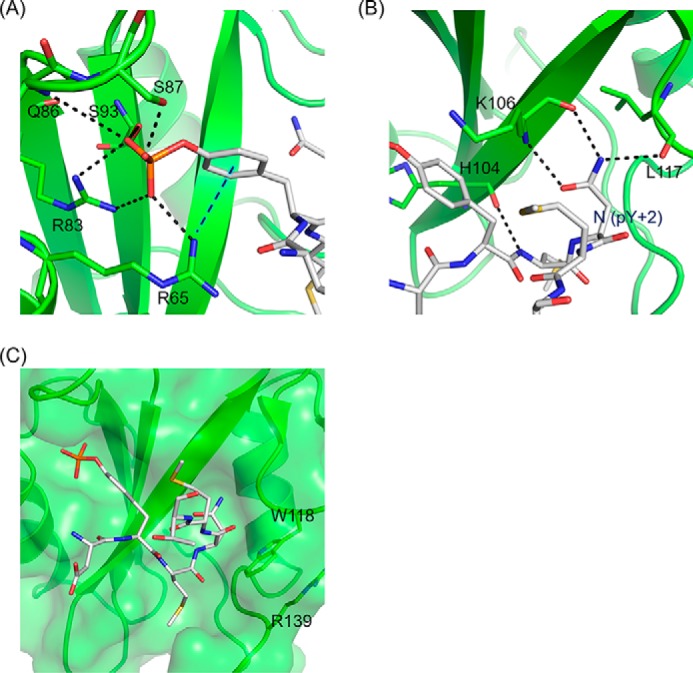
The site-specific recognition modes of Gads SH2. A, the interactions of phosphotyrosine of CD28-derived phosphopeptide with Gads SH2. B, the interactions (salt bridges and hydrogen bonds) of the conserved Asn (pY+2) of CD28-derived phosphopeptide and Gads SH2. C, SH2 domain is shown in ribbon and surface representation, and CD28-derived phosphopeptide is shown in stick representation. The black dashed lines indicate the salt bridges and hydrogen bonds. The blue dashed line indicates cation-π interaction between CD28-derived phosphopeptide and SH2.
In contrast, OctP is in a more extended conformation in the crystal structures of p85 nSH2 and cSH2 complexes as we modeled previously (16). The residue Asn at pY+2 is exposed to the solvent and does not interact with any residues in either p85 nSH2 or cSH2 (Fig. 4, A and B). Instead, the side chain of Met at pY+3, which is oriented toward the solvent in the structure of Gads SH2 complexed with OctP, sticks into the hydrophobic pocket of both p85 nSH2 and p85 cSH2 (Fig. 4, A and B). The pockets are formed by Ile381, Phe392, Leu413, Tyr416, and Leu420 in p85 nSH2 and corresponding residues of Cys670, Phe681, Leu703, His706, and Leu710 in p85 cSH2, respectively. Interestingly, Met at pY+3 occupies almost the same position for Trp118 of Gads SH2. There is neither a Trp nor a bulky residue at this position in both p85 nSH2 and cSH2 as the corresponding residue is Ser393 and Ala682 for p85 nSH2 and cSH2, respectively. The lack of bulky residue and slight conformational changes in this region allow Met at pY+3 to reach the hydrophobic pocket (Fig. 4, C and D). The absence of a bulky residue in p85 nSH2 and cSH2 also allows OctP to adopt extended conformations. The two SH2 domains in p85 show a similar molecular recognition toward the phosphate group of the phosphotyrosine; the salt bridge and hydrogen bonding to the phosphate group are formed with the side chains of Arg340, Arg358, Ser361, and Thr369 in p85 nSH2 (Fig. 4E) and with the side chains of Arg631, Arg649, Ser651, and Ser652 in p85 cSH2. However, p85 cSH2 has an additional unique cation-π interaction between the side chain of Arg631 and aromatic ring of the phosphotyrosine (Fig. 4F), which is not observed in p85 nSH2. This additional interaction seems to contribute to binding of the phosphotyrosine residue more tightly than in p85 nSH2.
FIGURE 4.
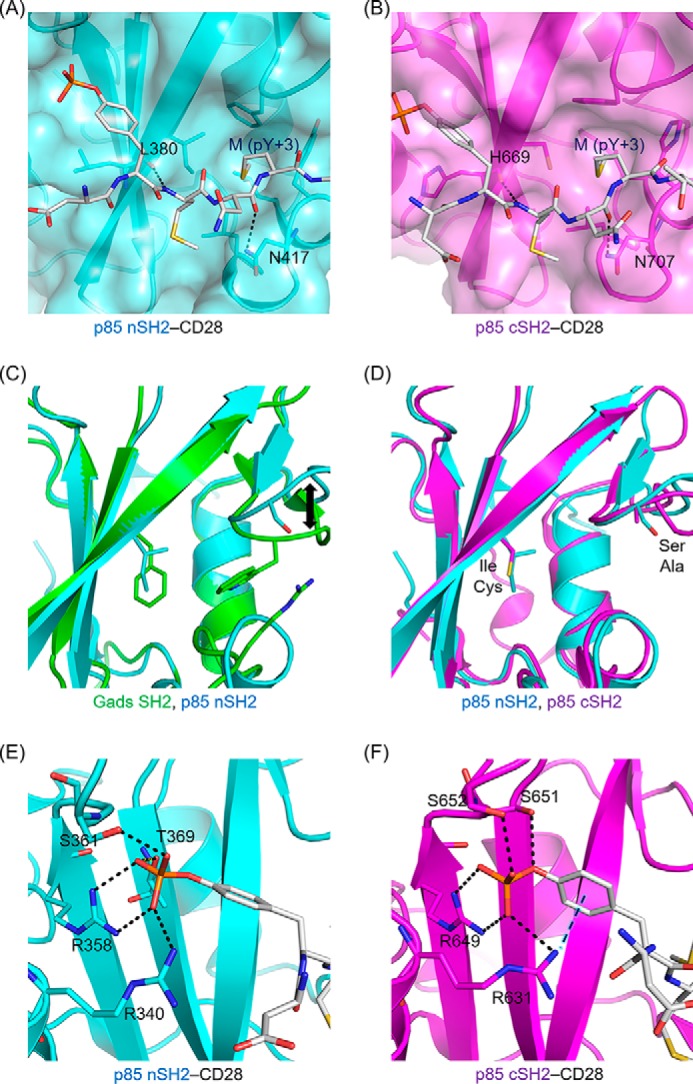
The site-specific recognition modes of p85 nSH2 and p85 cSH2. Shown are the interactions (salt bridges, hydrogen bonds, and hydrophobic contacts) of the conserved Met (pY+3) of CD28-derived phosphopeptide with p85 nSH2 (A) and p85 cSH2 (B). The superposition of Gads SH2 and p85 nSH2 (C) and p85 nSH2 and p85 cSH2 (D) are shown. The residues located near the entrance of hydrophobic core are shown in stick representations (see text). The interactions of the phosphotyrosine residue of CD28-derived phosphopeptide with p85 nSH2 (E) and p85 cSH2 (F) are shown. The SH2 domain is shown in ribbon and surface representation, and CD28-derived phosphopeptide is shown in stick representation. The black dashed lines indicate the salt bridges and hydrogen bonds. The blue dashed line indicates cation-π interaction between CD28-derived phosphopeptide and SH2. The backbone structures of Gads SH2, p85 nSH2, and p85 cSH2 are colored in green, cyan, and pink, respectively.
Binding Thermodynamics of CD28 Interaction with SH2 Domains
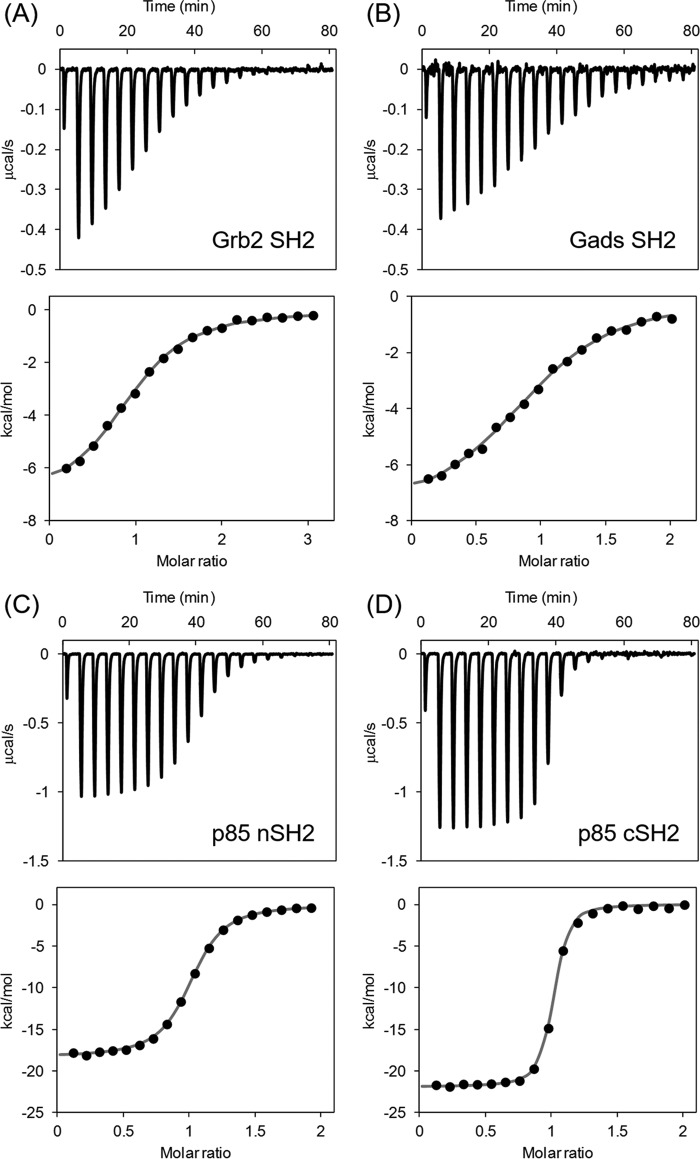
The interactions of CD28-derived 12-amino acid phosphopeptide SDpYMNMTPRRP, designated as DdcP, with SH2 domains of Gads, Grb2, and p85 were analyzed using isothermal titration calorimetry (ITC). Because of the low solubility of OctP, more soluble DdcP was used for the ITC experiments. Fig. 5, A–D, show the typical ITC profile of CD28 binding to SH2 domains, and the thermodynamic parameters are summarized in Table 2. All interactions between CD28 and SH2 domains showed exothermic reactions. The binding affinity of Gads SH2 was similar to that of Grb2 SH2, and their binding thermodynamics were comparable. The binding affinities of p85 nSH2 and cSH2 were higher than those of Grb2 SH2 and Gads SH2 mainly because of the favorable enthalpy change (ΔH). In comparison with the two SH2 domains of p85, the binding affinity of cSH2 was about 1 order of magnitude higher than that of nSH2 mainly because of the favorable ΔH. The heat capacity changes (ΔCp) for binding were successfully obtained from temperature-dependent ΔH changes in the range of 15–30 °C and are summarized in Table 2. The p85 cSH2 domain had the largest ΔCp value of the group.
FIGURE 5.
The typical isothermal titration calorimetric profiles of the interaction between CD28-derived peptide and Grb2-SH2 (A), Gads-SH2 (B), p85 nSH2 (C), and p85 cSH2 (D), respectively.
TABLE 2.
Thermodynamic parameters for interaction between CD28 phosphopeptide and SH2 domains of Grb2, Gads, and PI3K at 25 °C
| n | Ka | ΔG | ΔH | TΔS | ΔCpa | |
|---|---|---|---|---|---|---|
| /m | kcal/mol | kcal/mol K | ||||
| Grb2 SH2 | 0.97 | 4.39 × 105 | −7.76 | −7.25 | 0.51 | −0.202 |
| Gads SH2 | 0.96 | 4.04 × 105 | −7.64 | −7.75 | −0.10 | −0.149 |
| p85 nSH2 | 0.99 | 3.67 × 106 | −8.85 | −18.39 | −9.44 | −0.229 |
| nSH2 S393W | 0.97 | 1.75 × 106 | −8.51 | −16.61 | −8.10 | |
| p85 cSH2 | 0.97 | 2.15 × 107 | −10.00 | −21.86 | −11.86 | −0.244 |
| cSH2 A628W | 0.97 | 5.97 × 106 | −9.24 | −21.66 | −12.42 |
a The values were calculated from ΔH measured at four different temperatures between 15 and 30 °C.
The residues Ser393 in nSH2 and Ala628 in cSH2, corresponding to Trp118 in Gads, were mutated to Trp, and the binding thermodynamics of mutants to DdcP were analyzed using ITC (Table 2). Upon mutation, the binding affinities were slightly lower than those of wild types but still higher than those of Grb2 SH2 and Gads SH2.
Discussion
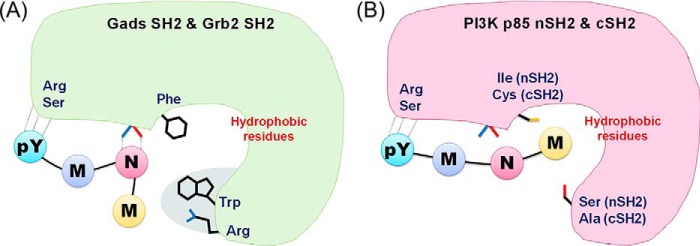
The crystal structure analyses of three SH2 domains, Gads SH2, p85 nSH2, and p85 cSH2, in complex with a CD28-derived peptide showed details of their peptide recognition, and the structures correlate well with their binding thermodynamics. The most striking result of the study is that the same CD28-peptide is recognized in a different manner, especially in the conformation of the bound peptide, despite the fact that they share high sequence similarity. In the case of p85 nSH2 and cSH2, the bound peptide showed an extended conformation, and Met at pY+3 position makes hydrophobic interactions with the protein (Fig. 6B). In contrast, Gads SH2 has a series of bulky residues, Phe105, Trp118, and Arg139, to force the phosphopeptide to adopt a bent conformation (Fig. 6A), similar to the conformation seen in Grb2 SH2 (16). As a result, Met at pY+3 loses contact with the protein, and Asn at pY+2, needed for the specific interaction, forms hydrogen bonds with the protein. Thr and Pro at pY+4 and pY+5 also make no contact with Grb2 SH2. The bent conformation and fewer contacts with less hydrophobic interaction between OctP peptide and Grb2/Gads SH2 would be the reason for the lower binding affinity than those in p85 nSH2 and cSH2 as determined by our ITC experiments (Table 2). The larger ΔCp values in p85 nSH2 and cSH2 also support the larger hydrophobic interactions (18, 19). Similar to the previous mutational study in Grb2 (20), our mutational study also supports the notion that the Trp residue, corresponding to Trp118 in Gads, is not the determining factor for the conformation of bound phosphopeptide (Table 2).
FIGURE 6.
Schematic views of recognition modes for Grb2 and Gads SH2 (A) and p85 nSH2 and cSH2 (B).
The binding affinity of p85 cSH2 was about 10-fold higher than that of p85 nSH2. This can be explained by the different recognition for the phosphotyrosine of CD28. In p85 cSH2, the side chain of Arg631 is located at the closed site of the phosphotyrosine side chain forming cation-π interaction (Fig. 4F). In p85 nSH2, the corresponding residue, Arg340, is located far from the phosphotyrosine (Fig. 4E). This additional interaction in cSH2 would contribute to the favorable binding enthalpy change. It should also be noted that the binding enthalpy change of p85 cSH2 is a significantly larger value compared with the other SH2 domains analyzed in this study and has been reported previously (21). The favorable ΔH is partially compensated by the unfavorable entropy change (ΔS), which is a general phenomenon of biomolecular interactions (22).
The binding thermodynamics of Grb2 SH2, p85 nSH2, and p85 cSH2 to phosphopeptides different from the CD28 peptide have been reported previously (21, 23, 24). The binding affinity of Grb2 SH2 to the Shc-derived phosphopeptide SpYVNVQ is about 10 times higher than that to DdcP02, SDpYMNMTPRRPG (24). The NMR structure of Grb2 SH2 in complex with Shc-derived peptide showed that the Shc peptide forms a β-turn (25), which is slightly different from the twisted conformation of CD28-peptide bound to Grb2 SH2. This would be mainly due to the residue at pY+3, Val of Shc peptide and Met of CD28 peptide. The side chain of Met would prevent the bound peptide from forming a canonical β-turn, resulting in the decreased binding affinity. The binding affinity of p85 nSH2 to platelet-derived growth factor receptor (PDGFR)-derived phosphopeptide SVDpYVDMSK is similar to that of the CD28 peptide (23), but the thermodynamic contribution is different from each other, which is also seen in case of p85 cSH2. In comparison with the binding of cSH2 to the PDGFR-derived phosphopeptide GGpYMDMSKDESVDYVPML, both enthalpy and entropy changes for the binding to the CD28 peptide are more negative, more favorable enthalpy change and less favorable entropy change (23). Within the consensus SH2-binding sequence, the residue at pY+2 is different in these peptides: Asp for both PDGFR peptides and Asn for CD28 peptide. Because the residue is exposed to the solvent in both PDGFR- and CD28-complexed structures (26), it can be excluded that their different thermodynamics is due to the difference in the interaction at the residue at pY+2. Instead, the difference in hydration might be because of the different numbers of water molecules located on the surface of SH2 and/or the peptide.
Based on binding affinities of respective SH2 domains to CD28, it is likely that interactions of p85 SH2 domains with CD28 could inhibit the binding of Grb2 and Gads to CD28. However, both interactions of Grb2 and Gads with CD28 are also important for the CD28-mediated co-stimulation. We recently showed that SH3 domains of Grb2 and Gads also contribute to CD28 binding, whereas that of p85 does not (15). In addition, we also should consider the possibility of formation of a CD28 dimer in which multivalent binding would increase the affinity relative to monovalent binding due to the effect of avidity (27). Nevertheless, the interaction of SH2 itself mainly contributes to CD28 binding, and therefore the high resolution structure information presented here will help to generate compounds to selectively inhibit the respective CD28 interactions (28). Such compounds would specifically modulate CD28 co-stimulatory signaling without affecting the function of other co-stimulatory molecules, such as inducible co-stimulator, CTLA-4, and PD-1 (29–31). It is also possible that they would lead to improvement in the treatment for immune diseases involving excessive T cell responses, such as allergy and autoimmune disease.
Experimental Procedures
Expression and Purification of Gads SH2, PI3K p85 nSH2, and cSH2
DNA fragments encompassing Gads SH2 domain (Trp58–His155), PI3K p85 nSH2 (Asn325–Lys430), and p85 cSH2 (Glu614–Ala720) domains were amplified by PCR using Gads and PI3K p85 full-length cDNA plasmids as the template and two synthetic primers (Table 3) to generate a BamHI site and an EcoRI site at the 5′- and the 3′-ends of the amplified fragment, respectively. After digestion with BamHI and EcoRI, the DNA fragments were cloned into plasmid pGEX-4T-2. The expression plasmid of Grb2 SH2 was constructed previously (16). Escherichia coli BL21(DE3) were transformed with the Grb2 SH2, Gads SH2, PI3K p85 nSH2, and cSH2 plasmids. Freshly precultivated cells were inoculated into growth medium containing 100 μg/ml ampicillin and grown at 37 °C. When the culture reached an A600 of about 0.6, isopropyl 1-thio-β-d-galactopyranoside was added to a final concentration of 0.1 mm. The cells were incubated at 22 °C for another 20 h. All SH2 domains were expressed as GST fusion proteins. The harvested cells were suspended in PBS (pH 7.4) and lysed by sonication at 4 °C. After the cell debris was removed by centrifugation, the supernatant was loaded to a glutathione column (glutathione-Sepharose 4B fast flow, GE Healthcare). To isolate the SH2 domains from GST, the thrombin protease was added to eluted fractions and incubated at 4 °C for 12 h followed by application onto a benzamidine-Sepharose column (HiTrap benzamidine fast flow, GE Healthcare) to remove thrombin. The purified fractions were pooled and loaded to a gel filtration column (HiPrep Sephacryl S-100 column, GE Healthcare), and the buffer was changed to PBS (pH 7.4) for ITC experiments and 10 mm potassium phosphate buffer (pH 7.5) for all crystallizations. The protein concentrations were determined from UV absorbance at 280 nm and were calculated by using the molar absorption coefficients of 1.43 × 104, 1.58 × 104, 1.99 × 104, and 1.53 × 104 m−1 cm−1 for Grb2 SH2, Gads SH2, PI3K p85 nSH2, and cSH2, respectively.
TABLE 3.
The primer sequences for Gads SH2, PI3K p85 nSH2, and cSH2 using PCR
| Primer | 5′ → 3′ |
|---|---|
| Gads-SH2-F | CGGGATCCTGGTTTCACGAAGGCCTCTCT |
| Gads-SH2-R | GGAATTCTTAGTGACCCTGGTCTTCTCG |
| p85-nSH2-F | CGGGATCCAATATGTCCTTACAAAATGCT |
| p85-nSH2-R | GGAATTCTCATTTGGATACTGGATAAAGTA |
| p85-cSH2-F | CGGGATCCGAAGATTTGCCCCATCATG |
| p85-cSH2-R | GGAATTCTCATGCATATACTGGGTAGG |
Synthesis of CD28-derived Phosphopeptide
The 8- and 12-residue phosphopeptides OctP and DdcP were synthesized by Fmoc solid-phase method as described previously (16). The C terminus is a carboxamide group prepared with Fmoc-NH-SAL-PEG resin (Watanabe Chemicals), and the N terminus is an amino group.
Crystallization, Data Collection, Model Building, and Refinement
Crystallization conditions were screened by the sparse matrix method using commercially available screening kits (Hampton Research) with the hanging drop vapor diffusion method at 20 °C (32). Prior to data collection, all the crystals were soaked in cryoprotectant solutions containing 20% (v/v) glycerol along with their respective reservoir buffers and flash frozen using nitrogen gas stream at −183 °C.
X-ray diffraction experiments were performed at the beamlines BL-17A at High Energy Accelerator Research Organization-Photon Factory (KEK-PF) and BL41XU at SPring-8. All data were processed and scaled using XDS (33) and truncated by the CCP4 program suite (34). For further refinement, 5% of independent, randomly selected reflections were set apart as a test set to calculate the Rfree values.
The structure of the SH2 domain of Gads complexed with a phosphopeptide (Protein Data Bank code 1R1Q) was used as the search model to determine the initial phases for Gads-OctP by the molecular replacement method using Phaser (35, 36). Several cycles of manual model rebuilding and refinement were performed using Coot (37) and PHENIX (38), respectively. The structure of cSH2-OctP was determined as described above by using the structure of the cSH2 complexed with PDGFR-derived peptide (Protein Data Bank code 1H9O) as a search model (39). The partially refined structure of cSH2-OctP was used as a search model for nSH2-OctP. The refined models were validated by MolProbity (40). The statistics for data collection and refinement are summarized in Table 1. The figures were prepared using PyMOL software. The coordinates and structural data for the complexes have been deposited in the Protein Data Bank (codes 5GJH, 5GJI, and 5AUL for Gads SH2-OctP, p85 nSH2-OctP, and p85 cSH2-OctP, respectively).
ITC Experiments
ITC experiments were performed on an MCS-ITC instrument (Malvern). The CD28-derived 12-amino acid peptide DdcP (100–200 μm) was titrated into the protein solution (10–20 μm) using a 250-μl syringe. Each titration consisted of a preliminary 5-μl injection followed by 19 subsequent 13-μl additions. The heat for each injection was subtracted from the heat of dilution of the injectant, which was measured by injecting the CD28-derived peptide solution into the experimental buffer. Each corrected heat was divided by the molar concentration of CD28-derived peptide injected and was analyzed on the basis of a “one set of sites” model with MicroCal Origin 5.0 software supplied by the manufacturer. The ΔCp was calculated from the slope of the regression line of linear fit of the enthalpies measured at four different temperatures between 15 and 30 °C.
Author Contributions
R. A., N. I., and M. O. designed the study. H. M. synthesized the CD28 phosphopeptides. S. I. expressed and purified the SH2 proteins. S. I. and N. N. crystallized the SH2 complexes with CD28 phosphopeptide. S. I., T. I., and N. N. determined the crystal structures. S. I. and M. O. characterized the binding thermodynamics. S. I., N. N., S. O., N. I., and M. O. wrote the paper. All authors analyzed the results and approved the final version of the manuscript.
Acknowledgments
We thank Chika Higashiura and Daiki Usui of Kyoto Prefectural University for technical support. The X-ray crystal structure analyses were performed with the approval of the Photon Factory Program Advisory Committee (Proposal 2014G585).
This work was supported by the Joint Usage/Research Program of Medical Research Institute, Tokyo Medical and Dental University. The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (codes 5GJH, 5GJI, and 5AUL) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- Grb2
- growth factor receptor-bound protein 2
- Gads
- Grb2-related adaptor downstream of Shc
- ITC
- isothermal titration calorimetry
- ΔCp
- heat capacity change
- ΔG
- Gibbs free energy change
- ΔH
- enthalpy change
- ΔS
- entropy change
- NFAT
- nuclear factor of activated T cells
- SH
- Src homology
- GAP
- GTPase-activating protein
- nSH2
- N-terminal SH2 domain
- cSH2
- C-terminal SH2 domain
- PDGFR
- platelet-derived growth factor receptor
- Fmoc
- N-(9-fluorenyl)methoxycarbonyl.
References
- 1. Rudd C. E., Taylor A., and Schneider H. (2009) CD28 and CTLA-4 coreceptor expression and signal transduction. Immunol. Rev. 229, 12–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Esensten J. H., Helou Y. A., Chopra G., Weiss A., and Bluestone J. A. (2016) CD28 costimulation: from mechanism to therapy. Immunity 44, 973–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ogawa S., Watanabe M., Sakurai Y., Inutake Y., Watanabe S., Tai X., and Abe R. (2013) CD28 signaling in primary CD4+ T cells: identification of both tyrosine phosphorylation-dependent and phosphorylation-independent pathways. Int. Immunol. 25, 671–681 [DOI] [PubMed] [Google Scholar]
- 4. Schneider H., Cai Y. C., Prasad K. V., Shoelson S. E., and Rudd C. E. (1995) T cell antigen CD28 binds to the GRB-2/SOS complex, regulators of p21ras. Eur. J. Immunol. 25, 1044–1050 [DOI] [PubMed] [Google Scholar]
- 5. Rudd C. E., and Schneider H. (2003) Unifying concepts in CD28, ICOS and CTLA4 co-receptor signalling. Nat. Rev. Immunol. 3, 544–556 [DOI] [PubMed] [Google Scholar]
- 6. Schneider H., and Rudd C. E. (2008) CD28 and Grb-2, relative to Gads or Grap, preferentially co-operate with Vav1 in the activation of NFAT/AP-1 transcription. Biochem. Biophys. Res. Commun. 369, 616–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Harada Y., Ohgai D., Watanabe R., Okano K., Koiwai O., Tanabe K., Toma H., Altman A., and Abe R. (2003) A single amino acid alteration in cytoplasmic domain determines IL-2 promoter activation by ligation of CD28 but not inducible costimulator (ICOS). J. Exp. Med. 197, 257–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Watanabe R., Harada Y., Takeda K., Takahashi J., Ohnuki K., Ogawa S., Ohgai D., Kaibara N., Koiwai O., Tanabe K., Toma H., Sugamura K., and Abe R. (2006) Grb2 and Gads exhibit different interactions with CD28 and play distinct roles in CD28-mediated costimulation. J. Immunol. 177, 1085–1091 [DOI] [PubMed] [Google Scholar]
- 9. Takeda K., Harada Y., Watanabe R., Inutake Y., Ogawa S., Onuki K., Kagaya S., Tanabe K., Kishimoto H., and Abe R. (2008) CD28 stimulation triggers NF-κB activation through the CARMA1-PKCθ-Grb2/Gads axis. Int. Immunol. 20, 1507–1515 [DOI] [PubMed] [Google Scholar]
- 10. Harada Y., Tanabe E., Watanabe R., Weiss B. D., Matsumoto A., Ariga H., Koiwai O., Fukui Y., Kubo M., June C. H., and Abe R. (2001) Novel role of phosphatidylinositol 3-kinase in CD28-mediated costimulation. J. Biol. Chem. 276, 9003–9008 [DOI] [PubMed] [Google Scholar]
- 11. Lowenstein E. J., Daly R. J., Batzer A. G., Li W., Margolis B., Lammers R., Ullrich A., Skolnik E. Y., Bar-Sagi D., and Schlessinger J. (1992) The SH2 and SH3 domain-containing protein GRB2 links receptor tyrosine kinases to ras signaling. Cell 70, 431–442 [DOI] [PubMed] [Google Scholar]
- 12. Liu S. K., and McGlade C. J. (1998) Gads is a novel SH2 and SH3 domain-containing adaptor protein that binds to tyrosine-phosphorylated Shc. Oncogene 17, 3073–3082 [DOI] [PubMed] [Google Scholar]
- 13. Asada H., Ishii N., Sasaki Y., Endo K., Kasai H., Tanaka N., Takeshita T., Tsuchiya S., Konno T., and Sugamura K. (1999) Grf40, A novel Grb2 family member, is involved in T cell signaling through interaction with SLP-76 and LAT. J. Exp. Med. 189, 1383–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Law C. L., Ewings M. K., Chaudhary P. M., Solow S. A., Yun T. J., Marshall A. J., Hood L., and Clark E. A. (1999) GrpL, a Grb2-related adaptor protein, interacts with SLP-76 to regulate nuclear factor of activated T cell activation. J. Exp. Med. 189, 1243–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Higo K., Oda M., Morii H., Takahashi J., Harada Y., Ogawa S., and Abe R. (2014) Quantitative analysis of CD28 interaction with cytoplasmic adapter molecules Grb2, Gads and p85 PI3K by surface plasmon resonance. Immunol. Invest. 43, 278–291 [DOI] [PubMed] [Google Scholar]
- 16. Higo K., Ikura T., Oda M., Morii H., Takahashi J., Abe R., and Ito N. (2013) High resolution crystal structure of the Grb2 SH2 domain with a phosphopeptide derived from CD28. PLoS One 8, e74482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu B. A., Engelmann B. W., and Nash P. D. (2012) The language of SH2 domain interactions defines phosphotyrosine-mediated signal transduction. FEBS Lett. 586, 2597–2605 [DOI] [PubMed] [Google Scholar]
- 18. Spolar R. S., Livingstone J. R., and Record M. T. Jr. (1992) Use of liquid hydrocarbon and amide transfer data to estimate contributions to thermodynamic functions of protein folding from the removal of nonpolar and polar surface from water. Biochemistry 31, 3947–3955 [DOI] [PubMed] [Google Scholar]
- 19. Murphy K. P., and Freire E. (1992) Thermodynamics of structural stability and cooperative folding behavior in proteins. Adv. Protein Chem. 43, 313–361 [DOI] [PubMed] [Google Scholar]
- 20. Papaioannou D., Geibel S., Kunze M. B., Kay C. W., and Waksman G. (2016) Structural and biophysical investigation of the interaction of a mutant Grb2 SH2 domain (W121G) with its cognate phosphopeptide. Protein Sci. 25, 627–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. O'Brien R., Rugman P., Renzoni D., Layton M., Handa R., Hilyard K., Waterfield M. D., Driscoll P. C., and Ladbury J. E. (2000) Alternative modes of binding of proteins with tandem SH2 domains. Protein Sci. 9, 570–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Oda M., Furukawa K., Ogata K., Sarai A., and Nakamura H. (1998) Thermodynamics of specific and non-specific DNA binding by the c-Myb DNA-binding domain. J. Mol. Biol. 276, 571–590 [DOI] [PubMed] [Google Scholar]
- 23. Ladbury J. E., Lemmon M. A., Zhou M., Green J., Botfield M. C., and Schlessinger J. (1995) Measurement of the binding of tyrosyl phosphopeptides to SH2 domains: a reappraisal. Proc. Natl. Acad. Sci. U.S.A. 92, 3199–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McNemar C., Snow M. E., Windsor W. T., Prongay A., Mui P., Zhang R., Durkin J., Le H. V., and Weber P. C. (1997) Thermodynamic and structural analysis of phosphotyrosine polypeptide binding to Grb2-SH2. Biochemistry 36, 10006–10014 [DOI] [PubMed] [Google Scholar]
- 25. Ogura K., Tsuchiya S., Terasawa H., Yuzawa S., Hatanaka H., Mandiyan V., Schlessinger J., and Inagaki F. (1997) Conformation of an Shc-derived phosphotyrosine-containing peptide complexed with the Grb2 SH2 domain. J. Biomol. NMR 10, 273–278 [DOI] [PubMed] [Google Scholar]
- 26. Nolte R. T., Eck M. J., Schlessinger J., Shoelson S. E., and Harrison S. C. (1996) Crystal structure of the PI3-kinase p85 amino-terminal SH2 domain and its phosphopeptide complexes. Nat. Struct. Biol. 3, 364–374 [DOI] [PubMed] [Google Scholar]
- 27. Oda M., and Azuma T. (2000) Reevaluation of stoichiometry and affinity/avidity in interactions between anti-hapten antibodies and mono- or multi-valent antigens. Mol. Immunol. 37, 1111–1122 [DOI] [PubMed] [Google Scholar]
- 28. Kraskouskaya D., Duodu E., Arpin C. C., and Gunning P. T. (2013) Progress towards the development of SH2 domain inhibitors. Chem. Soc. Rev. 42, 3337–3370 [DOI] [PubMed] [Google Scholar]
- 29. Linsley P. S., and Nadler S. G. (2009) The clinical utility of inhibiting CD28-mediated costimulation. Immunol. Rev. 229, 307–321 [DOI] [PubMed] [Google Scholar]
- 30. Teichmann L. L., Cullen J. L., Kashgarian M., Dong C., Craft J., and Shlomchik M. J. (2015) Local triggering of the ICOS coreceptor by CD11c+ myeloid cells drives organ inflammation in lupus. Immunity 42, 552–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schildberg F. A., Klein S. R., Freeman G. J., and Sharpe A. H. (2016) Coinhibitory pathways in the B7-CD28 ligand-receptor family. Immunity 44, 955–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jancarik J., and Kim S. H. (1991) Sparse matrix sampling: a screening method for crystallization of proteins. J. Appl. Crystallogr. 24, 409–411 [Google Scholar]
- 33. Kabsch W. (2010) XDS. Acta Crystallogr. D. Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D. Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cho S., Velikovsky C. A., Swaminathan C. P., Houtman J. C., Samelson L. E., and Mariuzza R. A. (2004) Structural basis for differential recognition of tyrosine-phosphorylated sites in the linker for activation of T cells (LAT) by the adaptor Gads. EMBO J. 23, 1441–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pauptit R. A., Dennis C. A., Derbyshire D. J., Breeze A. L., Weston S. A., Rowsell S., and Murshudov G. N. (2001) NMR trial models: experiences with the colicin immunity protein Im7 and the p85α C-terminal SH2-peptide complex. Acta Crystallogr. D. Biol. Crystallogr. 57, 1397–1404 [DOI] [PubMed] [Google Scholar]
- 40. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]