Abstract
Cell-penetrating peptides (CPPs) are well established as delivery agents for otherwise cell-impermeable cargos. CPPs can also theoretically be used to modulate intracellular processes. However, their susceptibility to proteolytic degradation often limits their utility in these applications. Previous studies have explored the consequences for cellular uptake of converting the residues in CPPs from l- to d-stereochemistry, but conflicting results have been reported and specific steps en route to intracellular activity have not been explored. Here we use dimeric fluorescence TAT as a model CPP to explore the broader consequences of l- to d-stereochemical conversion. We show that inversion of chirality provides protease resistance without altering the overall mode of cellular entry, a process involving endocytic uptake followed by endosomal escape and cytosolic access. However, whereas inversion of chirality reduces endocytic uptake, the d-peptide, once in the endosome, is significantly more prone to escape than its l-counterpart. Moreover, the d-peptide is retained in the cytosol of cells for several days, whereas the l-peptide is degraded within hours. Notably, while the l-peptide is relatively innocuous to cells, the d-peptide exerts a prolonged anti-proliferative activity. Together, our results establish connections between chirality, protease resistance, cellular penetration, and intracellular activity that may be useful for the development of future delivery agents with improved properties.
Keywords: cell permeabilization, cell proliferation, cell-penetrating peptide (CPP), drug delivery system, peptide chemical synthesis, peptides
Introduction
Cell-penetrating peptides (CPPs)2 have become important tools for the delivery of macromolecular cargoes inside cells (1–3). These delivery agents show promise in therapeutic applications and are useful reagents for cell biology assays (4–6). For instance, CPPs (TAT, penetratin, etc.) are currently being tested in several preclinical and clinical trials (7, 8). However, CPPs exposed to cells or serum are rapidly degraded, and this can consequently render these compounds less effective in vivo or in vitro (9–13). To protect CPPs from degradation, a common strategy has been to employ d-amino acids instead of their l-amino acid counterparts. d-Peptides are protease-resistant, and this approach has been applied to CPPs such as TAT, R9, penetratin, hLF, pVEC, and sweet arrow peptide (10, 14–17). In addition, the extended in vivo half-lives of d-peptides over l-peptides have contributed to the successful development of d-polyarginine CPPs as cancer contrast agents (18, 19). How inversion of chirality impacts the multifaceted functions of CPPs, however, remains unclear.
Several reports have indicated that cellular uptake of CPPs is independent of peptide backbone chirality (16, 20). Uptake of the CPPs studied was thought to involve direct plasma membrane translocation. This is because uptake persisted at 4 °C, a condition that typically abolishes endocytosis (16). In many cases, however, the penetration of CPPs into cells involves different routes of cellular entry (21). Instead of crossing the plasma membrane directly, certain CPPs are first internalized by endocytosis and accumulate inside endosomes. In a second step, CPPs escape from endosomes to penetrate the cytosolic space (22, 23). The effect of chirality on endocytosis has been reported for the CPPs R9, penetratin, and hLF. Preferential uptake of l-CPPs over d-CPPs was observed, suggesting that chirality is important for the interactions between these compounds and cell surface partners that induce endocytosis (24). Together, these studies reveal conflicting results that highlight how l- to d-amino acid conversion may lead to CPP-dependent results.
To date, several questions related to the effect of chirality on CPPs' activities remain unanswered. In particular, how chirality impacts endosomal escape during the cell penetration process and how an unnatural d-CPP affects cell physiology once inside a cell are still unclear (25). The issue of endosomal escape is important because this step is critical for the successful delivery of molecules of interest into a cell (26). Similarly, the issue concerning the physiological impact of cell penetration is of significance when one considers, for instance, using cellular delivery protocols as a tool for cell biology and therapeutic applications (i.e. most applications would benefit from delivery protocols that only minimally disrupt cells).
In this report, we address these questions by using dimeric fluorescent TAT (dfTAT) as a model CPP. dfTAT is a disulfide-bonded dimer of the prototypical CPP TAT. dfTAT is capable of escaping from endosomes with extremely high efficiency (27). For instance, whereas TAT escapes endosomes with an estimated efficiency of <1% (i.e. 1% of peptide present in a cell reaches the cytosol, whereas 99% remains trapped inside endosomes), the endosomal escape efficiency of dfTAT exceeds 90% (27–29). Mechanistically, cytosolic penetration of dfTAT involves egress from late endosomes (it does not involve escape from other endocytic organelles or direct plasma membrane translocation) (30). This egress appears to be mediated by the leaky fusion of late endosomal membranes and involves interactions between the positively charged CPP and the anionic lipid bis(monoacyl)glycerophosphate (BMP) (30). From a delivery point of view, the mode of cell penetration by dfTAT is useful because macromolecules, endocytosed along with dfTAT after co-incubation with cells, can also be released into the cytosol of cells (i.e. by leaking out of late endosomes along with the endosomolytic CPP). This approach has been used to deliver an enzyme, a transcription factor, peptides, and cell-impermeable small molecules with high efficiencies (27). Importantly, the leakage induced by dfTAT appears to be relatively innocuous to cells because dfTAT-mediated delivery does not lead to changes in cellular proliferation or transcriptional responses. dfTAT therefore represents a CPP with desirable properties and with an activity sufficiently high to enable mechanistic studies.
Herein, we investigate the cell penetration of a d-amino acid analog of dfTAT to establish the impact of chirality on cytosolic access. We establish that an l- to d-amino acid substitution, by inhibiting peptide degradation from endocytic proteases, promotes enhanced endosomal escape activity. We also establish that the d-CPP, after successful cytosolic entry, gives rise to perturbations in proliferation and transcription more severe than l-CPP. This can be attributed to the prolonged intracellular retention of the d-peptide. Therefore, for the first time, we report on an anti-proliferative effect of a d-CPP and reveal additional advantages and disadvantages inherent to conferring protease resistance to an endosomolytic agent. We envision that these findings are relevant to other cell-permeable molecules (e.g. stapled peptides, peptoids, CPPs, etc.), in particular those that, like dfTAT, have high arginine residue content.
Results
d-dfTAT Is Protease-resistant and Capable of Penetrating Cells
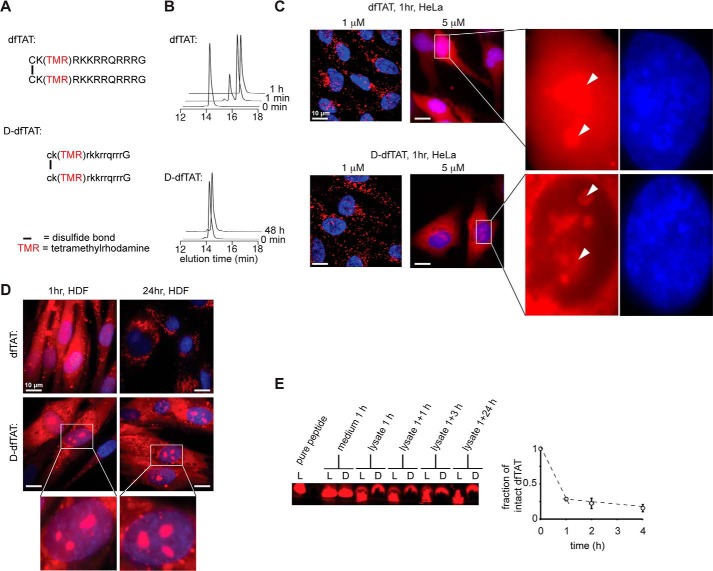
dfTAT (i.e. l-dfTAT) and d-dfTAT were prepared by synthesis of the peptide CK(TMR)RKKRRQRRRG or ck(TMR)rkkrrqrrrG, respectively, on solid phase followed by disulfide bond formation of the cysteine residue in solution (Fig. 1A and supplemental Figs. S1 and S2). TMR is the red fluorophore carboxytetramethylrhodamine. To confirm that the incorporation of d-amino acids confers protease resistance to d-dfTAT, the peptide was exposed to trypsin in vitro. Consistent with the high Arg and Lys content of the TAT sequence, the parent compound dfTAT was degraded by trypsin within minutes, as evidenced by HPLC analysis (Fig. 1B). In contrast, d-dfTAT remained intact for the duration of the experiment (48 h).
FIGURE 1.
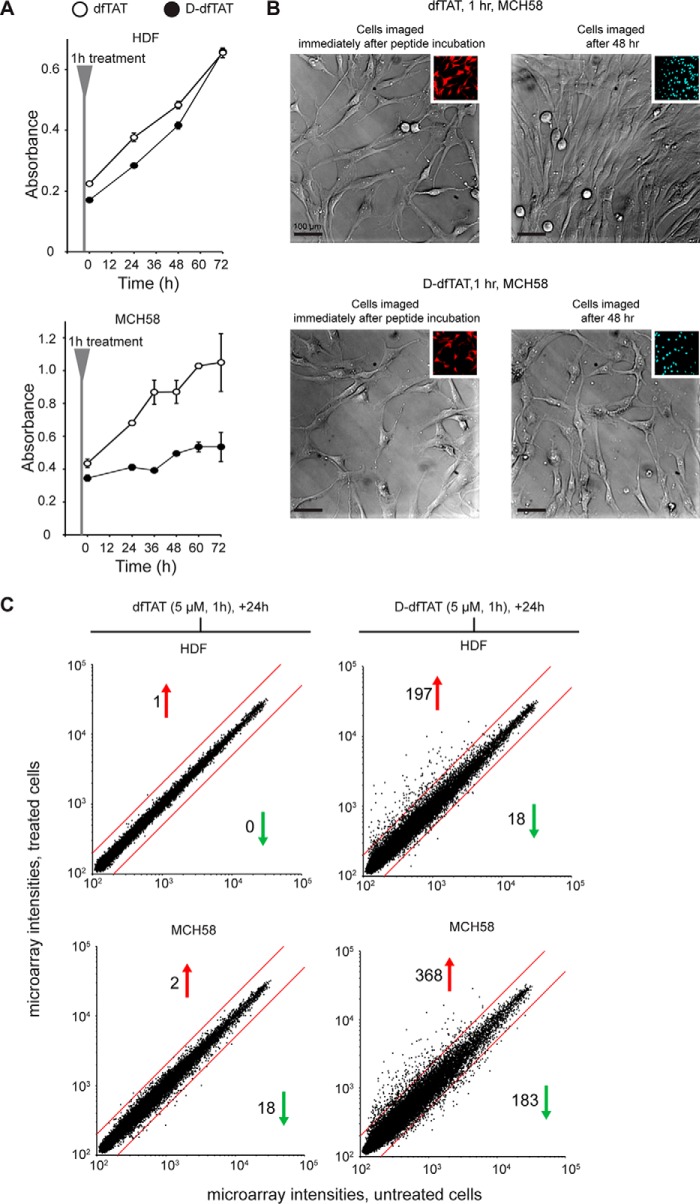
d-dfTAT is protease-resistant, whereas dfTAT is not. A, schematic representation of the amino acid sequence of dfTAT (top) and d-dfTAT (bottom). B, HPLC analysis of dfTAT and d-dfTAT (10 μm) before (0 min time point) and after treatment with trypsin (0.025%). Reactions were quenched at different time intervals with 0.01% TFA in water (1 min, 1 h, and 48 min). Peaks were detected by monitoring the absorbance of the TMR chromophore at 550.8 nm. C, cellular distribution of dfTAT and d-dfTAT after a 1-h incubation. HeLa cells were incubated with dfTAT or d-dfTAT at 1 or 5 μm for 1 h, washed, and then stained with the cell-permeable Hoechst nuclear stain (1 μg/ml). Live cells were imaged with a ×100 objective. Fluorescence images are overlays of the TMR emission at 560 nm (pseudocolored red) and Hoechst emission at 460 nm (pseudocolored blue). Insets to the left show a zoom into the nucleolar staining of each peptide (white arrowheads). Scale bars, 10 μm. D, cellular distribution of dfTAT and d-dfTAT immediately after delivery and after 24 h. HDF cells were incubated with dfTAT or d-dfTAT at 5 μm for 1 h and washed. Cells were imaged with a ×100 objective either immediately after treatment or after an additional 24-h incubation. Fluorescence images are overlays of the TMR emission at 560 nm (pseudocolored red) and Hoechst emission at 460 nm (DNA stain added during imaging, pseudocolored blue). Scale bars, 10 μm. E, analysis of peptide degradation by Tris-Tricine gel electrophoresis of cell lysates. HeLa cells were incubated with 5 μm dfTAT or d-dfTAT for 1 h and lysed either immediately (1 h) or at 1, 3, or 24 h after peptide incubation. Cell lysates were analyzed by electrophoresis along with an aliquot of the peptide solution incubated with cells (medium 1 h) and with a sample of pure peptide. The gel was imaged on a fluorescence scanner. The fluorescent bands were quantified by densitometry to generate the graph presented. The data reported represent the average and corresponding S.D. values (error bars) of biological triplicates.
To determine whether the cell penetration activity of dfTAT is retained after a l- to d-amino acid substitution, HeLa and human dermal fibroblast (HDF) cells were incubated with 1 or 5 μm dfTAT or d-dfTAT for 1 h and examined by fluorescence microscopy. At 1 μm, both peptides show a punctate distribution indicative of endosomal entrapment (as previously reported for dfTAT). In contrast, at 5 μm peptide incubation, cells displayed a diffuse cytoplasmic fluorescent signal accompanied with a nucleolar staining observable immediately after incubation (Fig. 1C). These fluorescence stainings are consistent with successful entry of each peptide into the cell's cytosolic space because cytosolic entry presumably precedes targeting to nucleoli (27, 31, 32). In addition, the fluorescence signal of d-dfTAT was low in the nucleus (apart from the nucleoli), and a sharp contrast was observed with the cytoplasmic/nucleolar signal. The fluorescence staining of cells was monitored for 24 h to establish how the fluorescence signal of each peptide changes overtime (Fig. 1D). The nucleolar staining of dfTAT disappeared within 2 h. The diffuse signal of dfTAT also progressively disappeared over time and was replaced by a punctate distribution in less than 24 h (Fig. 1D). In contrast, the fluorescence signal of d-dfTAT remained unchanged, and a strong nucleolar staining was retained throughout the duration of the experiment (Fig. 1D).
To assess the stability of each peptide in cellulo, an analysis of cellular extracts was performed using gel electrophoresis. A high percentage acrylamide gel was used to separate the full-length peptides from smaller degradation fragments (the peptides are reduced with DTT to simplify the analysis; consequently, the monomeric peptides fTAT and d-fTAT are detected in this assay). The fluorescence signal of TMR, a moiety presumably not readily degradable by cells, was used to detect all of the species generated upon cellular exposure. Cells were incubated with dfTAT or d-dfTAT (5 μm) for 1 h, washed, and incubated with fresh medium (medium not containing peptide) for up to 24 h. Incubation medium and cell lysates were extracted at various time points. The samples recovered from the medium immediately after incubation of cells with dfTAT and d-dfTAT display a single species comparable in size with that of the pure control peptide fTAT (reduced dfTAT) (Fig. 1E). The cell lysates obtained from cells incubated with d-dfTAT show a similar band, independent of incubation time. In contrast, the lysates of cells incubated with dfTAT for 1 h show a band corresponding to fTAT and a smear corresponding to smaller degradation species. Analysis of the band intensities by densitometry shows that ∼25% of the peptide is intact, whereas 75% is degraded. This ratio remains relatively unchanged after an additional 1 and 3 h of incubation in fresh medium. However, the band corresponding to fTAT is not present anymore at the 24 h time point. Overall, these data indicate that dfTAT is rapidly degraded during incubation with cells and during the delivery process. The degradation of dfTAT continues after cellular penetration but on a slower time scale. In contrast, d-dfTAT resists degradation throughout the duration of the experiment.
Both dfTAT and d-dfTAT Deliver Macromolecules inside Cells and Access the Cytosol of Cells by Using a Similar Endocytic Route
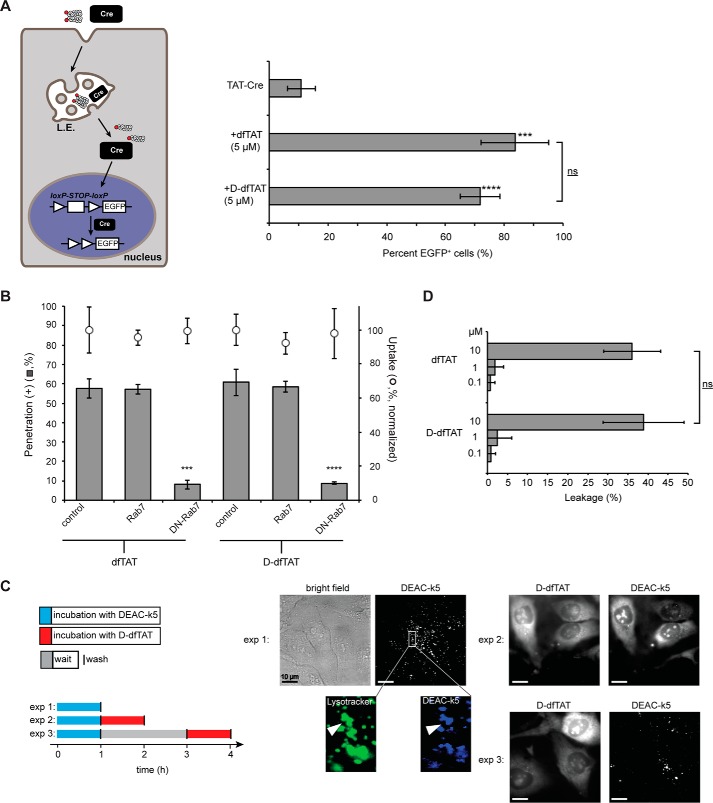
The utility of dfTAT is based on the fact that this reagent renders endosomes leaky and that, consequently, it can mediate the efficient cytosolic delivery of macromolecules into live cells by a simple co-incubation protocol (27). To determine whether d-dfTAT displays a similar activity, a Cre recombinase delivery assay was performed. In this assay, successful nuclear delivery of Cre recombinase (38 kDa) results in the expression of an EGFP reporter. We used TAT-Cre, a Cre recombinase fused to the protein transduction domain (PTD) TAT. TAT-Cre has previously been shown to enter cells, and we therefore used this reagent to assess how dfTAT-mediated delivery compares with the commonly used PTD approach (33). As shown in Fig. 2A, incubation of cells with TAT-Cre led to expression of EGFP in ∼10% of the cell population. In contrast, the addition of 5 μm dfTAT or d-dfTAT peptide during incubation led to >70% of cells positive for EGFP expression. These data suggest that both dfTAT and d-dfTAT are capable of delivering a macromolecule inside the cytosol and nucleus of live cells at a level superior to that observed with a PTD fusion alone.
FIGURE 2.
d-dfTAT enters cells by endocytosis followed by endosomal escape in a manner similar to dfTAT. A, dfTAT and d-dfTAT deliver Cre recombinase into live cells. HeLa cells were transfected with a plasmid containing EGFP upstream of a loxP-STOP-loxP sequence. The cells were then co-incubated with dfTAT or d-dfTAT (5 μm) and 4 μm Cre recombinase. Cells positive for EGFP expression were counted 24 h after the protein delivery treatment. ***, p ≤ 0.001; ****, p ≤ 0.0001 compared with control (TAT-Cre). The bracket indicates comparison of p values obtained using t test analysis of dfTAT and d-dfTAT treatments (ns, p > 0.05). B, cytosolic penetration of d-dfTAT is blocked by expression of dominant negative Rab7. HeLa cells were transfected with Rab7 or dominant negative Rab7 (DN-Rab7). dfTAT or d-dfTAT (3 μm) was then incubated with cells for 1 h, and cell penetration was quantified. The total fluorescence of cell lysates was also measured to assess the impact of each treatment on peptide endocytic uptake. The endocytic uptake of cells transfected with Rab7 or DN-Rab7 was normalized to the endocytic uptake of cells treated with dfTAT or d-dfTAT, respectively. ***, p ≤ 0.001; ****, p ≤ 0.0001 compared with control. C, d-dfTAT causes the release of a peptide preloaded into endosomes but not that of a peptide preloaded into lysosomes. In experiment 1 (exp 1), cells were incubated with DEAC-k5 (20 μm) for 1 h and washed. Cells were then incubated with LysoTracker during imaging to establish the accumulation of DEAC-k5 inside endosomes. In experiment 2 (exp 2), cells were incubated with DEAC-k5 (20 μm) for 1 h, washed, and then incubated with d-dfTAT (5 μm) for 1 h. Experiment 3 (exp 3) was performed as experiment 2 with the exception of a 2-h waiting time between incubation with DEAC-k5 and d-dfTAT. Fluorescence images represented are either monochromatic or pseudocolored green for LysoTracker and blue for DEAC-k5. Scale bars, 10 μm. D, d-dfTAT causes leakage of liposomes with a lipid composition mimicking that of late endosomes. Liposomes loaded with calcein (500 μm) were incubated with dfTAT or d-dfTAT (0.1, 1, or 10 μm). The fluorescence signal of free calcein was quantified after peptide treatment. ns, p > 0.05. The data reported in this figure represent the average and corresponding S.D. values (error bars) of biological triplicates.
dfTAT penetrates the cytosol of cells by escaping from late endosomes via interactions with the lipid BMP (30). To examine whether d-dfTAT follows the same route for cellular entry, d-dfTAT was incubated with cells transfected with a dominant negative Rab7 (DN-Rab7), a construct known to block endocytic trafficking between early and late endosomes (34, 35). The effect of Rab7 and DN-Rab7 on peptide endocytic uptake was established by measuring the overall fluorescence of cell lysates. The cytosolic penetration activity of d-dfTAT was established by counting the number of cells displaying a distinct nucleolar staining by the peptide. Notably, this assay does not directly measure how much peptide remains trapped inside endosomes versus how much escapes endosomes. Instead, it is a binary assay that establishes whether or not endosomal leakage is achieved above a detection threshold determined by our fluorescence microscopy set-up. As observed previously with dfTAT, the overall endocytic uptake of d-dfTAT was not significantly altered by overexpression of the Rab7 constructs, indicating that DN-Rab7 does not prevent d-dfTAT from accumulating inside endosomes. However, DN-Rab7 inhibited cytosolic penetration of d-dfTAT, whereas wild type did not (Fig. 2B). These results indicate that DN-Rab7 blocks the endosomal escape of d-dfTAT and that, consequently, d-dfTAT escapes from endocytic organelles that are downstream of early endosomes in the endocytic pathway. In addition, to further test the involvement of endocytic trafficking in cell penetration, cells were preincubated with anti-BMP (50 μg/ml), an antibody previously shown to block membrane fusion within late endosomes (36–38). As in the case of DN-Rab-7, the cell penetration of dfTAT and d-dfTAT was inhibited by this treatment to a similar extent (supplemental Fig. S3). Overall, these data support the model that both dfTAT and d-dfTAT reach the cytosol by an endocytic route in a majority of cells. It is important to note that penetration persisted in ∼5% of cells in the presence of DN-Rab7 or anti-BMP. On one hand, it is possible that the inhibitors did not work effectively in these cells (i.e. maybe because of low expression, degradation, etc.). On the other hand, these results may suggest that a secondary route of entry (e.g. direct plasma membrane translocation) is also plausible, albeit much less predominant than endosomal escape.
In principle, the previous results indicate that the escape of the peptide from either late endosomes or lysosomes is possible (39). In the case of dfTAT, we have previously ruled out the involvement of lysosomes by establishing that dfTAT does not release material loaded inside lysosomes (30), yet d-dfTAT might differ significantly from dfTAT in this instance. In particular, one can envision how dfTAT might be rendered inactive inside lysosomes because it is extensively degraded. In contrast, by being protease-resistant, d-dfTAT could remain membrane-disrupting in this milieu. To test this idea, the endosomes of HeLa cells were preloaded with DEAC-k5, a fluorescent and protease-resistant polylysine peptide (supplemental Fig. S4). As shown in experiment 1 (exp 1) of Fig. 2C, endosomes loaded with DEAC-k5 are observed by fluorescence microscopy as puncta inside cells (endosomal entrapment is confirmed by co-localization with LysoTracker). Cells were then washed and treated with d-dfTAT immediately (Fig. 2C, exp 2) or 2 h (Fig. 2C, exp 3) after DEAC-k5 incubation. The additional incubation time in the latter experiment was used to allow lysosomal accumulation of DEAC-k5. Microscopy images show that d-dfTAT is able to redistribute the fluorescence of DEAC-k5 inside cells after immediate incubation (like d-dfTAT, DEAC-k5 stains nucleoli because it is positively charged and presumably binds nucleolar nucleic acids). This is presumably because d-dfTAT is capable of causing the leakage of late endosomes that still contain DEAC-k5. In contrast, d-dfTAT was not able to cause cytosolic release of DEAC-k5 in experiment 3. In particular, d-dfTAT still displayed a nuclear/cytosolic distribution, but the DEAC-k5 fluorescent distribution remained punctate (Fig. 2C, exp 3). This, in turn, suggests that d-dfTAT, like dfTAT, does not cause the release into the cytosol of material that has accumulated inside lysosomes. Overall, these data also indicate that dfTAT and d-dfTAT use an identical route of cellular entry involving late endosomes. To further confirm that the membrane of these organelles is the site of peptide penetration, the activity of dfTAT and d-dfTAT toward liposomes mimicking late endosomes (i.e. BMP, PC, PE, cholesterol) was assessed in vitro. In particular, both peptides were capable of inducing liposome leakage with equivalent efficiencies (Fig. 2D). This establishes that the chirality of the peptide is not an important determinant in its interaction with lipids found in late endosomes.
l- to d-Substitution Negatively Impacts Endocytosis but Favors Endosomal Escape
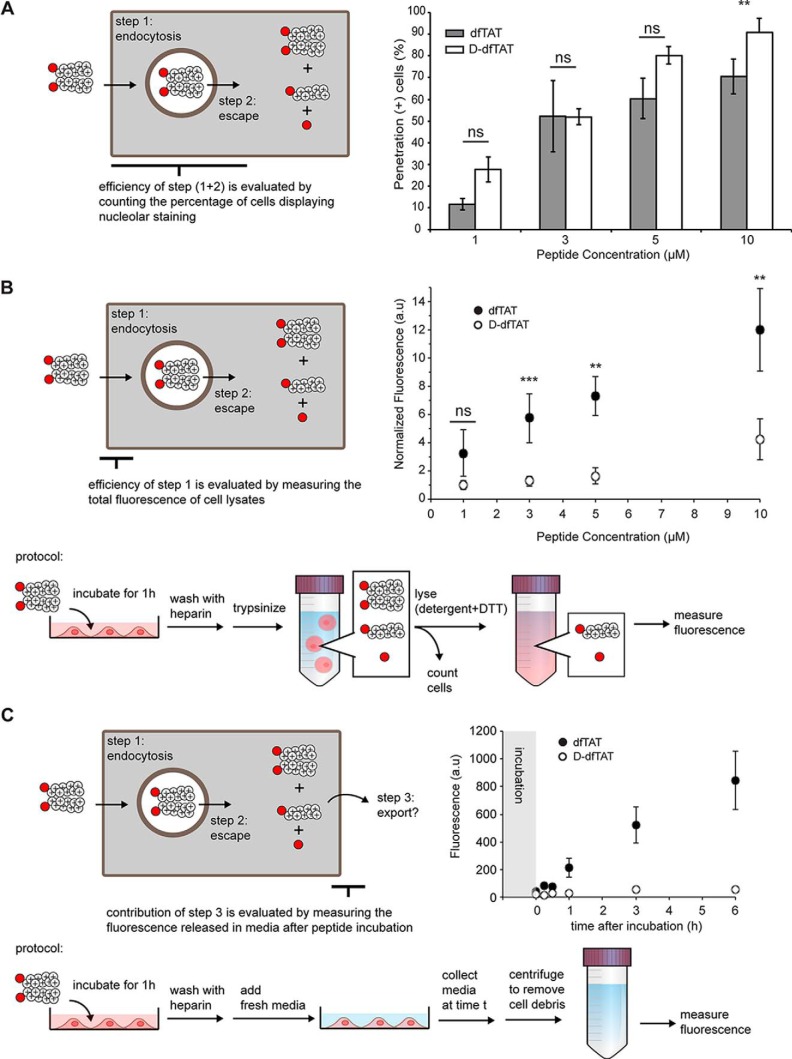
The previous experiments indicate that dfTAT and d-dfTAT share similar cell penetration properties, at least qualitatively. To establish whether the two peptides differ in their respective efficiencies quantitatively, the penetration activity of each compound was determined as a function of peptide concentration. First, cells were incubated with increasing concentrations of each peptide. Cells were then examined by fluorescence microscopy. Cells displaying a distinctive nucleolar staining are counted as positive for penetration (the cells counted are also negative for SYTOX Blue staining, indicating that their plasma membrane is not compromised and that these cells are not dead). At any given concentration of peptide in the incubation medium, the number of cells positive for penetration was similar between the two peptides (with the exception of 10 μm, where d-dfTAT is more active than dfTAT) (Fig. 3A). Cells negative for cell penetration typically display a punctate distribution consistent with endosomal entrapment of the peptide and weak endosomal escape (40). However, this assay is only binary in nature and only reveals whether penetration is achieved above a detection threshold dictated by our fluorescence microscope. To quantify the activity of each peptide in greater detail, overall peptide uptake was also measured (Fig. 3B). This was performed by measuring the bulk fluorescence emission of cell lysates. This assay was chosen because the fluorescence of the dimeric CPPs is partially quenched in comparison with that of their monomeric counterparts (supplemental Fig. S5). In addition, one may expect that once inside cells, the CPP might give rise to multiple species, including the intact dimer, the reduced monomer, and degradation fragments. Consequently, cell lysis enabled us to chemically reduce the pool of dimeric CPPs present in cells so as to generate unquenched species that can be better quantified (supplemental Fig. S5). This analysis shows that the overall cellular fluorescence is 3–4-fold higher for cells treated with dfTAT than for cells treated with d-dfTAT (Fig. 3B) (preferential uptake of the l-CPP over the d-CPP was corroborated by flow cytometry and fluorescence microscopy, although these assays suffer from the possible caveat of partial fluorescence quenching described above; supplemental Fig. S6). Control experiments indicated that none of the fluorescence detected was associated with extracellular membrane binding (supplemental Fig. S7). Notably, it is conceivable that there would be less d-dfTAT inside cells than dfTAT if the export of the peptides from inside to outside of cells was greater for the d-CPP than for the l-CPP. To test this possibility, cells were treated with both CPPs for 1 h and washed using protocols that remove extracellular peptides (supplemental Fig. S7). The media of cells incubated for up to 6 h were then collected, and the presence of the TMR signal was subsequently detected by fluorescence spectroscopy (Fig. 3C). Based on this assay, significantly more fluorescent species are released into the medium after delivery with dfTAT than after delivery with d-dfTAT (where little to no fluorescence is detected). Therefore, preferential export of d-dfTAT does not account for the differences observed in Fig. 3B. Overall, considering that intracellular fluorescence signals detected in Fig. 3B are proportional to initial endocytic uptake (i.e. step 1 in Fig. 3), these results indicate that an l- to d-conversion reduces endocytosis of the CPP.
FIGURE 3.
Comparison of the cellular uptake and penetration of dfTAT and d-dfTAT. A, evaluation of the cytosolic delivery efficiency of dfTAT and d-dfTAT as a function of peptide concentration in incubation medium. HeLa cells were incubated with peptides (1–10 μm) for 1 h. The percentage of cells detected as positive for penetration was established by microscopy (ns, p > 0.05; **, p ≤ 0.01). B, quantification of whole cell uptake by dfTAT and d-dfTAT as a function of the peptide concentration in incubation medium. HeLa cells were incubated with peptides (1–10 μm) as in A. The overall amount of peptide internalized by cells (endosomal + cytosolic) was assessed by measuring the bulk fluorescence of cell lysates. The illustration summarizes the protocol used for this assay. C, determination of the contribution of peptide export after dfTAT or d-dfTAT treatment. HeLa cells were incubated with 5 μm dfTAT or d-dfTAT for 1 h. The cells were washed, and the medium in all wells was replaced. The medium for each time point condition was removed at the indicated times (0, 0.25, 0.5, 1, 3, and 6 h postincubation). The fluorescence intensity of the medium collected form treated cells was measured and normalized to the medium obtained from untreated cells. All of the data reported in this figure represent the average and corresponding S.D. values (error bars) of biological triplicates.
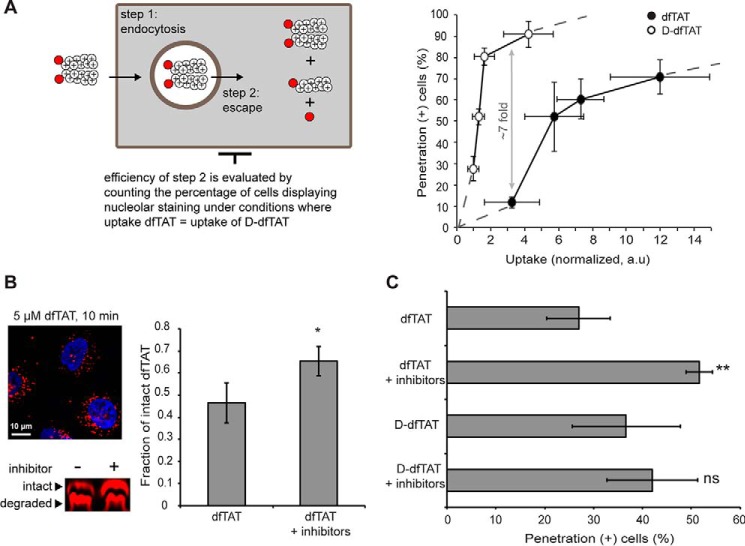
Given that d-dfTAT is endocytosed less efficiently than dfTAT and yet leads to equivalent numbers of cells positive for penetration, the previous data indicated that d-dfTAT might be able to escape from endosomes at lower concentrations than dfTAT. Using microscopy analysis of cells treated with LysoTracker Green, a fluorescent marker of late endosomes and lysosomes, we found that the number of endosomes present in cells treated with dfTAT is similar to that present in cells treated with d-dfTAT (supplemental Fig. S8). We therefore reasoned that the overall uptake measured in Fig. 3B is proportional to the concentration of peptide present inside endosomes before endosomal escape. To estimate the relative endosomal escape efficiency of each peptide, the penetration activities of d-dfTAT and dfTAT were therefore plotted as a function of overall uptake (Fig. 4A). These data show that the ability of d-dfTAT to escape from the endocytic pathway increases dramatically within a narrow range of intracellular concentrations. In contrast, dfTAT achieves similar cell penetration activities but requires a larger intracellular concentration to do so. Notably, the plot of dfTAT appears to plateau at high levels of peptide uptake, indicating that significant increases in intracellular concentration only lead to minor improvements in penetration. Moreover, under low intracellular uptake conditions, the percentage of penetration-positive cells for dfTAT is ∼7-fold less than in the case of d-dfTAT (e.g. uptake of ∼3). Overall, these data indicate that d-dfTAT is capable of escaping endosomes more effectively than dfTAT (for a discussion on the possible yet small contribution of peptide uptake by an endocytosis-independent process, see supplemental Fig. S3).
FIGURE 4.
Protection from degradation confers d-dfTAT with a greater endosomal escape activity when compared with dfTAT. A, evaluation of the endosomal escape activity of dfTAT and d-dfTAT as a function of the total amount of peptide internalized by cells (rationale highlighted in the scheme). The data represented combine the peptide penetration and uptake measured in Fig. 3, A and B. B, a protease inhibitor mixture protects dfTAT from proteolytic degradation before cytosol delivery. HeLa cells were preincubated (40 min) with E-64D (40 μm) and leupeptin (28 μm). Cells were then treated with dfTAT (5 μm) in the presence of E-64D for 10 min. Cell lysates were analyzed by gel electrophoresis. Bands corresponding to intact and degraded peptide, detected using a fluorescence scanner, were quantified by densitometry to generate the bar graph presented. The data reported represent the average and corresponding S.D. values (error bars) of biological triplicates. p values were obtained using t test analysis (*, p ≤ 0.01) between dfTAT with and without inhibitors. C, a protease inhibitor mixture increases the penetration activity of dfTAT but not of d-dfTAT. HDF cells were preincubated (40 min) with the protease inhibitor mixture (40 μm E-64D and 28 μm leupeptin). The cells were then treated with dfTAT or d-dfTAT (2 μm) in the presence of E-64D for 1 h. The percentage of cells detected as positive for penetration was established by microscopy. All data (A and B) represent the mean (mean of geometric means in the case of flow cytometry) of triplicate experiments performed on different days and the corresponding S.D. values. p values were obtained using t test analysis (ns, p > 0.05; **, p ≤ 0.01) between dfTAT and d-dfTAT with or without inhibitors (C). The data reported in this figure represent the average and corresponding S.D. values of biological triplicates.
To assess whether the differences in endosomal activity detected between the two peptides might arise from the partial endosomal proteolytic degradation of dfTAT, cell penetration activities were quantified in the presence of protease inhibitors. First, HeLa cells were preincubated with the cysteine-protease inhibitor E-64D and the cysteine, serine, and threonine protease inhibitor leupeptin (41, 42). To establish whether this mixture may protect the peptide from degradation before endosomal escape, cells were treated with 5 μm dfTAT for only 10 min and lysed immediately. This condition allows for peptide accumulation in the endocytic pathway (i.e. cells display a punctate peptide distribution under this condition) without providing sufficient time for endosomal escape. The cell lysates were analyzed by gel electrophoresis, and densitometry analysis was performed on bands representing intact and degraded peptide fractions. As shown in Fig. 4, a 20% increase in the amount of intact peptide was achieved in the presence of inhibitors. Although these results do not pinpoint the site of action of the inhibitors within the cell (e.g. plasma membrane versus endocytic organelles), they nonetheless indicated that an increase of the amount of intact peptide present within endosomes could be achieved. This inhibitor mixture was then used to determine whether protection against endosomal proteolytic degradation could impact the cell penetration activity of dfTAT. HDF cells were pretreated with E-64D and leupeptin and then treated with d-dfTAT and dfTAT (2 μm) for 1 h. The percentage of cells positive for penetration was then measured (Fig. 4B). The penetration activity of d-dfTAT was not impacted by the presence of inhibitors, indicating that these compounds do not indirectly affect endocytosis and endosomal escape. In contrast, the activity of dfTAT was significantly increased by the inhibitor mixture (∼2-fold increase at 2 μm). These data therefore suggest that proteolytic degradation along the trafficking pathway of the peptide might reduce its overall penetration activity.
Endosomal Escape Is Not Toxic to Cells, but Intracellular Retention of d-dfTAT Inhibits Cell Proliferation and Impacts Transcription
The viability of cells, determined using a SYTOX BLUE exclusion assay, was 95 and 98% after treatment with dfTAT and d-dfTAT (5 μm), respectively. dfTAT did not impact the proliferation of HDF or MCH58 cells. Likewise, d-dfTAT was innocuous to HDF. However, the proliferation of MCH58 cells was blocked by this peptide (Fig. 5A and supplemental Fig. S8). Consistent with these results, bright field images of MCH58 cells treated with d-dfTAT for 1 h show no appreciable change in confluency over a period of 48 h (Fig. 5B). To gain a more detailed insight into how cells respond to each peptide, a microarray analysis was performed. We have previously reported that, despite causing extensive leakage of endocytic organelles, dfTAT does not dramatically impact the transcriptional program of cells treated with the peptide for 1 h (analysis was performed 1 h after incubation) (27). Herein, we repeated this analysis 24 h after incubation to compare the effects of dfTAT and d-dfTAT. This choice was made based on the fact that dfTAT is completely degraded inside cells at this time point, whereas d-dfTAT remains intact (Fig. 1). As observed previously, HDF cells treated with dfTAT were virtually identical to untreated cells (1 of 47,000 transcripts being overexpressed over a 2-fold threshold). MCH58 cells were slightly more perturbed with a total of 20 transcripts dysregulated. In contrast, d-dfTAT caused the dysregulation of 215 and 551 transcripts in HDF and MCH58 cells, respectively (Fig. 5C). These data establish that the transcriptional response induced by d-dfTAT is cell-dependent yet consistently more severe than in the case of dfTAT. Notably, the level of transcriptional dysregulation induced by d-dfTAT also correlates with the effect of the peptide on proliferation (dysregulation of pro-apoptotic, as well as pro-survival genes, is identified in the microarray data; a list is provided in supplemental Table S1).
FIGURE 5.
d-dfTAT impacts cellular proliferation and transcription more dramatically than dfTAT after cytosolic delivery. A, proliferation assays. HDF and MCH58 cells were incubated with 5 μm dfTAT or d-dfTAT for 1 h. Cells were then incubated in growth medium (DMEM plus 10% FBS), and cellular proliferation was monitored over a period of 72 h using an MTT assay. Experiments were performed in triplicates with average and S.D. (error bars) indicated. B, the effect of dfTAT and d-dfTAT treatment on cell growth was monitored by microscopy. MCH58 cells were grown to 50–60% confluence and incubated with 5 μm dfTAT or d-dfTAT for 1 h. Cells were washed and imaged. The cells were then incubated in growth medium (DMEM plus 10% FBS) and imaged again at 48 h. Bright field images of live cells taken immediately after incubation and after 48 h are shown using a ×20 objective. Insets, fluorescence images of dfTAT and d-dfTAT treatment pseudocolored in red (emission 560 nm) and Hoechst emission pseudocolored in cyan (emission 480 nm). Scale bars, 100 μm. C, RNA-seq analysis of HDF and MCH58 cells treated with d-dfTAT or dfTAT (5 μm) in L-15 for 1 h. Untreated cells were incubated without peptide in L-15 for 1 h. After 24 h in growth medium (DMEM plus 10% FBS), cells were pelleted for total RNA extraction. The scatter plots shown display the microarray intensity values of 47,000 transcripts in treated cells (dfTAT or d-dfTAT; y axis) versus untreated cells (x axis). Analyses were performed in duplicate from two independent cell cultures. Both sets of microarray analyses yielded similar results. A representative set of results is shown (both sets are deposited in the GEO Gene Expression Omnibus). The red lines indicate the 2-fold intensity thresholds. The number of genes up- or down-regulated beyond these thresholds are indicated with a red or green arrow, respectively.
Discussion
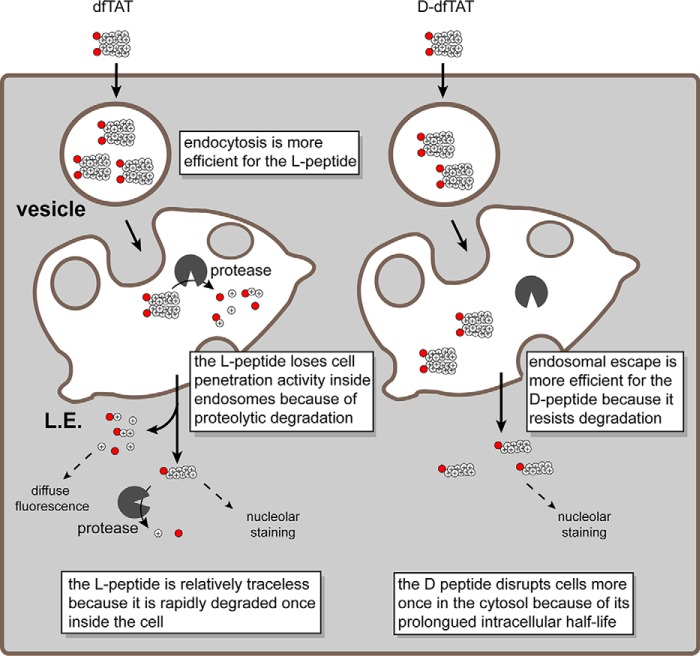
dfTAT is a multitasking molecule utilizing the endocytic pathway to access the cytosolic space of cells (Fig. 6) (27). On the cell surface, it interacts with cell components that facilitate endocytic uptake. In the lumen of late endosomes, it interacts with BMP to induce membrane leakage (30). Upon cytosolic access, dfTAT is reduced to monomeric fTAT. fTAT then accumulates at nucleoli. Throughout this pathway, dfTAT is presumably subjected to proteolysis. Our results clearly establish that substituting l-amino acids for their d-isomers protects the peptide from degradation. In particular, based on intracellular peptide distribution and gel analysis, d-dfTAT is stable for a minimum of 24 h after cell penetration. In contrast, dfTAT is already partially degraded after 1 h of incubation and fully degraded at 24 h. The degradation observed at 1 h is presumably a combination of proteolysis within the lumen of endosomes and cytosolic proteolysis after entry into cells.
FIGURE 6.
Schematic representation modeling the similarities and differences observed in the cellular penetration of dfTAT and d-dfTAT. dfTAT is internalized by endocytosis at a higher level than d-dfTAT. Along the endocytic pathway, dfTAT is partially degraded by proteolytic enzymes, whereas d-dfTAT is not. Upon reaching the late endosome, both dfTAT and d-dfTAT induce the leakage of BMP-containing membranes. Due to proteolytic degradation, the endosomal escape efficiency of dfTAT is, however, diminished compared with d-dfTAT. Upon escaping from late endosomes, d-dfTAT and intact dfTAT are represented as being reduced to their monomer counterparts in the cytosol (e.g. by the action of glutathione). The peptide then diffuses into the cytosolic space and accumulates at nucleoli. The degradation products of dfTAT contribute to a diffuse cytosolic fluorescent distribution. d-dfTAT remains intact inside cells for several days and impacts the physiology of cells negatively. In contrast, cytosolic enzymes degrade dfTAT in a matter of hours. This in turn leads to a dramatically diminished physiological impact.
Whereas inversion of chirality protects the delivery peptide from protease degradation, a change in stereochemistry should also impact the interaction of this compound with other chiral molecules (24). Our assays suggest that an inversion of chirality does not impact the overall mechanism of cell penetration. Like dfTAT, d-dfTAT enters cells by endocytosis, followed by escape from late endosomes via interactions with BMP. At first glance, both peptides also appear to enter cells with relatively equal efficiencies. Overall, this suggests that chirality is not a critical aspect of the interaction between the CPP and molecules involved in its intracellular transport. Yet, upon closer examination, our results reveal that the individual steps of endocytosis and endosomal escape are impacted in opposite ways by chirality; endocytosis is reduced, whereas endosomal escape is increased (Fig. 6). Based on our results, the reduction in endocytosis could not be explained by a preference in d-dfTAT export during incubation. The export of d-dfTAT was significantly lower than dfTAT during the time course of the experiment, presumably due to stability and retention of d-dfTAT at the nucleoli.
d-dfTAT is significantly less prone to internalization by endocytosis than dfTAT. The parent compound TAT is known to interact with negatively charged heparan sulfate proteoglycans on the cell surface (43, 44). The role played by these interactions with respect to endocytosis remains, however, unclear; proteoglycans might act as docking sites for the peptide and might or might not be directly involved in inducing endocytosis. In in vitro binding assays, Brock and co-workers (24) have found no differences in the binding of l-CPPs or d-CPPs to heparan sulfate, yet they have also reported a decrease in endocytic uptake by d-CPPs (CPPs tested were r9, d-penetratin, and d-hLF) when compared with their l-counterparts. Overall, these data suggest that interactions with chiral species other than heparan sulfate might play an important role in endocytosis. The identities of these chiral species, however, have yet to be established.
Given that dfTAT and d-dfTAT differ in the extent to which they accumulate in endocytic organelles, comparing these peptides with respect to the subsequent step of endosomal escape is challenging. Nonetheless, to address this issue, we compared the propensity of the peptides to reach the cytosol of cells under conditions where the same amount of peptide is endocytosed (rather than conditions where incubation is performed at the same concentration). Based on this analysis, d-dfTAT induces endosomal leakage at a luminal concentration significantly lower than that required for dfTAT, yet dfTAT reaches the cytosol of cells in a degraded form. In addition, protease inhibitors increase the endosomal escape activity of dfTAT. Finally, dfTAT and d-dfTAT have similar membrane leakage-inducing activities in vitro. Overall, our interpretation is that dfTAT is not necessarily less capable of inducing endosomal leakage per se but that instead it is degraded by proteases while trafficking within the endocytic pathway. As a result, the amount of membrane-active dfTAT present inside endosomes represents only a portion of what is measured in our fluorescence measurements (intact dfTAT might represent ∼25% of total signal based on Fig. 1C). Overall, d-dfTAT therefore appears to gain activity simply because, unaffected by proteolysis, it can accumulate inside late endosomes at concentrations higher than dfTAT.
We have shown previously that dfTAT is not toxic to cells (27). This is confirmed in this report because dfTAT does not significantly impact the proliferation of two cell lines. Cells treated with dfTAT are also virtually indistinguishable on a transcriptional level from untreated cells at 24 h. Overall, this indicates that dfTAT-mediated endosomal leakage is well tolerated by cells. This is in sharp contrast to d-dfTAT, which has a deleterious impact on both proliferation and transcription. Based on the notion that both dfTAT and d-dfTAT share a similar mechanism of cell penetration, we interpret the impact of d-dfTAT on cellular proliferation as not associated with the delivery process per se but instead a result of interactions with cytosolic and nucleolar partners that take place post-delivery. In particular, one can speculate that the positively charged peptide interacts with negatively charged nucleic acid in the nucleoli and disrupts ribosome-associated processes. Although dfTAT could potentially have similar interactions and lead to similar deleterious effects, this peptide is degraded rapidly, and its effects are minimized. In contrast, the protease-resistant d-dfTAT remains intact in the cytosol and nucleoli of cells for a prolonged time, and cellular responses become detectable.
Overall, our data reveal several insights relevant to the design of future cell-permeable peptide-based agents (e.g. stapled peptides, retro-inverso peptides, peptoids, CPPs, etc.). One is that conferring protease resistance to a CPP helps to promote its activity inside endosomes. This is important because such an approach might help generate agents that are cell-permeable at submicromolar concentrations and, thereby, more relevant to in vivo applications. Additionally, we infer that proteolytic degradation inside the endocytic pathway acts as a major determinant of peptide activity. For instance, one can envision how certain CPPs might appear relatively ineffective at escaping endosomes not because of low membrane permeation activity per se but rather because of a high propensity to be degraded and inactivated in this environment. Furthermore, for the first time, we establish that protease resistance has a significant impact on the cytosolic activity of CPPs. In particular, our results highlight that a protease-resistant CPP can reside in the cytoplasm and nucleus of cells for an extended period of time and consequently induce numerous cellular responses. This is problematic for in vitro applications that aim at delivering bioactive cargos to probe or manipulate biological processes (ideally, cells should not be perturbed by the delivery agent itself if one is to investigate the intracellular effects of a cargo). In addition, one can speculate that this could be equally problematic for in vivo applications, given that extended cellular retention of a delivery agent often leads to off-target effects (6). When considering these issues, it is apparent that the strategy of all-l- to all-d-residue conversion is not an ideal strategy to improve CPP properties. However, our results now point toward the idea of conferring protease resistance to endosomolytic agents in an environment-dependent manner. In particular, we envision that delivery agents that are protease-resistant inside endosomes but degradable in the cytosol should provide clear improvements. Therefore, establishing the feasibility of such strategy will be the focus of future studies.
Experimental Procedures
Peptide Design, Synthesis, and Purification
All peptides were synthesized on the rink amide 4-methylbenzhydrylamine (MBHA) resin (Novabiochem, San Diego, CA) by solid phase peptide synthesis. Fmoc-Lys(Mtt)-OH, Fmoc-Lys(Boc)-OH, Fmoc-d-Lys(Boc)-OH, Fmoc-d-Lys(Dde)-OH, Fmoc-Gly-OH, Fmoc-Arg(Pbf)-OH, Fmoc-d-Arg(Pbf)-OH, Fmoc-Gln(Trt)-OH, Fmoc-d-Gln(Trt)-OH, and Fmoc-Cys(Trt)-OH (Novabiochem) were used to assemble the peptides using standard Fmoc protocols. Boc-d-Cys(Trt)-OH (Novabiochem) was added to the N terminus of d-fTAT (single chain monomeric peptide), and the Boc protecting group was removed upon trifluoroacetic acid (TFA) cleavage. All reactions were performed at room temperature in a solid phase peptide synthesis vessel. Reactions were agitated using a stream of dry N2. For each amino acid coupling, the reaction was agitated for 4 h with a mixture of Fmoc-amino acid (1.6 mmol), HBTU (Novabiochem) (1.5 mmol), and diisopropylethylamine (DIEA) (sigma) (4.0 mmol) in dimethylformamide (DMF) (Fisher). After the addition of each amino acid, Fmoc deprotection was performed by the addition of 20% piperidine in DMF. The reaction was carried out twice: 1 × 5 min followed by 1 × 15 min with a DMF washing step between reactions. Upon completion of the linear chain assembly, the peptidyl resin was washed with DMF and dichloromethane (DCM) (Fisher). DEAC-k5 was synthesized by coupling five Fmoc-d-Lys(Boc)-OH. The DEAC (7-diethylaminocoumarin-3-carboxylic acid) (Anaspec) fluorophore was conjugated to the N terminus of the peptide by reaction of DEAC, HBTU, and DIEA (4, 3.9, and 10 eq) in DMF overnight. CK(ϵ-NH-TMR)TATG (l-fTAT) was synthesized as described earlier (27). Briefly, the MTT protecting group was removed using 1% TFA (Fisher) and 2% triisopropylsilane (Sigma) in DCM. The peptidyl resin was washed with both DMF and DCM between each reaction. For ck(ϵ-NH-Dde)rkkrrqrrrG (d-TAT), the Dde protecting group was removed using 2% hydrazine (Sigma) in DMF. The peptidyl resin for both peptides was then reacted with a mixture of TMR, HBTU, and DIEA (4, 3.9, and 10 eq) and agitated overnight. For l-fTAT, the resin was washed, and Fmoc deprotection was performed to remove the N-terminal Fmoc moiety. For d-fTAT and l-fTAT, the peptides (and remaining protecting groups) were then cleaved from the resin by treatment with 92.5% TFA, 2.5% H2O, 2.5% triisopropylsilane, and 2.5% ethanedithiol (EDT) (Sigma) for 3 h at room temperature. The crude peptide was precipitated in cold anhydrous ether (Fisher). The precipitate was resuspended in water and lyophilized. The lyophilized product was dissolved in 0.1% TFA plus water. The peptides were analyzed and purified by reverse-phase HPLC. HPLC analysis was performed on a Hewlett-Packard 1200 series instrument and an analytical Vydac C18 column (5 μm, 4 × 150 mm). The flow rate was 1 ml/min, and detection was at 214 and 550 nm. Semipreparative HPLC was performed on a Vydac C18 10 × 250-mm column. The flow rate was 4 ml/min, and detection was at 214 and 550 nm. All runs used linear gradients of 0.1% aqueous TFA (solvent A) and 90% acetonitrile, 9.9% water, and 0.1% TFA (solvent B). The correct identity of the peptides was confirmed by MALDI-TOF performed with a Shimadzu/Kratos instrument (AXIMA-CFR, Shimadzu, Kyoto). fTAT, expected mass: 2041.17, observed mass: 2040.66. DEAC-k5, expected mass: 900.59, observed mass: 900.84.
Generation of dfTAT and d-dfTAT by Dimerization of l-fTAT and d-fTAT, Respectively
fTAT or d-fTAT (0.3 mg, 1.5 × 10−4 mmol) was dissolved in 5 ml of aerated PBS at pH 7.4. The reaction was agitated on a nutator overnight (100% yield based on HPLC analysis). dfTAT and d-dfTAT were purified using analytical reverse-phase HPLC. Expected mass (MALDI-TOF): 4,078.27, observed mass dfTAT and d-dfTAT 4,080.87 and 4,078.87, respectively.
Cell Culture and Transfection
HeLa (ATCC CCL-2), MCH58, and HDF (ATCC PCS-201-010) cells were grown in DMEM (Fisher) supplemented with 10% FBS (Fisher) and 1× penicillin/streptomycin (Fisher) and incubated at 37 °C in a humidified atmosphere containing 5% CO2.
For transfection experiments, rab7 (GFP-rab7 WT, Addgene plasmid 12605) and DN-rab7 (GFP-rab7 DN, Addgene plasmid 12660) were a gift from Richard Pagano. For transfection of the TAT-Cre experiment, loxp-stop-loxp-EGFP (pCALNL-GFP, Addgene plasmid 13770) and gWiz GFP (ALDEVRON lot 35237) were used. Each plasmid (250 ng) was mixed with Lipofectamine 2000 (1 μl) in Opti-MEM medium and allowed to incubate for 10 min at room temperature. The DNA-Lipofectamine complex was then incubated for 18 h at 37 °C with HeLa cells seeded in an 8-well dish.
dfTAT and d-dfTAT Degradation
An in vitro proteolysis assay was performed by the addition of 1 μl of 0.5% trypsin to 19 μl of either dfTAT or d-dfTAT (10 μm) and incubated at room temperature. Reactions were quenched at specific time points by the addition of 80 μl of 0.1% TFA in water. The samples were analyzed on a reverse phase HPLC using a 15–40% gradient (0.1% TFA, 90% acetonitrile).
For the in cellulo assay, HeLa cells were seeded in a 24-well dish and allowed to grow to 90% confluency. The wells were washed three times with PBS and once with Leibovitz's L-15 medium that does not contain the amino acid cysteine (nonreducing L-15, nrL-15). nrL-15 is used for incubation because it lacks cysteine and avoids the reduction of the disulfide bond of dfTAT. Cells were then incubated with dfTAT or d-dfTAT (5 μm) for 1 h. The cells were washed three times with PBS and lysed at different time points as indicated, using a lysis buffer (50 mm Tris, pH 7.5, 2 mm EDTA, 2 mm DTT, 0.1% Triton X-100, and protein inhibitor mixture; 75 μl/well). The samples were then treated with DNase (3 μl; Genesse) for 15 min on ice. The samples were centrifuged at 4 °C and 13,000 rpm for 20 min. The supernatant (cell lysate) (39 μl) was added to 6 μl of loading dye. The samples were boiled for 10 min at 100 °C. The samples were run on a 16.5% Tris-Tricine gel (Bio-Rad, catalog no. 3450063) using a running buffer composed of 100 mm Tris, 100 mm Tricine, 0.1% SDS, pH 8.3.
Delivery of Peptides and Proteins into Live Cells
HeLa, HDF, and MCH58 cells were seeded in an 8-well dish. The cells were grown to 80–90% confluency in a 37 °C humidified atmosphere containing 5% CO2. The wells were washed three times with PBS and once with nrL-15. Cells were incubated with dfTAT or d-dfTAT (5 μm) at 37 °C for 1 h (unless otherwise specified). The cells were then washed three times with L-15 supplemented with heparin (1 mg/ml) and once with nrL-15. Cells were treated with the cell-impermeable nuclear stain SYTOX® BLUE or SYTOX® GREEN to identify cells with a compromised plasma membrane (i.e. dead cells). Alternatively, cells were incubated with cell-permeable Hoechst for nuclear staining. Cells were imaged using an inverted epifluorescence microscope (model IX81, Olympus, Center Valley, PA) equipped with a heating stage maintained at 37 °C. Images were collected using a Rolera-MGI Plus back-illuminated EMCCD camera (Qimaging, Surrey, Canada). Images were acquired using bright field imaging and three standard fluorescence filter sets: CFP (excitation = 436 ± 10 nm/emission = 480 ± 20 nm), RFP (excitation = 560 ± 20 nm/emission = 630 ± 35 nm), and FITC (excitation = 488 ± 10 nm/emission = 520 ± 20 nm). For viewing and processing individual fluorescent images, SlideBook version 4.2 software (Olympus) was used. Cells that displayed a cytosolic and nucleolar peptide staining were considered positive for cytosolic penetration. The percentage of penetration-positive cells was calculated by dividing the number of cells displaying a fluorescent nuclear staining (×20 images) by the total number of cells present (acquired using bright field imaging). Cells negative for cytosolic penetration typically display a punctate distribution consistent with endosomal entrapment of the peptides. Cells with plasma membrane compromised stained by SYTOX dyes were considered dead and were excluded from the analysis. For each experiment, an average of three ×20 pictures were taken, representing 300–400 cells. The reproducibility of all of the experiments was assessed by performing experiments with independent batches of cell cultures on three different days (i.e. biological triplicates).
To monitor the intracellular localization of dfTAT and d-dfTAT after cell delivery (1-h incubation), cells were washed and incubated in fresh growth medium (DMEM plus 10% FBS). The dish was placed at 37 °C in a humidified atmosphere containing 5% CO2. Cells were imaged after 24 and 48 h.
For delivery of TAT-Cre, HeLa cells were transfected with pCALNL-GFP or gWiz GFP (control plasmid for transfection efficiency) for 18 h. Cells were then washed and co-incubated with dfTAT or d-dfTAT and TAT-Cre (4 μm) for 1 h. The cells were washed again, and the medium was replaced with fresh DMEM. The cells were placed back in the incubator for 24 h. The cells were then imaged as described above. To quantify the results, EGFP+ cells were counted and divided by the total number of cells that were transfected (control cells that were transfected with gWiz GFP (250 ng)) in each ×20 image.
To investigate the process of cellular entry, HeLa cells were incubated in L-15 with DEAC-k5 (20 μm) for 1 h. Cells were then washed three times with PBS and incubated either immediately or after a 2 h wait in nrL-15 with d-dfTAT (5 μm) or without peptide (control cells) for 1 h. The cells were then washed three times with L-15 supplemented with heparin (1 mg/ml). Alternatively, the incubation steps were inverted. HeLa cells were first incubated with d-dfTAT for 1 h and then washed as described above. Cells were then incubated in nrL-15 for 20 min. The cells were then incubated with DEAC-k5 (20 μm in L-15) for 1 h. Cells were washed and imaged as described earlier.
To establish the involvement of BMP in cellulo, HeLa cells were incubated with 50 μg/ml anti-BMP mAb (mouse, Z-PLBPA) or anti-IgG mAb (mouse, ab99763) in L-15 for 30 min at 37 °C. Cells were washed and incubated with d-dfTAT (3 μm). Cells were then imaged as described previously.
To assess whether endosomal proteolytic degradation could play a role in the difference in cell penetration activity observed between dfTAT and d-dfTAT, protease inhibitors were used. In particular, HDF cells were preincubated with 40 μm E-64D (Sigma) and 28 μm leupeptin (Sigma) in nrL-15 for 40 min. The treatment was removed, and the cells were incubated with d-dfTAT or dfTAT (2 μm) in the presence of 40 μm E-64D. The cells were then washed and imaged as described above. For determination of the amount of intact peptide remaining before cytosolic access, HeLa cells were preincubated with 40 μm E-64D (Sigma) and 28 μm leupeptin (Sigma) in nrL-15 for 40 min. The treatment was removed, and the cells were incubated with dfTAT (5 μm) in the presence of 40 μm E-64D for 10 min. The control cells were treated with dfTAT (5 μm) for 10 min in the absence of inhibitors. The cells were then washed, lysed, and processed as described above for preparation to run on a 16.5% Tricine gel (Bio-Rad, catalog no. 3450063).
Quantitative Determination of Peptide Uptake
HeLa cells were seeded in a 48-well dish and grown to 80% confluency. Cells were treated with dfTAT and d-dfTAT for 1 h at various concentrations (range, 1–10 μm). Total peptide uptake was measured in two ways: fluorescence measurements of a cell lysate assay or flow cytometry. For whole cell lysate analysis, cells were washed with heparin (1 mg/ml) and then trypsinized (50 μl) for 3 min. Cells were resuspended in nrL-15 (350 μl) and centrifuged at 4 °C and 2,500 rpm for 10 min. The supernatant was removed, and the cell pellet was resuspended in nrL-15 (50 μl). An aliquot of the resuspended cells (3 μl) was removed and used through flow cytometry to normalize for the total number of cells/sample. The remaining resuspended cells were lysed by the addition of 50 μl of a lysis buffer (50 mm Tris, pH 7.5, 2 mm EDTA, 4 mm DTT, 20% Triton X-100, and protein inhibitor mixture) and vortexing. For the whole cell lysates in the absence of DTT, the lysis buffer prepared and used lacked DTT (50 mm Tris, pH 7.5, 2 mm EDTA, 20% Triton X-100, and protein inhibitor mixture). The samples were vortexed, and a volume of 80 μl of supernatant for each condition was placed in a 96-well plate. The fluorescence emission intensity was measured using a plate reader equipped with a fluorescence module (excitation = 525, emission = 580–640 nm) (GloMax®-Multi+ Detection System, Promega, Fitchburg, WI). To normalize, the aliquot of cells (3 μl) was resuspended in nrL-15 (97 μl), and the total amount of cells/sample was determined using flow cytometry.
For fluorescence measurement using the flow cytometer, cells were trypsinized and resuspended in nrL-15 medium. Cells were analyzed using a BD Accuri C6 flow cytometer equipped with the FL2 filter (excitation = 488 nm/emission = 533 ± 30 nm). All data were acquired at a flow rate of 66 μl/min with detection of a minimum of 40,000 events. The geometric mean of the FL2 signal for each experiment was determined using Flowjo software. The data reported represent the average and corresponding S.D. values of three independent experiments for each peptide concentration.
For export measurements, HeLa cells were seeded in a 48-well dish and grown to 80% confluency. Cells were treated with dfTAT and d-dfTAT at 5 μm for 1 h. Cells were washed according to protocol, and the medium was replaced with fresh medium (200 μl, DMEM + FBS + penicillin/streptomycin). The extracellular medium from separate wells was removed at specific time points. The medium was centrifuged at 8,000 rpm for 8 min. The medium from each time point was aliquoted into a 96-well plate. The fluorescence intensity was measured as mentioned above using a plate reader. The data were normalized by comparison with the fluorescence of the extracellular medium of the untreated cells.
To determine whether the peptide remained bound to the cell surface after the initial wash, HeLa cells were seeded in a 48-well dish and grown to 80% confluency. Cells were treated with dfTAT and d-dfTAT at 5 μm for 1 h. The cells were then washed in three different ways. For all conditions, cells were first washed three times with L-15 supplemented with heparin (1 mg/ml) and once with nrL-15. For sample 1, control cells were then trypsinized and resuspended in nrL-15 for flow cytometry. For sample 2, the cells were then washed again three times with L-15 supplemented with heparin (1 mg/ml) and once with nrL-15. For sample 3, the cells were then trypsinized and resuspended in nrL-15 and centrifuged at 4 °C and 2,500 rpm for 10 min. The supernatant was removed, and the cell pellet was trypsinized (50 μl) for 3 min. The trypsinized cells were then resuspended in nrL-15 for flow cytometry analysis.
Liposome Preparation
Lipids used in our experiments consisted of 1,2-dioleoyl-S-glycero-3-phosphocholine, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, BMP, and cholesterol (Avanti Polar Lipids). Liposomes were prepared by first transferring various volumes of lipids dissolved in chloroform (stock solutions of known concentrations) into scintillation vials. For liposomes mimicking the intraluminal late endosome vesicles, the molar ratios of lipids consisted of 77:19:4 BMP/PC/PE. For liposomes mimicking the plasma membrane, the lipid mixture was 65:15:20 PC/PE/cholesterol. The lipid film was prepared by removing the chloroform from the lipid mixture using a stream of N2 and then placing the vial in a desiccator for 24 h. A buffer containing 100 mm NaCl, 10 mm NaH2PO4, pH 7.4, and 60 mm calcein was added to the lipid film for hydration. The lipids were then mixed vigorously and swelled for 1 h at 42 °C under N2 to obtain multilamellar vesicles. For production of unilamellar vesicles, the multilamellar vesicles were extruded (20 passes) through a 100-nm pore size polycarbonate membrane (Whatman) using a Mini-Extruder (Avanti Polar Lipids). Dynamic light scattering was used to determine the average diameter size distribution of the liposomes using a Zeta Sizer (Malvern Instruments). The liposomes were purified by gel filtration using a Sephadex G-50 (GE Healthcare) column (2.5 × 17.5 cm) to separate the liposomes from free calcein. The eluate was collected in a 96-well plate, and the plate was read using a Promega GloMax-Multi plate reader (Promega) at 450 and 750 nm corresponding to the wavelength of detection for calcein and liposome, respectively.
Liposome Leakage Assays
Purified calcein-loaded unilamellar vesicles were mixed with d-dfTAT or dfTAT at different ratios for 1 h at room temperature in 100 mm NaCl, 10 mm NaH2PO4, pH 5.5. Samples were centrifuged for 1 min at 4,000 rpm. To measure the amount of leaked calcein and separate soluble liposomes from released calcein, the supernatants were purified using an Illustra NAP-10 Sephadex G-25 column (GE Healthcare) (the elution volumes of liposomes and free calcein were determined independently with pure samples). Fractions were collected in a 96-well plate, and the fluorescence of calcein was measured using a Promega GloMax-Multi plate reader (excitation = 490 nm, emission = 520–560 nm). The percentage of leakage was calculated using the equation,
| (Eq. 1) |
where Flt is the free calcein fluorescence intensity of a sample at a specific peptide/lipid ratio measured after 1 h, Fl0 is the free calcein fluorescence intensity of an untreated sample, and Flmax is the free calcein fluorescence intensity of a sample after treatment with 0.2% Triton X-100.
Cell Proliferation Assays
An MTT assay was performed to compare the effect of dfTAT to d-dfTAT on the proliferation rate of MCH58 and HDF. Cells were seeded and grown in a 6-well dish and treated as described earlier. Each well was incubated with either 5 μm dfTAT or d-dfTAT at 37 °C for 1 h. Cells were washed, trypsinized, and seeded into 96-well dishes containing 100 μl of DMEM plus 10% FBS. The cells were then allowed to adhere to the bottom of the dish for 12 h. The DMEM was removed and replaced with 100 μl of fresh DMEM plus 10% FBS, and 10 μl of a 12 mm MTT stock solution was added to the wells. The 96-well dish was incubated at 37 °C for 4 h, followed by the addition of 100 μl of SDS-HCl (10 mm). The cells were then allowed to incubate with this solution for 12 h. After the incubation, each sample was mixed, and the absorbance at 560 nm was measured. Controls included a negative control, where 10 μl of MTT was added to 100 μl of DMEM alone (no cells). A second control consisted of cells treated with the delivery peptide but to which no MTT was added to subtract the contribution of TMR from the measured absorbance. The absorbance of the negative control was subtracted from all samples. Assays were performed at multiple time points to obtain a cellular proliferation curve.
Whole Genome Microarray Analysis
MCH58 and HDF cells were seeded in 24-well dishes and incubated with dfTAT or d-dfTAT (5 μm) or nrL-15 (untreated cells) for 1 h. Cells were then washed, and the medium was replaced by DMEM. The cells were then placed back into the 37 °C humidified atmosphere containing 5% CO2 and incubated for 24 h. After 24 h, the cells were then trypsinized after washing with PBS or nrL-15 (1 h + 24 h time point). Cells were pipetted into an Eppendorf tube and spun down at 12,000 rpm for 4 min at 4 °C. The cell pellets were preserved in 1 ml of TRIzol (Ambion®, Life Technologies, Inc.) and shipped frozen to the University of Texas Southwestern microarray facility for total RNA extraction and analysis using the Illumina Human HT-12 v4 sequencer. The protocol is described on the University of Texas Southwestern website. Data sets have been deposited in the GEO Gene Expression Omnibus under accession number GSE90575.
Author Contributions
K. N. and J.-P. P. designed experiments. K. N. and A. E.-O. generated data. K. N. processed data. K. N., A. E.-O., D. J. B., and T.-Y. W. contributed reagents. K. N. and J.-P. P. wrote, edited, and approved the final manuscript.
Supplementary Material
Acknowledgments
We thank Richard Pagano (Mayo Clinic) for GFP-rab7 WT and GFP-rab7 DN and C. Cepko (Harvard Medical School) for pCALNL-GFP. We also thank J. Sacchettini (Texas A&M University) for HDF cells and E. Shoubridge (Montreal Neurological Institute and Hospital) for MCH58 cells.
This article was supported by National Institutes of Health Grant R01GM110137 and Cancer Prevention and Research Institute of Texas Grant RP100819. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Table S1 and Figs. S1–S8.
- CPP
- cell-penetrating peptide
- HBTU
- 2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate
- TMR
- carboxytetramethylrhodamine
- dfTAT
- dimeric fluorescence TAT
- DEAC
- 7-diethylaminocoumarin-3-carboxylic acid
- BMP
- bis(monoacyl)glycerophosphate
- HDF
- human dermal fibroblast
- EGFP
- enhanced green fluorescent protein
- PTD
- protein transduction domain
- DN
- dominant negative
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- Fmoc
- N-(9-fluorenyl)methoxycarbonyl
- DIEA
- diisopropylethylamine
- DMF
- dimethylformamide
- DCM
- dichloromethane
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- EDT
- ethanedithiol
- nrL-15
- nonreducing L-15
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- Boc
- t-butoxycarbonyl
- Mtt
- 4-methyltrityl
- Trt
- trityl
- Pbf
- 2,2,4,6,7-pentamethyldihydrobenzofurane-5-sulfonyl
- Dde
- 1-(4,4-dimethyl-2,6-dioxocyclohex-1-ylidene)ethyl.
References
- 1. Bechara C., and Sagan S. (2013) Cell-penetrating peptides: 20 years later, where do we stand? FEBS Lett. 587, 1693–1702 [DOI] [PubMed] [Google Scholar]
- 2. Zorko M., and Langel U. (2005) Cell-penetrating peptides: mechanism and kinetics of cargo delivery. Advanced Drug Deliv. Rev. 57, 529–545 [DOI] [PubMed] [Google Scholar]
- 3. Gupta B., Levchenko T. S., and Torchilin V. P. (2005) Intracellular delivery of large molecules and small particles by cell-penetrating proteins and peptides. Adv. Drug Deliv. Rev. 57, 637–651 [DOI] [PubMed] [Google Scholar]
- 4. Krosl J., Austin P., Beslu N., Kroon E., Humphries R. K., and Sauvageau G. (2003) In vitro expansion of hematopoietic stem cells by recombinant TAT-HOXB4 protein. Nat. Med. 9, 1428–1432 [DOI] [PubMed] [Google Scholar]
- 5. Morris M. C., Depollier J., Mery J., Heitz F., and Divita G. (2001) A peptide carrier for the delivery of biologically active proteins into mammalian cells. Nat. Biotechnol. 19, 1173–1176 [DOI] [PubMed] [Google Scholar]
- 6. Stewart M. P., Sharei A., Ding X., Sahay G., Langer R., and Jensen K. F. (2016) In vitro and ex vivo strategies for intracellular delivery. Nature 538, 183–192 [DOI] [PubMed] [Google Scholar]
- 7. Rizzuti M., Nizzardo M., Zanetta C., Ramirez A., and Corti S. (2015) Therapeutic applications of the cell-penetrating HIV-1 Tat peptide. Drug Discov. Today 20, 76–85 [DOI] [PubMed] [Google Scholar]
- 8. Munyendo W. L. L., Lv H., Benza-Ingoula H., Baraza L. D., and Zhou J. (2012) Cell penetrating peptides in the delivery of biopharmaceuticals. Biomolecules 2, 187–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Palm C., Jayamanne M., Kjellander M., and Hällbrink M. (2007) Peptide degradation is a critical determinant for cell-penetrating peptide uptake. Biochim. Biophys. Acta 1768, 1769–1776 [DOI] [PubMed] [Google Scholar]
- 10. Elmquist A., and Langel U. (2003) In vitro uptake and stability study of pVEC and its all-D analog. Biol. Chem. 384, 387–393 [DOI] [PubMed] [Google Scholar]
- 11. Lindgren M. E., Hällbrink M. M., Elmquist A. M., and Langel U. (2004) Passage of cell-penetrating peptides across a human epithelial cell layer in vitro. Biochemical Journal 377, 69–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pujals S., Sabidó E., Tarragó T., and Giralt E. (2007) all-D proline-rich cell-penetrating peptides: a preliminary in vivo internalization study. Biochem. Soc. Trans. 35, 794–796 [DOI] [PubMed] [Google Scholar]
- 13. Youngblood D. S., Hatlevig S. A., Hassinger J. N., Iversen P. L., and Moulton H. M. (2007) Stability of cell-penetrating peptide-morpholino oligomer conjugates in human serum and in cells. Bioconjug. Chem. 18, 50–60 [DOI] [PubMed] [Google Scholar]
- 14. Wender P. A., Mitchell D. J., Pattabiraman K., Pelkey E. T., Steinman L., and Rothbard J. B. (2000) The design, synthesis, and evaluation of molecules that enable or enhance cellular uptake: peptoid molecular transporters. Proc. Natl. Acad. Sci. U.S.A. 97, 13003–13008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mitchell D. J., Kim D. T., Steinman L., Fathman C. G., and Rothbard J. B. (2000) Polyarginine enters cells more efficiently than other polycationic homopolymers. J. Pept. Res. 56, 318–325 [DOI] [PubMed] [Google Scholar]
- 16. Derossi D., Calvet S., Trembleau A., Brunissen A., Chassaing G., and Prochiantz A. (1996) Cell internalization of the third helix of the Antennapedia homeodomain is receptor-independent. J. Biol. Chem. 271, 18188–18193 [DOI] [PubMed] [Google Scholar]
- 17. Pujals S., Fernández-Carneado J., Ludevid M. D., and Giralt E. (2008) d-SAP: a new, noncytotoxic, and fully protease resistant cell-penetrating peptide. ChemMedChem 3, 296–301 [DOI] [PubMed] [Google Scholar]
- 18. Jiang T., Olson E. S., Nguyen Q. T., Roy M., Jennings P. A., and Tsien R. Y. (2004) Tumor imaging by means of proteolytic activation of cell-penetrating peptides. Proc. Natl. Acad. Sci. U.S.A. 101, 17867–17872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Olson E. S., Aguilera T. A., Jiang T., Ellies L. G., Nguyen Q. T., Wong E. H., Gross L. A., and Tsien R. Y. (2009) In vivo characterization of activatable cell penetrating peptides for targeting protease activity in cancer. Integr. Biol. 1, 382–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brugidou J., Legrand C., Méry J., and Rabié A. (1995) The retro-inverso form of a homeobox-derived short peptide is rapidly internalized by cultured neurons: a new basis for an efficient intracellular delivery system. Biochem. Biophys. Res. Commun. 214, 685–693 [DOI] [PubMed] [Google Scholar]
- 21. Mueller J., Kretzschmar I., Volkmer R., and Boisguerin P. (2008) Comparison of cellular uptake using 22 CPPs in 4 different cell lines. Bioconjug. Chem. 19, 2363–2374 [DOI] [PubMed] [Google Scholar]
- 22. Erazo-Oliveras A., Muthukrishnan N., Baker R., Wang T. Y., and Pellois J. P. (2012) Improving the endosomal escape of cell-penetrating peptides and their cargos: strategies and challenges. Pharmaceuticals 5, 1177–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Madani F., Lindberg S., Langel U., Futaki S., and Gräslund A. (2011) Mechanisms of cellular uptake of cell-penetrating peptides. J. Biophys. 2011, 414729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Verdurmen W. P., Bovee-Geurts P. H., Wadhwani P., Ulrich A. S., Hällbrink M., van Kuppevelt T. H., and Brock R. (2011) Preferential uptake of l- versus d-amino acid cell-penetrating peptides in a cell type-dependent manner. Chem. Biol. 18, 1000–1010 [DOI] [PubMed] [Google Scholar]
- 25. Tünnemann G., Ter-Avetisyan G., Martin R. M., Stöckl M., Herrmann A., and Cardoso M. C. (2008) Live-cell analysis of cell penetration ability and toxicity of oligo-arginines. J. Pept. Sci. 14, 469–476 [DOI] [PubMed] [Google Scholar]
- 26. Pan C., Lu B., Chen H., and Bishop C. (2010) Reprogramming human fibroblasts using HIV-1 TAT recombinant proteins OCT4, SOX2, KLF4 and c-MYC. Mol. Biol. Rep. 37, 2117–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Erazo-Oliveras A., Najjar K., Dayani L., Wang T.-Y., Johnson G. A., and Pellois J.-P. (2014) Protein delivery into live cells by incubation with an endosomolytic agent. Nat. Methods 11, 861–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee Y. J., Datta S., and Pellois J. P. (2008) Real-time fluorescence detection of protein transduction into live cells. J. Am. Chem. Soc. 130, 2398–2399 [DOI] [PubMed] [Google Scholar]
- 29. Wadia J. S., Stan R. V., and Dowdy S. F. (2004) Transducible TAT-HA fusogenic peptide enhances escape of TAT-fusion proteins after lipid raft macropinocytosis. Nat. Med. 10, 310–315 [DOI] [PubMed] [Google Scholar]
- 30. Erazo-Oliveras A., Najjar K., Truong D., Wang T. Y., Brock D. J., Prater A. R., and Pellois J. P. (2016) The late endosome and its lipid BMP act as gateways for efficient cytosolic access of the delivery agent dfTAT and its macromolecular cargos. Cell Chem. Biol. 23, 598–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fretz M. M., Penning N. A., Al-Taei S., Futaki S., Takeuchi T., Nakase I., Storm G., and Jones A. T. (2007) Temperature-, concentration- and cholesterol-dependent translocation of l- and d-octa-arginine across the plasma and nuclear membrane of CD34+ leukaemia cells. Biochem. J. 403, 335–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vivès E., Brodin P., and Lebleu B. (1997) A truncated HIV-1 Tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus. J. Biol. Chem. 272, 16010–16017 [DOI] [PubMed] [Google Scholar]
- 33. Peitz M., Pfannkuche K., Rajewsky K., and Edenhofer F. (2002) Ability of the hydrophobic FGF and basic TAT peptides to promote cellular uptake of recombinant Cre recombinase: a tool for efficient genetic engineering of mammalian genomes. Proc. Natl. Acad. Sci. U.S.A. 99, 4489–4494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Colpitts T. M., Moore A. C., Kolokoltsov A. A., and Davey R. A. (2007) Venezuelan equine encephalitis virus infection of mosquito cells requires acidification as well as mosquito homologs of the endocytic proteins Rab5 and Rab7. Virology 369, 78–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bucci C., Thomsen P., Nicoziani P., McCarthy J., and van Deurs B. (2000) Rab7: a key to lysosome biogenesis. Mol. Biol. Cell 11, 467–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kobayashi T., Stang E., Fang K. S., de Moerloose P., Parton R. G., and Gruenberg J. (1998) A lipid associated with the antiphospholipid syndrome regulates endosome structure and function. Nature 392, 193–197 [DOI] [PubMed] [Google Scholar]
- 37. Kobayashi T., Beuchat M.-H., Lindsay M., Frias S., Palmiter R. D., Sakuraba H., Parton R. G., and Gruenberg J. (1999) Late endosomal membranes rich in lysobisphosphatidic acid regulate cholesterol transport. Nat. Cell Biol. 1, 113–118 [DOI] [PubMed] [Google Scholar]
- 38. Kobayashi T., Startchev K., Whitney A. J., and Gruenber J. (2001) Localization of lysobisphosphatidic acid-rich membrane domains in late endosomes. Biol. Chem. 382, 483–485 [DOI] [PubMed] [Google Scholar]
- 39. Piper R. C., and Katzmann D. J. (2007) Biogenesis and function of multivesicular bodies. Annu. Rev. Cell Dev. Biol. 23, 519–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Duchardt F., Fotin-Mleczek M., Schwarz H., Fischer R., and Brock R. (2007) A comprehensive model for the cellular uptake of cationic cell-penetrating peptides. Traffic 8, 848–866 [DOI] [PubMed] [Google Scholar]
- 41. Tamai M., Matsumoto K., Omura S., Koyama I., Ozawa Y., and Hanada K. (1986) In vitro and in vivo inhibition of cysteine proteinases by EST, a new analog of E-64. J. Pharmacobiodyn. 9, 672–677 [DOI] [PubMed] [Google Scholar]
- 42. Vidard L., Rock K. L., and Benacerraf B. (1991) The generation of immunogenic peptides can be selectively increased or decreased by proteolytic enzyme inhibitors. J. Immunol. 147, 1786–1791 [PubMed] [Google Scholar]
- 43. Poon G. M., and Gariépy J. (2007) Cell-surface proteoglycans as molecular portals for cationic peptide and polymer entry into cells. Biochem. Soc. Trans. 35, 788–793 [DOI] [PubMed] [Google Scholar]
- 44. Nakase I., Tadokoro A., Kawabata N., Takeuchi T., Katoh H., Hiramoto K., Negishi M., Nomizu M., Sugiura Y., and Futaki S. (2007) Interaction of arginine-rich peptides with membrane-associated proteoglycans is crucial for induction of actin organization and macropinocytosis. Biochemistry 46, 492–501 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.