Abstract
The membrane protein RFT1 is essential for normal protein N-glycosylation, but its precise function is not known. RFT1 was originally proposed to translocate the glycolipid Man5GlcNAc2-PP-dolichol (needed to synthesize N-glycan precursors) across the endoplasmic reticulum membrane, but subsequent studies showed that it does not play a direct role in transport. In contrast to the situation in yeast, RFT1 is not essential for growth of the parasitic protozoan Trypanosoma brucei, enabling the study of its function in a null background. We now report that lack of T. brucei RFT1 (TbRFT1) not only affects protein N-glycosylation but also glycosylphosphatidylinositol (GPI) anchor side-chain modification. Analysis by immunoblotting, metabolic labeling, and mass spectrometry demonstrated that the major GPI-anchored proteins of T. brucei procyclic forms have truncated GPI anchor side chains in TbRFT1 null parasites when compared with wild-type cells, a defect that is corrected by expressing a tagged copy of TbRFT1 in the null background. In vivo and in vitro labeling experiments using radiolabeled GPI precursors showed that GPI underglycosylation was not the result of decreased formation of the GPI precursor lipid or defective galactosylation of GPI intermediates in the endoplasmic reticulum, but rather due to modifications that are expected to occur in the Golgi apparatus. Unexpectedly, immunofluorescence microscopy localized TbRFT1 to both the endoplasmic reticulum and the Golgi, consistent with the proposal that TbRFT1 plays a direct or indirect role in GPI anchor glycosylation in the Golgi apparatus. Our results implicate RFT1 in a wider range of glycosylation processes than previously appreciated.
Keywords: endoplasmic reticulum (ER), glycosylphosphatidylinositol (GPI anchor), Golgi, N-linked glycosylation, Trypanosoma brucei
Introduction
Trypanosoma brucei is a human and animal parasite endemic in sub-Saharan Africa causing sleeping sickness in humans and nagana in livestock. The life cycle of the extracellular living parasite comprises stages in the midgut and salivary gland of the tsetse fly vector and in the blood of the mammalian host. Alternating between the two organisms, the parasite not only adapts its energy metabolism to the respective environment but also its cell surface protein coat, which is a crucial determinant of the parasite's virulence. Apart from being a deadly pathogen affecting the socio-economic development in endemic areas, the parasite has emerged as an interesting model organism for basic research. Two proliferative stages of T. brucei, bloodstream trypomastigotes (bloodstream form parasites) and the insect-stage procyclic trypomastigotes (procyclic form parasites), can easily be cultured in vitro. Biological features such as trans-splicing (1), RNA editing (2), and antigenic variation (3) were first described in Trypanosomatids and only later also found in other eukaryotes. In addition, T. brucei was one of the first organisms in which glycosylphosphatidylinositol (GPI)3 anchoring of cell surface proteins was described and extensively explored (4, 5). Protein-linked GPI anchors consist of the conserved core structure ethanolamine-HPO4-Manα1-2Manα1-6Manα1-4GlcNα1-6-myo-inositol phospholipid with the amino group of ethanolamine linked to the C terminus of the protein (4, 5). A wide variety of linear and branched glycosyl substituents and additional ethanolamine phosphate moieties can be attached to this core, depending on the protein to which the anchor is attached and the organism in which it is synthesized. The best-studied and most abundant GPI-anchored proteins of T. brucei are the variant surface glycoproteins (VSGs) in bloodstream form parasites (6) and the procyclins in procyclic forms (7–9). Although the variant surface glycoprotein GPI core is modified by rather simple galactosyl side chains (5), the GPI anchors of procyclins are the largest and most complex anchors known, comprising large branched N-acetyllactosamine (Galβ1-4GlcNAc) and lacto-N-biose (Galβ1-3GlcNAc)-containing side chains often capped with α2-3-linked sialic acid residues (10, 11). Based on their C-terminal amino acid sequences containing di- or pentapeptide tandem repeats, procyclins are divided into two classes: EP (rich in Glu-Pro repeats) and GPEET (rich in Gly-Pro-Glu-Glu-Thr repeats) procyclins (11). Two of the three subclasses of EP procyclins, EP1 and EP3, contain a single N-glycosylation site (11, 12), whereas EP2 and GPEET procyclins are not N-glycosylated. Interestingly, EP1 and EP3 procyclins are modified exclusively by a triantennary Man5GlcNAc2 moiety (11), transferred to protein by oligosaccharyltransferase TbSTT3B, which is expressed in procyclic forms (13) and specifically uses mature Man9GlcNAc2-PP-dolichol (mDLO) for transfer to N-glycosylation sites (14). Due to the lack of a Golgi α-mannosidase in procyclic form trypanosomes, Man9GlcNAc2 glycans can only be trimmed to triantennary Man5-GlcNAc2 that are not further modified (15). Fig. 1A shows a schematic representation of a typical N-glycosylated EP procyclin.
FIGURE 1.
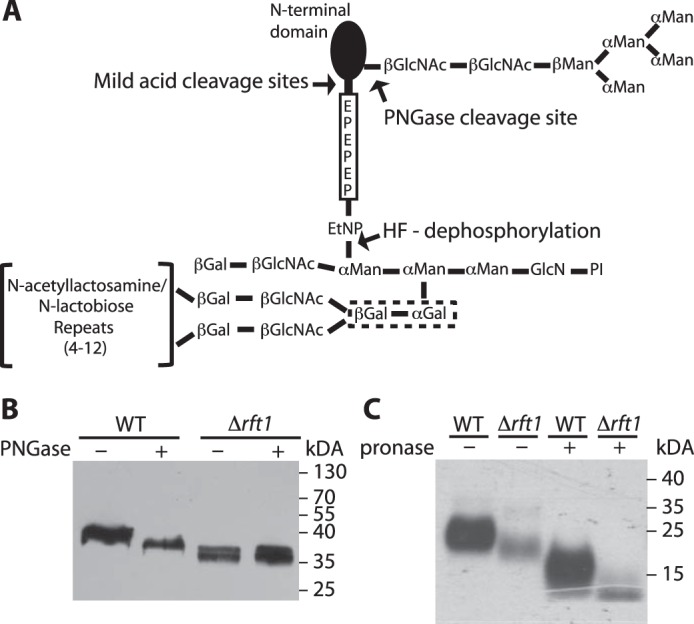
Schematic representation of procyclin glycosylation and analysis of GPI-anchored proteins in TbRFT1 null cells. A, most procyclin isoforms (except EP2 and GPEET) are modified by a homogeneous triantennary Man5GlcNAc2 glycan near the N terminus. The GPI anchor of all procyclins is modified with several N-acetyllactosamine or lacto-N-biose repeats, which may be capped with sialic acids depending on the presence of blood sialoglycoconjugates. These repeats are linked to the middle mannose of the GPI anchor core via two consecutive galactose residues (dotted box), which are probably added in the ER as suggested in Fig. 2. The C-terminal regions of EP procyclins consist of 22–30 Glu-Pro repeats. PI, phosphatidylinositol. B, immunoblotting analysis of EP procyclins isolated from WT and TbRFT1 null (Δrft1) cells. Denatured proteins were treated with (+) or without (−) PNGase F to remove N-glycans and then separated by SDS-PAGE. After electrotransfer to membranes, EP was visualized by enhanced chemiluminescence using anti-EP antibody and HRP-conjugated anti-mouse IgG. C, [3H]ethanolamine labeling and fluorography of GPI-anchored proteins from WT and TbRFT1 null (Δrft1) cells. Trypanosomes were grown in the presence of [3H]ethanolamine, and GPI-anchored proteins were extracted from the delipidated protein pellet using 9% butan-1-ol. Extracts incubated in the absence (−) or presence (+) of Pronase to remove the protein portions of GPEET and EP were separated by SDS-PAGE and analyzed by fluorography.
RFT1 was first described in Saccharomyces cerevisiae as a protein “requiring fifty-three,” i.e. human p53, in a screen of mutants that could be rescued by heterologous expression of p53 (16). Only later, yeast RFT1 (Rft1p) was found to play an essential role in protein N-glycosylation (17). The multi-pass transmembrane protein was reported to be localized in the endoplasmic reticulum (ER) of yeast and human cells (17, 18), although no corresponding localization data have been published. The accumulation of the dolichol-linked oligomannose intermediate Man5GlcNAc2-PP-dolichol (M5-DLO) in S. cerevisiae cells depleted of Rft1p but having intact O-glycosylation and GPI anchoring suggested that RFT1 is a flippase enabling translocation of M5-DLO across the ER membrane (17). However, this interpretation was challenged by subsequent biochemical studies, where flipping of M5-DLO was assayed in vitro using proteoliposomes containing Triton X-100-extracted yeast ER membrane proteins (19, 20). Flipping of M5-DLO occurred robustly in the absence of Rft1p; e.g. when proteoliposomes were reconstituted with fractionated ER membrane proteins, M5-DLO translocation activity was found in fractions devoid of Rft1p (19). Similar experiments using sealed microsomes confirmed these findings (21). Hence, it was postulated that S. cerevisiae Rft1p has only an indirect involvement in the translocation of M5-DLO (19–21).
More recently, the role of RFT1 was revisited using T. brucei as a model organism. By complementation of yeast lacking RFT1 function, T. brucei RFT1 (TbRFT1; Tb927.11.11670) was shown to represent a functional homolog of S. cerevisiae Rft1p (22). TbRFT1 null procyclic trypanosomes grew nearly normally and had normal steady-state levels of mDLO and reduced but still significant N-glycosylation, indicating robust M5-DLO flippase activity. Nevertheless, TbRFT1 null parasites had 30–100-fold greater steady-state levels of M5-DLO when compared with wild-type trypanosomes. Fluorophore-assisted carbohydrate electrophoresis analysis of N-glycans released from N-linked glycoproteins showed that all N-glycans in the TbRFT1 null cells originate from mDLO, indicating that the M5-DLO excess is not used for glycosylation. Together, these results suggested that rather than facilitating M5-DLO flipping, RFT1 appears to promote conversion of M5-DLO to mDLO by another mechanism, possibly by acting as an M5-DLO chaperone (22).
We now report that the lack of TbRFT1 in T. brucei procyclic forms not only affects N-glycosylation but also GPI anchor glycosylation. In addition, we unexpectedly localize TbRFT1 to both the ER and the Golgi. These results suggest that RFT1 has a pleiotropic influence on protein glycosylation.
Results and Discussion
Procyclins of TbRFT1 Null Cells Exhibit Reduced Apparent Molecular Masses
Based on our previous observation that the lysosomal marker protein p67 is underglycosylated in TbRFT1 null mutants (22), we investigated whether a similar glycosylation phenotype could also be observed for the major surface coat protein of T. brucei procyclic forms, EP procyclin. EP procyclins are encoded by three different genes, EP1–3, with EP1 and EP3 proteins each containing a single N-glycosylation site, whereas EP2 and the other subclass of procyclin, GPEET, are not N-glycosylated (11, 23, 24). The relative abundance of EP1–3 and GPEET varies among trypanosome strains and culture conditions (11, 12, 25) and during tsetse infection (26, 27). In the strain used in this study, T. brucei Lister 427, GPEET represents the predominant surface protein, but EP is also expressed (25). Analysis by SDS-PAGE and immunoblotting revealed a smaller apparent molecular mass of EP in TbRFT1 null cells when compared with WT trypanosomes (Fig. 1B). Treatment of EP from WT cells with protein N-glycosidase F (PNGase) to release protein N-glycans (Fig. 1A) reduced its apparent molecular mass. However, the apparent size of de-N-glycosylated EP from WT trypanosomes was larger than that of EP from TbRFT1 null cells, which was unaffected by PNGase treatment (Fig. 1B). Together, these results indicate that the altered molecular mass of EP is not only due to altered N-glycosylation in TbRFT1 null parasites but involves other modifications caused by lack of TbRFT1.
TbRFT1 Null Procyclins Have Truncated GPI Anchor Side Chains
Because all EP isoforms, as well as GPEET, are GPI-anchored (23, 28), we hypothesized that decreased glycosylation of the GPI anchor side chain may contribute to the observed phenotype. The procyclin GPI anchors are modified by a large heterogeneous, branched side chain comprising poly-N-acetyllactosamine and lacto-N-biose units that may be capped with sialic acid residues (10, 11) (Fig. 1A). Partial or complete loss of this glycan moiety would lead to reduced apparent molecular masses of GPEET and EP. Hence, we analyzed the procyclin GPI anchors by in vivo labeling of trypanosomes with [3H]ethanolamine ([3H]Etn), which gets incorporated into the GPI core structure (23, 25). SDS-PAGE and fluorography showed that GPEET, migrating with an apparent molecular mass of 22–29 kDa, was readily labeled with [3H]Etn in WT trypanosomes (Fig. 1C). Labeling of GPEET was also observed in TbRFT1 null parasites; however, the protein migrated with a lower molecular mass when compared with WT trypanosomes (Fig. 1C). To eliminate molecular mass differences present in the protein part of GPEET, extracts were treated with Pronase, which has been shown to digest the entire protein portion of GPEET down to the C-terminal glycine residue (25). The resulting GPI anchor was again analyzed by SDS-PAGE and fluorography and showed a clear difference in molecular mass between WT and TbRFT1 null parasites (Fig. 1C).
To confirm that the GPEET polypeptide itself is not truncated in TbRFT1 null parasites, its mass was analyzed by MALDI-TOF-MS after sequential treatment with aqueous hydrofluoric acid (to remove the GPI anchor, leaving ethanolamine attached to the C-terminal amino acid (Fig. 1A)) and mild trifluoroacetic acid (to cleave Asp-Pro bonds within the EP sequence (Fig. 1A)) (12). The results in Fig. 2, A and B, show that both WT and ΔTbrft1 cells express the same GPEET fragments GP-4 and GP-13, representing proteins lacking 4 and 13, respectively, N-terminal amino acids. Furthermore, the masses are consistent with the presence of ethanolamine linked to Gly, further corroborating that ΔTbrft1 cells are not defective in the transfer of GPIs to proteins. In addition, a series of EP procyclin C-terminal fragments was also detected.
FIGURE 2.
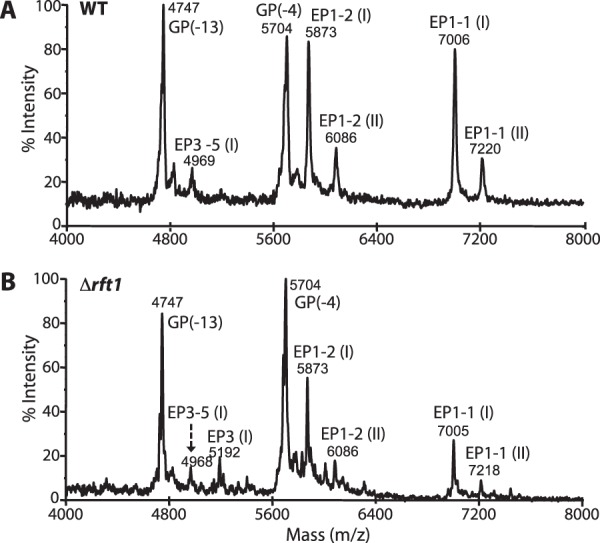
Negative-ion MALDI-TOF-MS analysis of procyclins after removal of the GPI anchors. A and B, butan-1-ol extracts from WT (A) and Δrft1 (B) cells were subjected to 48% aqueous hydrofluoric acid dephosphorylation followed by mild trifluoroacetic acid hydrolysis to remove the GPI anchor and generate EP procyclin peptides. The resulting polypeptides, corresponding to the C-terminal portions of procyclins, were analyzed by negative-ion MALDI-TOF-MS. GP(−4) and GP(−13) refer to GPEET fragments lacking 4 and 13, respectively, amino acids at the N terminus. EP isoforms EP1-1 (I) (P(EP)nG-Etn) and EP1-1 (II) (PDP(EP)nG-Etn) represent C-terminal mild acid fragments (12). EP3-5 is an unusual form containing 21 EP repeats (49).
To determine the degree of GPI underglycosylation in ΔTbrft1 cells, butan-1-ol extracts (rich in procyclins) were analyzed by GC-MS. As expected, although the WT sample yielded a composition of Man:Gal:GlcNAc:Sia of 1.0:1.4:0.4:0.2, ΔTbrft1 cells showed an ∼7-fold reduction in the overall GPI sugar content, resulting in a ratio of Man:Gal:GlcNAc:Sia of 1.0:0.2:0.1:∼0 (Sia were not detectable). A similar reduction in the overall sugar composition of another ΔTbrft1 mutant (i.e. B1 cells (22)) was also observed by GC-MS (not shown). Collectively, these results show that trypanosomes lacking TbRFT1 express procyclins with truncated GPI anchor side chains containing fewer poly-N-acetyllactosamine/lacto-N-biose repeats and sialic acids, thus explaining the observed reductions in apparent molecular masses after SDS-PAGE.
Procyclin GPI Anchor Size Is Restored by Ectopically Expressed TbRFT1
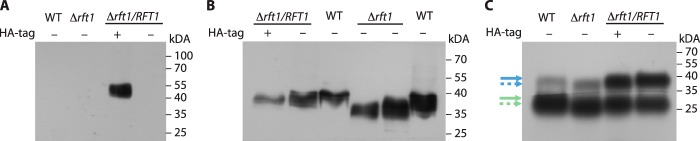
To study whether the observed differences in EP and GPEET molecular masses between WT and TbRFT1 null mutants are indeed due to the lack of TbRFT1, we generated add-back mutants by expressing HA-tagged or untagged copies of TbRFT1 in the ΔTbrft1 background. If functional, these ectopically expressed proteins are expected to restore the molecular masses of EP and GPEET to wild-type sizes. Immunoblotting using antibodies against the HA epitope demonstrated that RFT1-HA was expressed in the respective clones (Fig. 3A). In addition, the results showed that in TbRFT1 null parasites expressing HA-TbRFT1 or untagged TbRFT1, the apparent molecular mass of EP was comparable with wild-type EP procyclin (Fig. 3B), indicating that both tagged and untagged TbRFT1 are functional and restored EP glycosylation as well as GPI glycan maturation. A similar result was obtained by analyzing [3H]Etn-labeled EP and GPEET using SDS-PAGE and fluorography (Fig. 3C).
FIGURE 3.
Ectopic expression of TbRFT1 and TbRFT1-HA. EP and/or GPEET from WT cells, TbRFT1 null cells (Δrft1), and TbRFT1 null cells expressing a HA-tagged (+) or untagged (−) ectopic copy of TbRFT1 (Δrft1/RFT1) were analyzed by SDS-PAGE and immunoblotting. A and B, enhanced chemiluminescence was used to visualize HA (A) or EP (B) using corresponding first and secondary antibodies. C, EP and GPEET in WT, TbRFT1 null cells (Δrft1), and TbRFT1 null cells expressing a HA-tagged (+) or untagged (−) ectopic copy of TbRFT1 (Δrft1/RFT1) were labeled with [3H]ethanolamine, extracted, and analyzed as described in the legend for Fig. 1. The sizes of wild-type (solid lines) and mutant (dashed lines) proteins are marked (blue, EP; green, GPEET).
GPI Precursor Synthesis and in Vitro GPI Galactosylation Are TbRFT1-independent
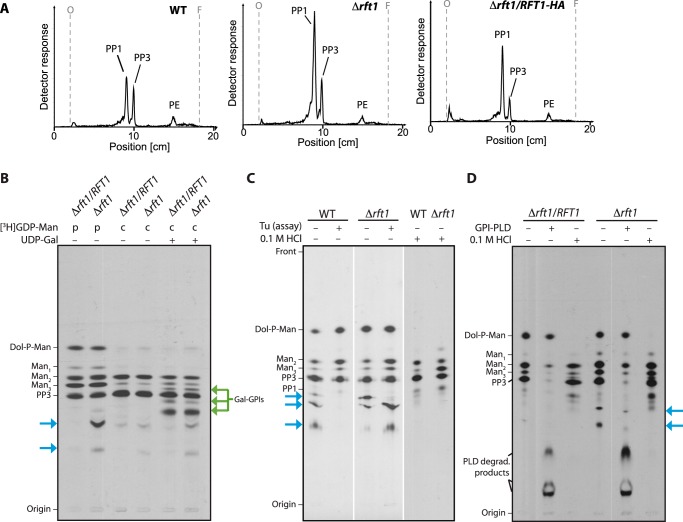
To study whether the lack of TbRFT1 affects the formation of PP1, the GPI precursor added to protein in the ER (29), trypanosomes were cultured in the presence of [3H]Etn, which becomes incorporated into all ethanolamine-capped GPI precursors. Analysis of extracts from WT, TbRFT1 null, and TbRFT1 addback parasites by TLC revealed that PP1 was the major [3H]Etn-labeled lipid irrespective of the presence or absence of TbRFT1 (Fig. 4A).
FIGURE 4.
Analysis of GPI precursor formation. A, analysis of in vivo [3H]Etn-labeled GPI precursors PP3 and PP1 extracted from WT, TbRFT1 knock-out (Δrft1), and addback (Δrft1/RFT1) cells. Trypanosome densities were adjusted before the addition of [3H]Etn to the cultures. After 4 h of labeling, GPI precursors were extracted, separated by TLC, and visualized using a radioactivity TLC scanner. The migration of PP1, PP3 (50), and phosphatidylethanolamine (PE) is indicated. B–D, in vitro [3H]GDP-mannose (GDP-Man) labeling of GPI precursors. B, membranes from hypotonically lysed TbRFT1 knock-out (Δrft1) and addback (Δrft1/RFT1) cells were pulse-labeled (p) with [3H]GDP-Man, followed by a chase (c) with non-radioactive GDP-Man in the presence (+) or absence of (−) UDP-galactose. 3H-labeled glycolipids were extracted, separated by TLC, and visualized by fluorography. Galactosylated GPI intermediates are indicated with green arrows. Blue arrows indicate the additional [3H]GDP-mannose-containing species formed in Δrft1 extracts. C, analysis of 3H-labeled glycolipids from wild-type and Δrft1 after a 30-min labeling with [3H]GDP-Man in the presence (+) or absence (−) of tunicamycin (Tu) (lanes 1–4). Lanes 5 and 6 show aliquots labeled in absence of tunicamycin that were treated with 0.1 m HCl before TLC. D, biochemical analysis of 3H-labeled glycolipids from Δrft1 and Δrft1/RFT1 after a 30-min labeling with [3H]GDP-Man in the presence (+) of tunicamycin. Primary lipid extracts were split and treated with GPI-specific phospholipase D (GPI-PLD) or 0.1 m HCl as indicated. Lipids were re-extracted after treatment and separated by TLC along with an aliquot of untreated primary extract.
Assembly of the GPI core structure and attachment to protein occurs in the ER (30–32). At present, it is unclear whether the glycan modification at the central mannose residue starts before or after GPI attachment to protein in the ER, or on protein-bound GPI anchors in the Golgi. It is known that galactose is added in vitro to GPI anchor precursors in bloodstream form T. brucei (33), suggesting the presence of ER-resident galactosyltransferases that are able to act on GPI precursors. To date, only two T. brucei glycosyltransferases involved in GPI processing are known: GT8 and GT3. However, both seem to localize to the Golgi, with GT8 mediating the transfer of the first GlcNAc moiety to the terminal digalactose moiety of the immature GPI anchor (34, 35) and GT3 attaching a galactose residue to GlcNAc (36). To study whether decreased galactosylation in the ER may cause underglycosylation of the GPI anchors in TbRFT1 null cells, we pulse-labeled crude membrane preparations from TbRFT1 add-back and TbRFT1 null parasites with [3H]GDP-mannose ([3H]GDP-Man) in the presence of UDP-GlcNAc and chased the labeled GPI precursors with non-radiolabeled GDP-Man and UDP-Gal. Tunicamycin was added to the assays to inhibit formation of the N-glycan precursor dolichyl-pyrophosphate GlcNAc (Dol-PP-GlcNAc). TLC analysis of 3H-labeled GPI lipids after extraction with chloroform/methanol/water followed by butan-1-ol-water partitioning showed that additional more polar species were formed during the chase with UDP-Gal (Fig. 4B, green arrows). However, we observed no differences in the galactosylation pattern between wild-type and TbRFT1 null cells, indicating that galactosylation of GPI precursors in the ER is not affected by lack of TbRFT1.
The ER lumenal mannose donor dolichol-phosphate mannose (Dol-P-Man) is one of the most prominent radiolabeled lipid species after pulse labeling membranes with [3H]GDP-Man (Fig. 4B). As expected, Dol-P-Man as well as the early GPI intermediate Manα1–4GlcNα1–6-myo-inositol phospholipid (Man1-GlcN-PI) almost completely disappeared during the chase. Interestingly, extracts from both pulse-labeled and chase-labeled ΔTbrft1 membranes contained additional polar mannose-containing lipids (Fig. 4B, blue arrows). A comparison with extracts from membranes labeled in the absence of tunicamycin (Fig. 4C) shows that compounds with similar Rf values are also formed by WT membranes as long as N-glycan synthesis is enabled. Treatment of the labeled lipids with mild acid led to degradation of Dol-P-Man as well as the unknown polar species from both wild-type and mutant extracts (Fig. 4, C and D). Conversely, the acid-sensitive lipids were not cleaved by the GPI-specific phospholipase D, whereas the known GPI intermediates were readily hydrolyzed (Fig. 4D). Together, these results suggest that the unknown species made by mutant membranes are tunicamycin-insensitive dolichol-linked oligomannose species that most likely originate from a pool of preformed (early) intermediates. We conclude that side-chain glycosylation of newly synthesized GPI precursors occurs normally in ΔTbrft1 membranes.
Possible Roles of TbRft1 in GPI Anchor Modification
The defects in GPI anchor maturation of TbRFT1 null cells can be interpreted in two ways. TbRFT1 may have a direct role in GPI anchor glycan modification that is independent of its function in N-glycosylation. Alternatively, the defect may result from incomplete N-glycosylation of a glycosyltransferase involved in GPI anchor modification, leading to decreased activity and thus incomplete glycan modification.
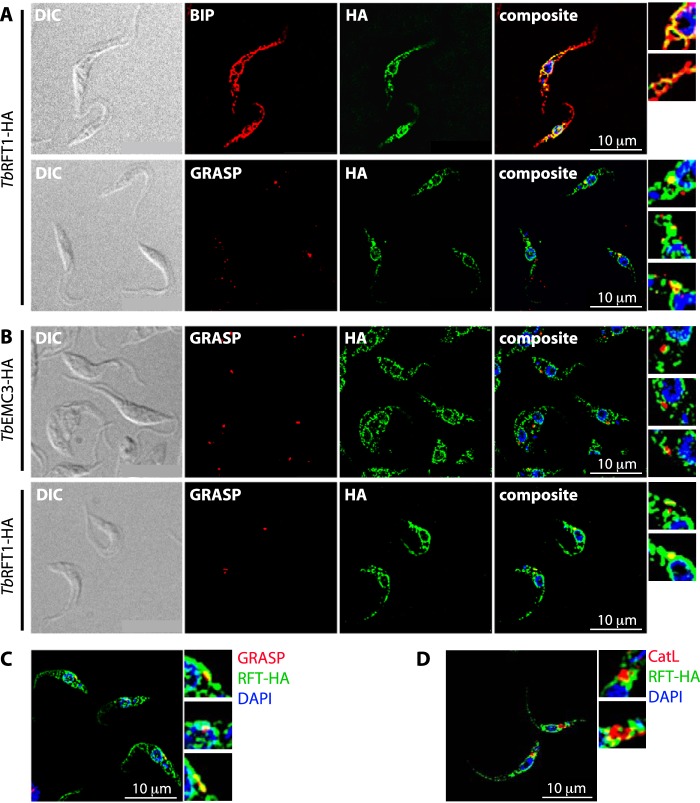
First, we considered the possibility that the appearance of truncated GPI anchors may be caused by a glycosylation defect occurring in the Golgi. To study whether TbRFT1 is present exclusively in the ER, as suggested for yeast (17), we analyzed the localization of the functional HA-tagged copy of TbRFT1 in the TbRFT1 null background using immunofluorescence microscopy. Co-staining of TbRFT1-HA with an antibody against the ER lumenal chaperone BiP confirmed the expected localization of TbRFT1 in the ER, where it was predominantly found in the perinuclear region (Fig. 5A). Interestingly, however, co-staining of TbRFT1-HA with an antibody against the Golgi resident protein TbGRASP (37) also showed co-localization (Fig. 5A). In >70% of parasites examined (n > 100), TbRFT1-HA co-stained with a spot located between the nucleus and the kinetoplast and representing the Golgi. To exclude the possibility that the observed co-localization of TbRFT1 with TbGRASP is unspecific, we analyzed the localization of an HA-tagged membrane-bound member of the EMC family of proteins, TbEMC3 (Tb927.10.4760), which has been shown in yeast to localize to the ER (38). The results showed <35% co-localization of TbEMC3-HA with TbGRASP (Fig. 5B). Although we cannot completely exclude the possibility that a portion of TbRFT1 is localized to the Golgi as a result of saturation of the retention system for ER membrane proteins, we consider it unlikely for the following reasons. (i) TbRFT1-HA shows a similar dual localization when expressed by a different, tetracycline-inducible expression vector in T. brucei 427 wild-type cells (Fig. 5C); (ii) overexpression of another ER-localized membrane protein, TbEPT (T. brucei ethanolamine phosphotransferase), using the same tetracycline-inducible vector showed no Golgi localization and was specifically targeted to the perinuclear ER (39); and (iii) there is no precedent in trypanosomes for mislocalization of ER proteins on overexpression of a single ER resident. Dual localization of trypanosome proteins in both the ER and the Golgi is not unique to TbRFT1 but has been reported before (40). In contrast, no co-localization was observed between TbRFT1-HA and cathepsin L, a marker for the T. brucei lysosome (41) that also localizes between the nucleus and the kinetoplast (Fig. 5D). The role of TbRFT1 in glycosylation in the Golgi remains speculative. In a recent publication, RNAi-mediated silencing of the nucleotide sugar transporter TbNST4 responsible for import of UDP-GlcNAc, GDP-mannose, and UDP-GalNAc into the Golgi resulted in production of underglycosylated EP procyclin in T. brucei procyclic forms (42). In addition, defective forms of GT3 (36) or GT8 (35, 43) resulted in impaired GPI glycan maturation and reduced protein N-glycosylation. It is possible that a lack of TbRFT1 affects Golgi resident proteins involved in glycosylation, by directly interacting with these proteins or by affecting their glycosylation status.
FIGURE 5.
Localization of TbRFT1 in procyclic form parasites. A, functional TbRFT1-HA was co-stained with the ER marker protein BIP (upper panels) or the Golgi marker TbGRASP (lower panels). The merged channels (composite) show overlap (yellow color) of the BIP and TbRFT1-HA signals mainly in the perinuclear zone, with some weaker signal distributed throughout the rest of the cells. Co-staining with the Golgi marker TbGRASP shows that the brightest spots of the HA signal co-localize with the Golgi signal. Some areas were zoomed for better visibility (panels on the right). DIC, differential interference contrast. B, trypanosomes expressing TbEMC3-HA were co-stained with TbGRASP (upper panels). The merged channels (composite) show little overlap (yellow color) of the signals. Golgi-stained areas were zoomed for better visibility (panels on the right). The lower panels show co-staining of TbRFT1-HA with TbGRASP done in parallel with the staining shown in the upper panels. Again, TbRFT1 co-localized with TbGRASP. DNA was stained with DAPI (blue). C, co-staining of TbRFT1-HA (RFT-HA) with the Golgi marker TbGRASP in cells transiently expressing TbRFT1-HA under the control of a tetracycline operator. Tetracycline (1 μg/ml) was added to the growth medium for 20 h prior to preparation of slides. D, functional TbRFT1-HA was co-stained with the lysosomal marker protein cathepsin L (CatL) and DAPI as indicated. The composite shows little overlap (yellow color) of the signals. Cathepsin L-stained areas were zoomed for better visibility (panels on the right).
To study the second possibility, we analyzed GPEET procyclin in cells grown in the presence of tunicamycin during 10 days to inhibit N-linked glycosylation. The results show that the molecular mass of GPEET was reduced by tunicamycin treatment to that of GPEET in ΔTbrft1 cells (Fig. 6A). Binding of FITC-labeled concanavalin A to parasites followed by analysis by FACS demonstrated that tunicamycin effectively inhibited N-glycosylation (Fig. 6B).
FIGURE 6.
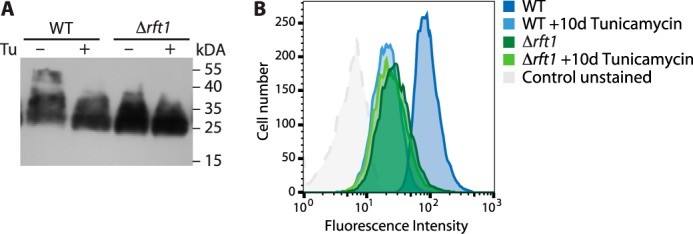
Tunicamycin-mediated inhibition of N-glycosylation in vivo. Cells were grown in the presence (+) or absence (−) of 1 μg/ml tunicamycin (Tu) during 10 days. A, proteins extracted from 2 × 107 cells were separated by SDS-PAGE, and GPEET procyclin was detected by immunoblotting using anti-GPEET antibody 5H3. B, cell surface N-glycosylation levels were assessed by lectin binding and flow cytometry using FITC-conjugated concanavalin A. The shift in cell surface fluorescence intensity reflects inhibition of N-glycosylation by tunicamycin.
Concluding Remarks
We report several new findings that implicate RFT1 in a wider range of glycosylation processes than previously demonstrated. Procyclic form trypanosomes lacking TbRFT1 not only have decreased protein N-glycosylation but also produce underglycosylated GPI anchors. The defect is not associated with the synthesis of the anchor but with GPI processing/maturation steps that likely occur in the Golgi apparatus. A role for TbRFT1 in the Golgi would be consistent with our observation that TbRFT1-HA is dually localized in the ER and Golgi. Our observations open new questions regarding the enigmatic function of TbRFT1 and its orthologs in yeast and mammalian cells.
Experimental Procedures
Materials
Unless otherwise stated, all reagents were of analytical grade and purchased from Sigma-Aldrich (Buchs, Switzerland) or Merck (Darmstadt, Germany). Restriction enzymes were from Fermentas (St. Leon-Rot, Germany), and antibiotics were from Sigma-Aldrich, InvivoGen (Nunningen, Switzerland), or Invitrogen (Basel, Switzerland). [1-3H]Etn (40–60 Ci/mmol) and [3H]GDP-Man were from American Radiolabeled Chemicals Inc. (St. Louis, MO). BioMax MS and MXBE films were from GE Healthcare (Buckinghamshire, UK) or Carestream Health (Rochester, NY).
Trypanosome Cultures
T. brucei strain Lister 427 procyclic forms were cultured at 27 °C in SDM-79 containing 5% heat-inactivated fetal bovine serum. TbRFT1 knock-out trypanosomes (ΔTbrft1, TbRFT1 null) (22) were grown under the same conditions, but in the presence of 1 μg/ml G418 for the single allele knock-out and an additional 10 μg/ml blasticidin for the double allele knock-out clones. TbRFT1 addback mutants in ΔTbrft1 double allele knock-out cells were selected and grown in the same medium, with an additional 2 μg/ml puromycin. T. brucei strain Lister 427 29-13 (TetR T7RNAP) procyclic forms for tetracycline-inducible gene expression were cultured at 27 °C in SDM-79 containing 10% heat-inactivated fetal bovine serum, 25 μg/ml hygromycin, and 15 μg/ml G418.
Generation of T. brucei RFT1 Addback Procyclic Forms
The generation of the T. brucei strain Lister 427 procyclic form TbRFT1 null cell line ΔTbrft1::NEO/ΔTbrft1::BLAST has been described previously (22). For the generation of procyclic trypanosomes constitutively expressing TbRFT1 or 2× HA-tagged TbRFT1 (TbRFT1-HA) in the ΔTbrft1 background (ΔTbrft1/RFT1 and ΔTbrft1/RFT1-HA), the ORF of TbRFT1 (Tb927.11.11670) was PCR-amplified with primers TbRFT1β_fwd (tgtagcaagcttgaattcatggacttcaaacgacagctg) and TbRFT1β_rev (ccccgcagatctctactcgccgcttctttttga) or TbRFT1_fwd and TbRFTβ-HA rev (ccccgcagatctctatgcatagtctggtacgtcataagggtatgcatagtctggtacgtcataagggtatgcatagtctggtacgtcataagggtactcgagctcgccgcttctttttgagct), respectively, and ligated into vector pG-EGFPΔLII β (44) (kindly provided by Isabel Roditi, University of Bern, Bern, Switzerland), previously digested with HindIII and BglII. The resulting vectors pG-RFT1ΔLII β and pG-RFT1-HAΔLII β were digested with NotI prior to transfection into ΔTbrft1 cells. Clones were obtained by limiting dilution under antibiotic selection using 2 μg/ml puromycin. For the expression of tetracycline-inducible TbRFT1-HA, the gene was PCR-amplified using primers TbRFT1-HA_fwd (tgtagcgggcccatggacttcaaacgacagctg) and TbRFT1-HA_rev (cctcatgcatctagactcgccgcttctttttgagct), digested with ApeI and XbaI, and ligated into equally digested vector pALC14-HA (original vector pALC14 kindly provided by André Schneider, University of Bern), containing a 3× HA tag downstream of the C-terminal restriction site. The resulting vector pALC14-TbRFT1-HA was linearized with NotI prior to transfection into T. brucei, strain Lister 427 29-13 procyclic forms. Clones were obtained by limiting dilution under antibiotic selection using 2 μg/ml puromycin.
Immunofluorescence Microscopy of Trypanosomes
2 × 106 cells were harvested at mid-log phase, washed twice in cold PBS (137 mm NaCl2, 2.7 mm KCl, 10 mm Na2PO4, 2 mm KH2PO4, pH 7.4), and spotted on microscope slides. After adherence, cells were fixed with 4% paraformaldehyde in PBS for 5 min, washed with cold PBS, and permeabilized with 0.2% (w/v) Triton X-100 in PBS for 5 min. Subsequently, cells were blocked with 2% (w/v) bovine serum albumin in PBS prior to incubation with blocking solution containing primary antibodies (mouse monoclonal anti-HA 11, 16B12 (1:200; Enzo Life Sciences; catalog number ENZ-ABS118-0500, lot 04211508), rabbit polyclonal anti-BiP (1:2500) and rabbit polyclonal anti-cathepsin L (1:500) (both kindly provided by J. Bangs, University of Buffalo, Buffalo, NY), and rabbit polyclonal anti-TbGRASP (1:1500; kindly provided by G. Warren, Vienna Biocenter, Vienna, Austria)). Fluorescent secondary antibodies with different excitation and emission maxima were used to visualize TbRFT1-HA separately from BiP, TbGRASP, and cathepsin L, respectively (Alexa Fluor goat anti-mouse 488 (Invitrogen, catalog number A11001, lot 1170048) and goat anti-rabbit 594 (Invitrogen, catalog number A11005, lot 1750828), diluted 1:1000 in blocking solution). Coverslips were mounted on dried slides with VECTASHIELD® containing DAPI (Vector Laboratories) to visualize the nuclei. Pictures were taken with a Leica DM 16000 B inverted microscope combined with a Leica DFC360 FX camera. Image deconvolution (3D deconvolution) and further processing were performed using Leica LAS X software and ImageJ (National Institutes of Health), respectively.
[3H]Etn Labeling of GPI Precursors and GPI-anchored Proteins
For in vivo radiolabeling of procyclins, procyclic form trypanosomes were cultured in the presence of [3H]Etn (2 μCi/ml) during 16 h to a density of ∼1.5 × 107 cells/ml, as described before (25). Cells were counted, harvested by centrifugation, and washed twice in cold TBS (10 mm Tris-HCl, pH 7.5, 144 mm NaCl). Bulk lipids from up to 2.5 × 108 cells were extracted from the cell pellets using 2 × 10 ml chloroform/methanol (2:1, v/v), and GPI precursors and free GPIs were extracted using 3 × 5 ml of chloroform/methanol/water (10:10:3, v/v/v) (25). GPI-anchored proteins were extracted from the remaining protein pellet using 2 × 1 ml of 9% (v/v) butan-1-ol in water during 2 h on ice, followed by 10 min of centrifugation at 17,000 × g. The resulting supernatants were pooled, dried under a stream of nitrogen, and dissolved in electrophoresis sample buffer containing 2.5% (w/v) SDS. Butan-1-ol-insoluble material was further extracted with 0.1% (w/v) Triton X-100 in 20 mm Tris (pH 7.4) for 10 min at 95 °C (25). Radioactivity in the butan-1-ol and Triton X-100 extracts was determined by liquid scintillation counting of small aliquots. For the analysis of [3H]Etn-labeled GPI precursors, all cultures were adjusted to 1.5 × 107 cells/ml prior to incubation with 4 μCi/ml [3H]Etn during 4 h. After washing with TBS and extraction of bulk lipids using 2 × 10 ml chloroform/methanol (2:1, v/v), GPI precursors were extracted by 3 × 5 ml of chloroform/methanol/water (10:10:3, v/v/v). The resulting supernatants were pooled, dried under a stream of nitrogen, and partitioned between 0.5 ml of butan-1-ol and water. After a second extraction of the resulting water phase with 0.5 ml of water-saturated butan-1-ol, butanol phases were pooled and dried using a SpeedVac apparatus. Dry GPI lipids were resuspended in 50 μl of chloroform/methanol/water (10:10:3, v/v/v) and separated by TLC as described below.
Protein Analysis
Proteins from butan-1-ol and Triton X-100 extracts were separated by SDS-PAGE using 12% polyacrylamide gels. [3H]Etn-labeled proteins were analyzed by soaking the fixed gel in AmplifyTM for 1 h, drying at 80 °C for 2 h, and exposing to MXBE film at −70 °C. HA-tagged TbRFT1, EP, and GPEET procyclins were analyzed by immunoblotting onto polyvinylidene difluoride membranes and enhanced chemiluminescence using mouse anti-HA antibody (HA11, diluted 1:3000 in TBS containing 5% skimmed milk powder; Enzo Life Sciences), mouse anti-EP antibody (mouse monoclonal anti-EP 247, diluted 1:1000 in TBS, 5% milk; Cedarlane Labs, Burlington, Ontario Canada; catalog number CLP001A, lot P115), and mouse anti-GPEET 5H3 antibody (diluted 1:5000 in TBS, 5% milk; Cedarlane Labs, lot number 991109 (45)), respectively, followed by HRP-conjugated anti-mouse IgG (diluted 1:5000 in TBS, 5% milk; Dako, Baar, Switzerland). MXBE films were exposed to ECL-activated membranes (SuperSignal West Pico, Pierce-Thermo Fisher) for 30 s to a few minutes.
Mass Spectrometry Analysis
For the analysis of procyclin C-terminal polypeptides, total procyclins were purified by sequential extraction with chloroform/methanol/water (10:10:3, by volume) and 9% (v/v) butan-1-ol as described above. Butan-1-ol extracts were then dried, dephosphorylated with 48% anhydrous hydrofluoric acid for 24 h at 0 °C, hydrolyzed with mild (40 mm) trifluoroacetic acid, and then analyzed by negative-ion MALDI-TOF on an ABI Voyager DE-STR instrument using sinapinic acid as the matrix (12). To determine the total monosaccharide composition of procyclins, samples were mixed with 200 pmol of scyllo-inositol (as internal standard) and analyzed by GC-MS as described elsewhere (46).
Enzyme Treatments
For PNGase treatment, parasites were lysed by boiling 10 min at 100 °C in denaturing buffer (0.5% SDS, 40 mm DTT) and incubated with PNGase F (New England Biolabs) according to the manufacturer's instructions in a buffer containing 1% Nonidet P-40 during 1 h at 37 °C. For Pronase treatment, 3H-labeled GPI extracts were dried under a stream of nitrogen, re-dissolved in a buffer containing 20 mm Tris (pH 7.5) and 5 mm CaCl2, and incubated with Pronase (0.3 mg/ml) during 16 h at 37 °C.
Cell-free GPI Glycosylation Analysis
Membranes for cell-free labeling of GPI precursors were collected from trypanosome cultures grown to a density of 107 cells/ml by hypotonic lysis in water containing 0.1 mm tosyl-l-lysyl-chloromethane hydrochloride (TLCK) and 1.25 μg/ml leupeptin. Pulse-chase radiolabeling of GPI precursors with [3H]GDP-Man was performed essentially according to the protocol developed by Masterson et al. (47) as described by Leal et al. (48). If indicated, experiments were performed in the presence of 0.4 mg/ml tunicamycin in the assay buffer to inhibit the formation of dolichol-linked N-glycan precursors. In pulse-chase experiments, membranes from 1.5 × 108 cells were labeled with 3 μCi of [3H]GDP-Man and 1 mm UDP-GlcNAc for 8 min at 37 °C, followed by a 75-min chase with 2 mm non-radioactive GDP-Man. For the analysis of GPI galactosylation, 8 mm UDP-Gal was added during the chase with GDP-Man. GPI lipids were extracted as described (47). Butan-1-ol extracts were separated by TLC on Silica Gel 60 plates (Merck Millipore) using chloroform/methanol/water (10:10:3, v/v/v) as solvent system and analyzed using a radioactivity TLC scanner (Berthold Technologies, Regensburg, Switzerland). For autoradiography, TLC plates were treated with EN3HANCETM spray (Perkin Elmer) prior to exposure to film at −70 °C.
Flow Cytometry
Trypanosomes were grown in the presence or absence of tunicamycin (1 μg/ml) during 10 days. Approximately 1 × 107 parasites were harvested by centrifugation at 4 °C for 10 min at 1500 × g in 15-ml centrifuge tubes, washed twice in ice-cold SDM-79, and resuspended in 200 μl of SDM-79 containing 0.5 mm MnCl2. Concanavalin A-FITC conjugate was added to a final concentration of 1.5 μg/ml. After 1 h of incubation in the dark on ice, the cells were diluted with ice-cold PBS to a volume of 5 ml, pelleted, resuspended in a final volume of 2 ml (final concentration 5 × 106/ml PBS), and passed through a cell-filter cap prior to analysis by FACSCalibur (BD Biosciences). Data were analyzed using flow cytometry software FlowJo.
Author Contributions
P. G. designed, performed, and analyzed the experiments shown in Figs. 1C and 3–6 and prepared all figures. A. G. S. generated RFT1 addback-strains and performed the experiment shown in Fig. 1B. Y. C. L. and A. A. S. developed the protocol for in vitro galactosylation of GPI precursors. A. A. S. designed the scheme in Fig. 1A and designed and performed mass spectrometry analyses presented in Fig. 2. P. B. conceived and coordinated the study, and P. B. and P. G. wrote the paper. A. A. S. and A. K. M. provided scientific and experimental advice and critically revised the paper.
Acknowledgments
We thank Luce Farine for generating a T. brucei procyclic form cell line expressing TbEMC3-HA and Jennifer Jelk and Monika Rauch for excellent technical assistance during parts of the study. We also thank the Dundee University Proteomics Facility and Mike Ferguson for generous use of the MALDI Voyager and GC-MS, respectively, and Aitor Casas-Sánchez for the tunicamycin protocol. We thank William White for stimulating discussions.
The work was supported by Swiss National Science Foundation Grants Sinergia CRSII3_141913 and 149353 (to P. B.), National Institutes of Health Grant R21NS093457 (to A. K. M.), and GlycoPar-EU FP7 Marie Curie Initial Training Network Grant GA. 608295 (to A. A. S.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- GPI
- glycosylphosphatidylinositol
- VSG
- variant surface glycoprotein
- mDLO
- Man9GlcNAc2-PP-dolichol
- M5-DLO
- Man5GlcNAc2-PP-dolichol
- PNGase
- protein N-glycosidase F
- Etn
- ethanolamine
- Dol
- dolichol
- Dol-P-Man
- dolichol-phosphate mannose
- TbRFT1
- Trypanosoma brucei RFT1
- ER
- endoplasmic reticulum
- GRASP
- Golgi reassembly and stacking protein
- EMC
- endoplasmic reticulum membrane protein complex.
References
- 1. Sutton R. E., and Boothroyd J. C. (1986) Evidence for Trans splicing in trypanosomes. Cell 47, 527–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Blum B., Bakalara N., and Simpson L. (1990) A model for RNA editing in kinetoplastid mitochondria: RNA molecules transcribed from maxicircle DNA provide the edited information. Cell 60, 189–198 [DOI] [PubMed] [Google Scholar]
- 3. Cross G. A. (1977) Antigenic variation in trypanosomes. Am. J. Trop. Med. Hyg. 26, 240–244 [DOI] [PubMed] [Google Scholar]
- 4. Ferguson M. A., Homans S. W., Dwek R. A., and Rademacher T. W. (1988) Glycosyl-phosphatidylinositol moiety that anchors Trypanosoma brucei variant surface glycoprotein to the membrane. Science 239, 753–759 [DOI] [PubMed] [Google Scholar]
- 5. Ferguson M. A. J. (1999) The structure, biosynthesis and functions of glycosylphosphatidylinositol anchors, and the contributions of trypanosome research. J. Cell Sci. 112, 2799–2809 [DOI] [PubMed] [Google Scholar]
- 6. Allen G., Gurnett L. P., and Cross G. A. M. (1982) Complete amino acid sequence of a variant surface glycoprotein (VSG 117) from Trypanosoma brucei. J. Mol. Biol. 157, 527–546 [DOI] [PubMed] [Google Scholar]
- 7. Roditi I., Carrington M., and Turner M. (1987) Expression of a polypeptide containing a dipeptide repeat is confined to the insect stage of Trypanosoma brucei. Nature 325, 272–274 [DOI] [PubMed] [Google Scholar]
- 8. Mowatt M. R., and Clayton C. E. (1988) Polymorphism in the procyclic acidic repetitive protein gene family of Trypanosoma brucei. Mol. Cell. Biol. 8, 4055–4062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Roditi I., Schwarz H., Pearson T. W., Beecroft R. P., Liu M. K., Richardson J. P., Bühring H.-J., Pleiss J., Bülow R., Williams R. O., and Overath P. (1989) Procycline gene expression and loss of the variant surface glycoprotein during differentiation of Trypanosoma brucei. J. Cell Biol. 108, 737–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ferguson M. A. J., Murray P., Rutherford H., and McConville M. J. (1993) A simple purification of procyclic acidic repetitive protein and demonstration of a sialylated glycosyl-phosphatidylinositol membrane anchor. Biochem. J. 291, 51–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Treumann A., Zitzmann N., Hülsmeier A., Prescott A. R., Almond A., Sheehan J., and Ferugson M. A. J. (1997) Structural characterisation of two forms of procyclic acidic repetitive protein expressed by procyclic forms of Trypanosoma brucei. J. Mol. Biol. 269, 529–547 [DOI] [PubMed] [Google Scholar]
- 12. Acosta-Serrano A., Cole R. N., Mehlert A., Lee M. G.-S., Ferguson M. A. J., and Englund P. T. (1999) The procyclin repertoire of Trypanosoma brucei: identification and structural characterization of the Glu-Pro-rich polypeptides. J. Biol. Chem. 274, 29763–29771 [DOI] [PubMed] [Google Scholar]
- 13. Izquierdo L., Mehlert A., and Ferguson M. A. J. (2012) The lipid-linked oligosaccharide donor specificities of Trypanosoma brucei oligosaccharyltransferases. Glycobiology. 22, 696–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Acosta-Serrano A., O'Rear J., Quellhorst G., Lee S. H., Hwa K.-Y., Krag S. S., and Englund P. T. (2004) Defects in the N-linked oligosaccharide biosynthetic pathway in a Trypanosoma brucei glycosylation mutant. Eukaryot. Cell 3, 255–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Manthri S., Güther M. L. S., Izquierdo L., Acosta-Serrano A., and Ferguson M. A. J. (2008) Deletion of the TbALG3 gene demonstrates site-specific N-glycosylation and N-glycan processing in Trypanosoma brucei. Glycobiology. 18, 367–383 [DOI] [PubMed] [Google Scholar]
- 16. Koerte A., Chong T., Li X., Wahane K., and Cai M. (1995) Suppression of the yeast mutation rft1-1 by human p53. J. Biol. Chem. 270, 22556–22564 [DOI] [PubMed] [Google Scholar]
- 17. Helenius J., Ng D. T. W., Marolda C. L., Walter P., Valvano M. A., and Aebi M. (2002) Translocation of lipid-linked oligosaccharides across the ER membrane requires Rft1 protein. Nature 415, 447–450 [DOI] [PubMed] [Google Scholar]
- 18. Haeuptle M. A., Pujol F. M., Neupert C., Winchester B., Kastaniotis A. J., Aebi M., and Hennet T. (2008) Human RFT1 deficiency leads to a disorder of N-linked glycosylation. Am. J. Hum. Genet. 82, 600–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sanyal S., Frank C. G., and Menon A. K. (2008) Distinct flippases translocate glycerophospholipids and oligosaccharide diphosphate dolichols across the endoplasmic reticulum. Biochemistry 47, 7937–7946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Frank C. G., Sanyal S., Rush J. S., Waechter C. J., and Menon A. K. (2008) Does Rft1 flip an N-glycan lipid precursor? Nature 454, E3–4; discussion E4–5 [DOI] [PubMed] [Google Scholar]
- 21. Rush J. S., Gao N., Lehrman M. A., Matveev S., and Waechter C. J. (2009) Suppression of Rft1 expression does not impair the transbilayer movement of Man5GlcNAc2-P-P-dolichol in sealed microsomes from yeast. J. Biol. Chem. 284, 19835–19842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jelk J., Gao N., Serricchio M., Signorell A., Schmidt R. S., Bangs J. D., Acosta-Serrano A., Lehrman M. A., Bütikofer P., and Menon A. K. (2013) Glycoprotein biosynthesis in a eukaryote lacking the membrane protein Rft1. J. Biol. Chem. 288, 20616–20623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Clayton C. E., and Mowatt M. R. (1989) The procyclic acidic repetitive proteins of Trypanosoma brucei: purification and post-translational modification. J. Biol. Chem. 264, 15088–15093 [PubMed] [Google Scholar]
- 24. Hwa K.-Y., Acosta-Serrano A., Khoo K.-H., Pearson T., and Englund P. T. (1999) Protein glycosylation mutants of procyclic Trypanosoma brucei: defects in the asparagine-glycosylation pathway. Glycobiology 9, 181–190 [DOI] [PubMed] [Google Scholar]
- 25. Bütikofer P., Ruepp S., Boschung M., and Roditi I. (1997) “GPEET” procyclin is the major surface protein of procyclic culture forms of Trypanosoma brucei brucei strain 427. Biochem. J. 326, 415–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vassella E., Van Den Abbeele J., Bütikofer P., Renggli C. K., Furger A., Brun R., and Roditi I. (2000) A major surface glycoprotein of Trypanosoma brucei is expressed transiently during development and can be regulated post-transcriptionally by glycerol or hypoxia. Genes Dev. 14, 615–626 [PMC free article] [PubMed] [Google Scholar]
- 27. Acosta-Serrano A., Vassella E., Liniger M., Kunz Renggli C., Brun R., Roditi I., and Englund P. T. (2001) The surface coat of procyclic Trypanosoma brucei: programmed expression and proteolytic cleavage of procyclin in the tsetse fly. Proc. Natl. Acad. Sci. 98, 1513–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ferguson M. A. J., Low M. G., and Cross G. A. (1985) Glycosyl-sn-1,2-dimyristylphosphatidylinositol is covalently linked to Trypanosoma brucei variant surface glycoprotein. J. Biol. Chem. 260, 14547–14555 [PubMed] [Google Scholar]
- 29. Field M. C., Menon A. K., and Cross G. A. (1991) A glycosylphosphatidylinositol protein anchor from procyclic stage Trypanosoma brucei: lipid structure and biosynthesis. EMBO J. 10, 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Orlean P., and Menon A. K. (2007) Thematic review series: Lipid Posttranslational Modifications. GPI anchoring of protein in yeast and mammalian cells, or: how we learned to stop worrying and love glycophospholipids. J. Lipid Res. 48, 993–1011 [DOI] [PubMed] [Google Scholar]
- 31. Sanyal S., and Menon A. K. (2009) Flipping lipids: why an' what's the reason for? ACS Chem. Biol. 4, 895–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hong Y., and Kinoshita T. (2009) Trypanosome glycosylphosphatidylinositol biosynthesis. Korean J. Parasitol. 47, 197–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mayor S., Menon A. K., and Cross G. A. M. (1992) Galactose-containing glycosylphosphatidylinositols in Trypanosoma brucei. J. Biol. Chem. 267, 754–761 [PubMed] [Google Scholar]
- 34. Izquierdo L., Nakanishi M., Mehlert A., Machray G., Barton G. J., and Ferguson M. A. J. (2009) Identification of a glycosylphosphatidylinositol anchor-modifying β1-3 N-acetylglucosaminyl transferase in Trypanosoma brucei. Mol. Microbiol. 71, 478–491 [DOI] [PubMed] [Google Scholar]
- 35. Nakanishi M., Karasudani M., Shiraishi T., Hashida K., Hino M., Ferguson M. A. J., and Nomoto H. (2014) TbGT8 is a bifunctional glycosyltransferase that elaborates N-linked glycans on a protein phosphatase AcP115 and a GPI-anchor modifying glycan in Trypanosoma brucei. Parasitol. Int. 63, 513–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Izquierdo L., Acosta-Serrano A., Mehlert A., and Ferguson M. A. (2015) Identification of a glycosylphosphatidylinositol anchor-modifying β1–3 galactosyltransferase in Trypanosoma brucei. Glycobiology. 25, 438–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. He C. Y., Ho H. H., Malsam J., Chalouni C., West C. M., Ullu E., Toomre D., and Warren G. (2004) Golgi duplication in Trypanosoma brucei. J. Cell Biol. 165, 313–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lahiri S., Chao J. T., Tavassoli S., Wong A. K. O., Choudhary V., Young B. P., Loewen C. J. R., and Prinz W. A. (2014) A conserved endoplasmic reticulum membrane protein complex (EMC) facilitates phospholipid transfer from the ER to mitochondria. PLoS Biol. 12, e1001969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Farine L., Niemann M., Schneider A., and Bütikofer P. (2015) Phosphatidylethanolamine and phosphatidylcholine biosynthesis by the Kennedy pathway occurs at different sites in Trypanosoma brucei. Sci. Rep. 5, 16787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Martin K. L., and Smith T. K. (2006) Phosphatidylinositol synthesis is essential in bloodstream form Trypanosoma brucei. Biochem. J. 396, 287–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mbawa Z. R., Gumm I. D., Shaw E., and Lonsdale-Eccles J. D. (1992) Characterisation of a cysteine protease from bloodstream forms of Trypanosoma congolense. Eur. J. Biochem. 204, 371–379 [DOI] [PubMed] [Google Scholar]
- 42. Liu L., Xu Y.-X., Caradonna K. L., Kruzel E. K., Burleigh B. A., Bangs J. D., and Hirschberg C. B. (2013) Inhibition of nucleotide sugar transport in Trypanosoma brucei alters surface glycosylation. J. Biol. Chem. 288, 10599–10615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Izquierdo L., Schulz B. L., Rodrigues J. A., Güther M. L. S., Procter J. B., Barton G. J., Aebi M., and Ferguson M. A. J. (2009) Distinct donor and acceptor specificities of Trypanosoma brucei oligosaccharyltransferases. EMBO J. 28, 2650–2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Furger A., Schürch N., Kurath U., and Roditi I. (1997) Elements in the 3′ untranslated region of procyclin mRNA regulate expression in insect forms of Trypanosoma brucei by modulating RNA stability and translation. Mol. Cell Biol. 17, 4372–4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bütikofer P., Vassella E., Ruepp S., Boschung M., Civenni G., Seebeck T., Hemphill A., Mookherjee N., Pearson T. W., and Roditi I. (1999) Phosphorylation of a major GPI-anchored surface protein of Trypanosoma brucei during transport to the plasma membrane. J. Cell Sci. 112, 1785–1795 [DOI] [PubMed] [Google Scholar]
- 46. Ferguson M. A. J. (1993) Glycobiology: A Practical Approach (Fukuda M., and Kobata A. eds), pp. 349–383, Oxford University Press, Oxford [Google Scholar]
- 47. Masterson W. J., Doering T. L., Hart G. W., and Englund P. T. (1989) A novel pathway for glycan assembly: biosynthesis of the glycosyl-phosphatidylinositol anchor of the trypanosome variant surface glycoprotein. Cell 56, 793–800 [DOI] [PubMed] [Google Scholar]
- 48. Leal S., Acosta-Serrano A., Morris J., and Cross G. A. M. (2004) Transposon mutagenesis of Trypanosoma brucei identifies glycosylation mutants resistant to concanavalin A. J. Biol. Chem. 279, 28979–28988 [DOI] [PubMed] [Google Scholar]
- 49. Hall B. S., Pal A., Goulding D., Acosta-Serrano A., and Field M. C. (2005) Trypanosoma brucei: TbRAB4 regulates membrane recycling and expression of surface proteins in procyclic forms. Exp. Parasitol. 111, 160–171 [DOI] [PubMed] [Google Scholar]
- 50. Field M. C., Menon A. K., and Cross G. A. M. (1991) Developmental variation of glycosylphosphatidylinositol membrane anchors in Trypanosoma brucei. J. Biol. Chem. 266, 8392–8400 [PubMed] [Google Scholar]