Abstract
Ricin toxin is a heterodimer consisting of RTA, a ribosome-inactivating protein, and RTB, a lectin that facilitates receptor-mediated uptake into mammalian cells. In previous studies, we demonstrated that toxin-neutralizing antibodies target four spatially distinct hot spots on RTA, which we refer to as epitope clusters I–IV. In this report, we identified and characterized three single domain camelid antibodies (VHH) against cluster II. One of these VHHs, V5E1, ranks as one of the most potent ricin-neutralizing antibodies described to date. We solved the X-ray crystal structures of each of the three VHHs (E1, V1C7, and V5E1) in complex with RTA. V5E1 buries a total of 1,133 Å2 of surface area on RTA and makes primary contacts with α-helix A (residues 18–32), α-helix F (182–194), as well as the F-G loop. V5E1, by virtue of complementarity determining region 3 (CDR3), may also engage with RTB and potentially interfere with the high affinity galactose-recognition element that plays a critical role in toxin attachment to cell surfaces and intracellular trafficking. The two other VHHs, E1 and V1C7, bind epitopes adjacent to V5E1 but display only weak toxin neutralizing activity, thereby providing structural insights into specific residues within cluster II that may be critical contact points for toxin inactivation.
Keywords: antibody, antigen, crystal structure, epitope mapping, toxin, neutralizing, ricin
Introduction
Ricin toxin, a 65-kDa heterodimeric glycoprotein from the castor bean plant (Ricinus communis), is one of the most potent biological toxins known. The toxin's A subunit (RTA)4 is a ribosome-inactivating protein that cleaves the N-glycosidic bond of a single conserved adenosine residue within the sarcin-ricin loop of 28S rRNA that is required for ribosomal elongation (1, 2). RTA (267 residues) is joined via a single disulfide bond to RTB (262 residues), a galactose-specific lectin that attaches to glycoproteins and glycolipids on the surface of mammalian cells (3). RTB promotes the uptake and retrograde trafficking of the toxin to the endoplasmic reticulum (ER), where RTA is liberated from RTB through the action of protein-disulfide isomerase (4). RTA is then dislocated (retro-translocated) across the ER membrane with the assistance of the ER-associated protein degradation system. Once in the cytoplasm, RTA triggers ribosome inactivation and programmed cell death (5).
Structurally, RTA is a globular protein that can be divided into three distinct folding domains (6). Folding domain I (residues 1–117) is dominated by a six-stranded β-sheet that terminates in a prominent solvent-exposed α-helix, known as α-helix B (residues 97–108). Folding domain II (residues 118–210) is dominated by five α-helices (C–G), with α-helix E being at the core of the molecule. Folding domain III (residues 211–267) forms a protruding element that associates with RTB through a disulfide bond as well as extensive non-polar interactions (6). Five residues, Tyr-80, Tyr-123, Glu-177, Arg-180, and Trp-211, define RTA's active site, which consists of a shallow pocket in the central portion of the protein (7). With the exception of the region spanning α-helices F (residues 182–194) and G (residues 202–219), which is rich in arginine residues and thought to promote ribosome binding through electrostatic interactions, no other functionality has been ascribed to RTA's secondary elements or solvent-exposed surface.
Establishing a comprehensive molecular B cell epitope map of RTA is of importance in terms of facilitating design of more effective RTA-based immunotoxins for cancer therapy (8–10) and ricin toxin subunit vaccine antigens for use in the military and in certain civilian populations in the event that ricin were to be used as a biological weapon (11–13). Toward this end, we have generated a large collection of murine monoclonal antibodies (mAbs), as well as single domain heavy chain-only camelid antibodies (VHHs) against linear and discontinuous epitopes on RTA (14–16). The camelid library (referred to as “HobJo”) has an estimated diversity of >107 independent clones and was generated from two animals that had been repeatedly immunized with a mixture of recombinant non-toxic RTA, RTB, and ricin toxoid (16). Analysis to date has suggested that there are (at least) four immunodominant regions on RTA, which we refer to as epitope clusters I–IV. The four clusters are represented by the following toxin-neutralizing mAbs: PB10 (I), SyH7 (II), IB2 (III), and GD12 (IV) (14). Competition ELISAs with the four representative mAbs enabled us to readily categorize new mAbs and VHHs into their respective clusters (14–17).
To date, we have solved the X-ray crystal structures of seven different cluster I VHHs in complex with RTA (18) and complemented these studies with epitope mapping of three different cluster I mAbs by hydrogen deuterium exchange.5 Another group has reported the X-ray crystal structure of a Fab fragment in complex with cluster I (19). At the structural level, cluster I epitopes encompass α-helix B (residues 98–106), β-strand h (residues 113–117), and the C terminus of α-helix D (residues 154–156). In general, we found that within cluster I, ricin toxin neutralizing activity correlated with the degree of interaction between the VHH and RTA, particularly α-helix B (17, 18). Moreover, the interactions between the different VHHs and RTA were largely dictated by the CDR3 elements. Finally, our original assumption that neutralizing and non-neutralizing antibodies would occupy spatially distinct structural epitopes on the surface of RTA was proven incorrect. Instead, we found several examples in which non-neutralizing antibody epitopes were nested within the footprints of neutralizing antibodies.
In this study, we have embarked on an effort to define structural B cell epitopes within cluster II. As noted above, cluster II is defined by SyH7, a toxin-neutralizing mAb that was first identified based on its reactivity with a peptide spanning RTA residues 187–198 (15). In addition to its in vitro activities, SyH7 is able to passively protect mice against systemic 10× lethal dose 50 (LD50) ricin challenge. We subsequently identified three additional toxin-neutralizing mAbs, TB12, PA1, and PH12, that recognize discontinuous epitopes within cluster II (14). The epitopes recognized by SyH7, TB12, PA1, and PH12 are distinct from each other, as determined by HX-MS.5 Because all four cluster II mAbs neutralize ricin in vitro and in vivo, we have proposed that their epitopes represent a particular site of vulnerability on the toxin. We report here the X-ray crystal structures of three different cluster II VHHs in complex with RTA, including one VHH whose epitope possibly spans the interface between RTA and RTB. This study affords the first structural insight into the nature of epitope cluster II and reveals possible mechanisms by which antibodies directed against this region of RTA neutralize the toxin.
Results
Isolation of VHHs against Epitope Cluster II on RTA
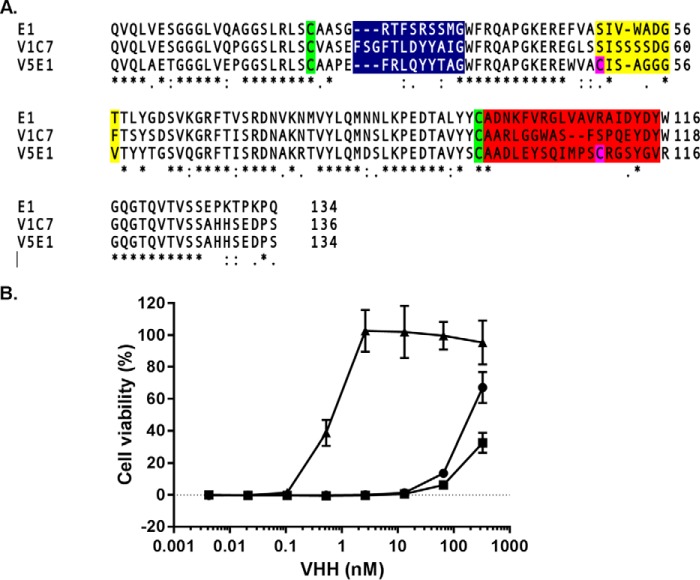
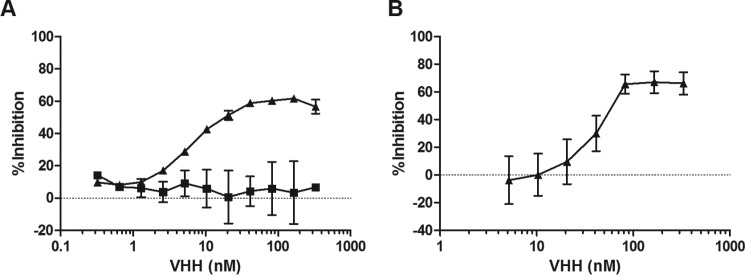
As part of this study, the HobJo phage-displayed single domain VHH library was panned against several different ricin-derived targets, including RTA directly immobilized on polystyrene plates and ricin holotoxin captured via the surrogate receptor (ASF). However, to specifically enrich for cluster II-specific antibodies, the library was panned against ricin captured via JIV-F5 to broadly mask epitope cluster I and in the presence of saturating amounts of JIV-F6 to occlude epitope cluster III (16).6 The screening approaches yielded three different RTA-specific VHHs as follows: E1, V1C7, and V5E1. The antibodies were unique from all other previously identified VHHs based on the primary amino acid sequence of their respective CDR1–3 elements (Fig. 1A). The VHHs also differed in binding affinities (KD) for ricin toxin, as determined by SPR; V5E1 had the highest affinity (KD 20 pm), followed by E1 (690 pm) and V1C7 (3.17 nm) (Table 1). In a cell-based toxin-neutralizing assay, V5E1 was a potent inhibitor of ricin (IC50 <1 nm), whereas E1 and V1C7 were classified as weak inhibitors (IC50 >100 nm) (Fig. 1B).
FIGURE 1.
Amino acid sequences and toxin neutralizing activities associated with E1, V1C7, and V5E1. A, primary amino acid sequence alignments of VHHs E1, V1C7, and V5E1. Highlighted are the conserved cysteine residues (green), two additional cysteine residues in V5E1 (pink), CDR1 (blue), CDR2 (yellow), and CDR3 (red). B, ricin toxin neutralizing activities associated with E1 (circles), V1C7 (squares), and V5E1 (triangles), as determined in a Vero cell cytotoxicity assay (see “Experimental Procedures”). The values shown are the average (with standard deviation) of two separate experiments each done in triplicate.
TABLE 1.
Summary of RTA-VHH binding data and interface information
| VHH | KDa | Kaa | Kda | IC50a | CDR3b | H-bondsc |
BSA | |
|---|---|---|---|---|---|---|---|---|
| Total | CDR1/2/3 | |||||||
| Å2 | ||||||||
| E1 | 0.7 | 3.5 × 105 | 2.4 × 10−4 | 170 | 19 | 16d | 4/0/10 | 1634 |
| 22d | 8/0/12 | 2029 | ||||||
| V1C7 | 3.2 | 9.1 × 104 | 2.9 × 10−4 | NDe | 17 | 12 | 0/5/7 | 1440 |
| V5E1 | 0.02 | 2.2 × 105 | 4.2 × 10−6 | 0.5 | 19 | 10 | 2/0/5 | 1133 |
a KD (× 10−9 m); Ka (1/ms); Kd (1/s); IC50 (× 10−9 m).
b Amino acid length of CDR3 is given.
c H-bonds are between RTA and VHH (total) and CDR1, CDR2, and CDR3, respectively.
d 16 and 22 refer to different RTA-E1 copies in the asymmetric units, as shown in the supplemental material.
e An actual IC50 value is not reported for V1C7 because at 330 nm (the highest antibody concentration tested) it displayed only 30–40% toxin neutralizing activity, as shown in Fig. 1.
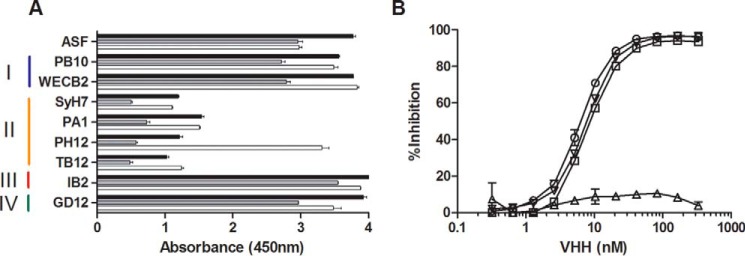
The three VHHs, E1, V1C7, and V5E1, each recognized receptor-bound ricin, as well as ricin that had been captured with representative cluster I (PB10 and WECB2), III (IB2), and IV (GD12) mAbs (Fig. 2). However, the VHHs were inhibited from binding to ricin that had been captured by cluster II-specific antibodies SyH7, PA1, PH12, and TB12 (Figs. 2A and 3). V5E1 was interesting in this regard because binding to ricin holotoxin was inhibited by SyH7, PA1, and TB12 but not by PH12 (Fig. 2B), revealing a previously unrecognized spatial complexity within cluster II. In pairwise competition ELISAs, E1 and V1C7 inhibited each other from binding to ricin but had little or no effect on V5E1 (Fig. 4). Conversely, V5E1 had little or no effect on E1 or V1C7, although it did (of course) inhibit itself from binding ricin. In summary, the three cluster II-specific VHHs, E1, V1C7, and V5E1, likely represent two distinct structural epitopes within cluster II as follows: E1 and V1C7 bind epitope(s) associated with weaker toxin neutralizing activity and V5E1 binds an epitope associated with stronger neutralizing activity.
FIGURE 2.
Competition ELISAs demonstrating that E1, V1C7, and V5E1 recognize epitopes within cluster II. A, ricin was captured on microtiter plates via ASF (top row) or mAbs representing epitope clusters I–IV, as indicated by colored vertical bars (left) and then probed with E1 (black bars), V1C7 (gray bars), and V5E1 (white bars), as described under the “Experimental Procedures.” The values shown are the average (with standard deviation) of a single representative experiment done in triplicate. B, competition ELISAs in which biotinylated ricin was mixed with indicated concentrations (x axis) of V5E1 in solution and then applied to microtiter plates coated with the four cluster II mAbs as follows: SyH7 (circles); PA1 (squares); PH12 (triangles); TB12 (inverted triangles). The plate was then washed and probed with avidin-HRP to detect bound ricin. The values shown are the % inhibition (y axis) as compared with mAb capture of biotinylated ricin without addition of VHH.
FIGURE 3.
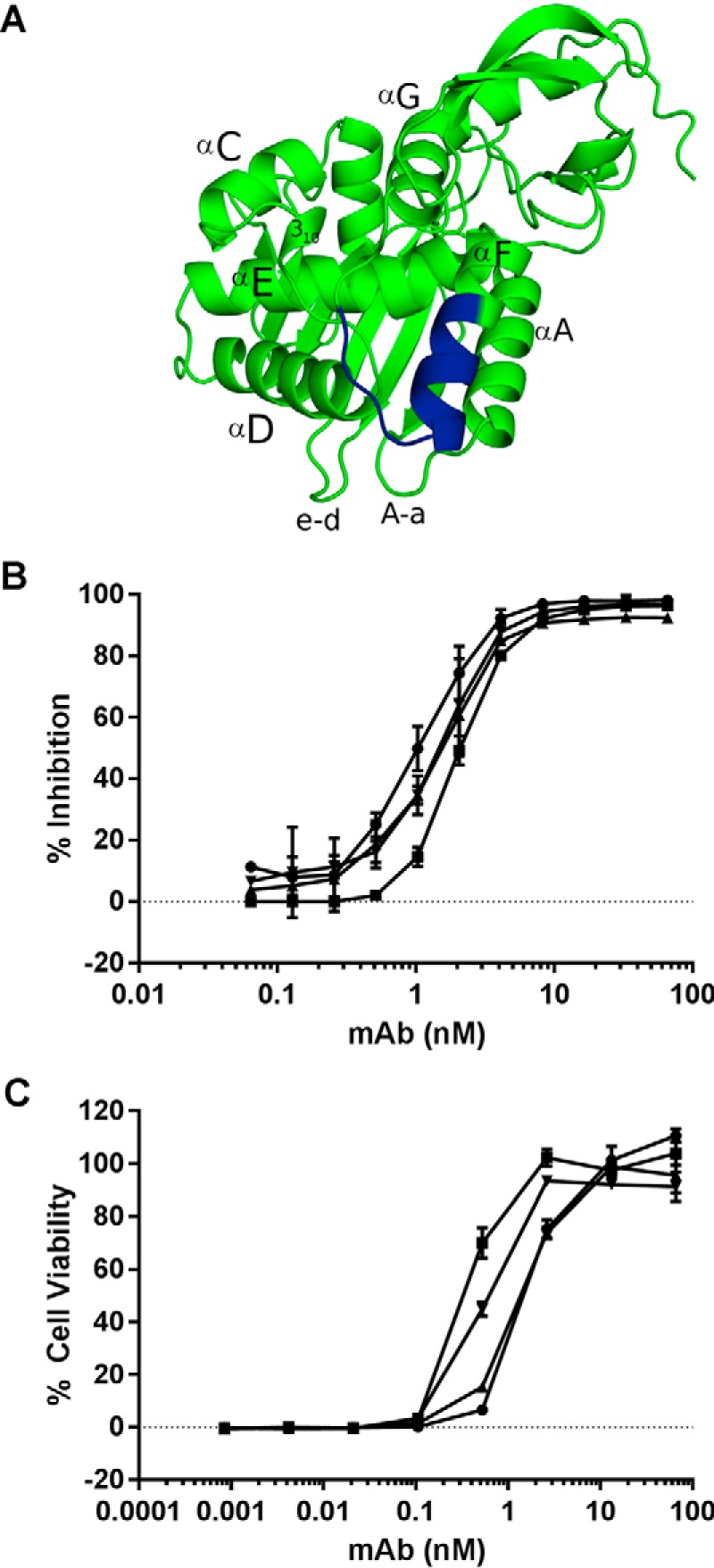
Characterization of epitope cluster II mAbs. A, ribbon diagram of RTA (PDB 1RTC) with relevant secondary structures labeled. The linear epitope recognized by SyH7 is shown in blue. B, competition ELISA in which SyH7 (at indicated concentrations) was mixed with biotinylated ricin in solution and then applied to microtiter plates coated with mAbs SyH7 (circles), PA1 (squares), PH12 (triangles), and TB12 (inverted triangles). The plates were then probed with avidin-HRP, and the amount that SyH7 that blocked biotin-ricin capture by the plate-bound mAbs is indicated on y axis (% inhibition). C, ricin toxin neutralizing activity of SyH7 (circles), PA1 (squares), PH12 (triangles), and TB12 (inverted triangles), as described under the “Experimental Procedures.”
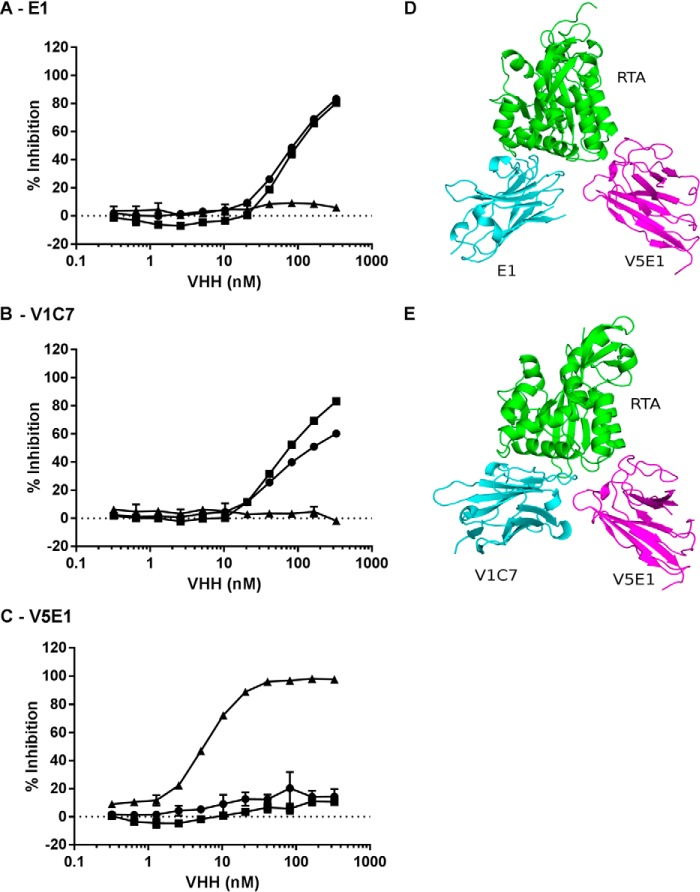
FIGURE 4.
V5E1 recognizes an epitope on RTA that is spatially distinct from E1 and V1C7's epitopes. Competition ELISA in which E1 (A), V1C7 (B), or V5E1 (C), at indicated concentrations, was mixed with biotinylated ricin in solution and then applied to microtiter plates coated with E1 (circles), V1C7 (squares), or V5E1 (triangles). The plates were then probed with avidin-HRP, and the amount that the soluble VHH blocked biotin-ricin capture by the plate bound antibody is indicated on y axis (% inhibition). D and E, structure of RTA-V5E1 complex was superpositioned onto the RTA-E1 or RTA-V1C7 structures to demonstrate the distinct binding profiles. RTA is colored green; V5E1 is magenta, and E1 and V1C7 are in cyan.
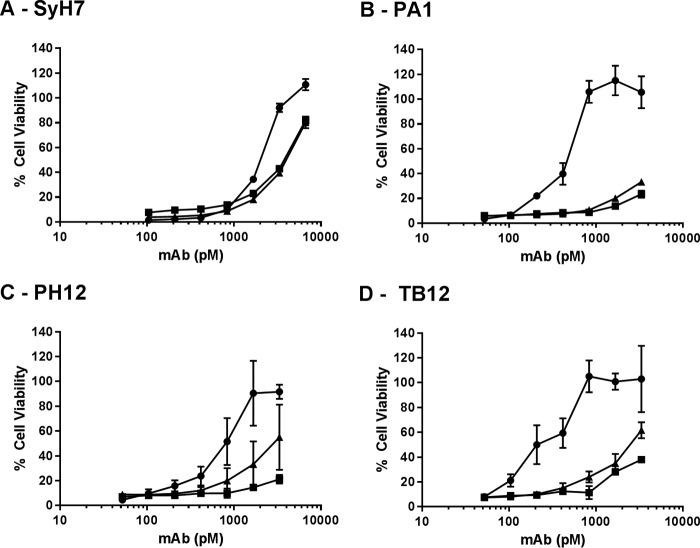
The observation that two VHHs (E1 and V1C7) with weak toxin neutralizing activities recognized epitopes within cluster II prompted us to investigate whether they might competitively interfere with the ability of cluster II mAbs to neutralize ricin. To test this hypothesis, SyH7, PA1, PH12, or TB12 was mixed with a molar excess of E1 or V1C7 and then tested in a cell-based toxin neutralizing assay. We observed that E1 and V1C7 interfered (to varying degrees) with the neutralizing activities of PA1, PH12, and TB12 but had only a marginal effect on SyH7 (Fig. 5). For example, Vero cell viability was 100% in the presence of ricin and 0.8 nm PA1. In contrast, upon addition of molar excess amounts of E1 and V1C7, Vero cell viability in the presence of 0.8 nm PA1 declined to <20% (Fig. 5B). For reasons that are not immediately apparent, SyH7 was largely refractory to the effects of E1 and V1C7, as evidenced by only a slight change in SyH7's IC50 value upon addition of E1 and V1C7.
FIGURE 5.
VHHs E1 and V1C7 interfere with neutralizing activity of cluster II mAbs. VHH E1 (squares) and V1C7 (triangles) mixed with indicated concentrations (x axis) of SyH7 (A), PA1 (B), PH12 (C), or TB12 (D) were incubated with ricin toxin and then applied to Vero cells, as described under the “Experimental Procedures.” For each panel, the circles represent mAb without the addition of E1 or V1C7. The VHHs were at a constant concentration of 133 nm (2 μg/ml). Cell viability was determined 48 h later. The values shown are the average (with standard deviation) of a single representative experiment done in triplicate.
X-ray Crystallography of VHH-RTA Complexes
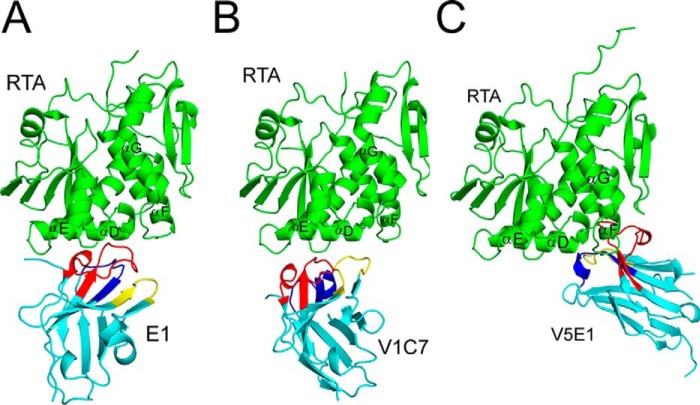
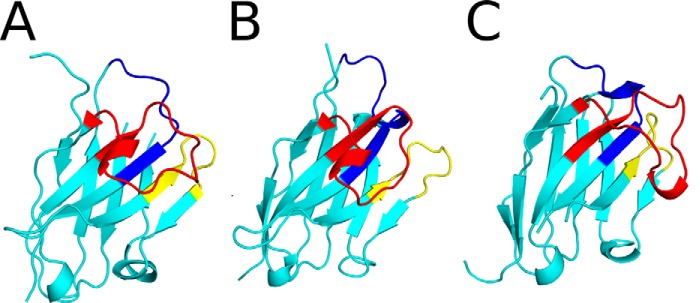
To elucidate the exact epitopes recognized by the three cluster II VHHs, we solved the X-ray crystal structures of E1, V1C7, and V5E1 in complex with RTA, at 3.1, 1.8, and 1.7 Å, respectively (Tables 1 and 2; Fig. 6). In all three complexes, the VHHs assumed a classical immunoglobulin fold consisting of nine β-strands arranged in two β-sheets with all three CDRs on one face of the molecule (Fig. 7). E1 displayed the canonical disulfide bond between Cys-22 and Cys-96 that links FR1 to FR3 (20), whereas V1C7 did not (Figs. 1 and 7). We postulate that the absence of this disulfide bond in V1C7 is due to the low pH (pH 4.6) of the crystallization conditions of the V1C7-RTA complex. Low pH is known to favor the reduction of disulfide bonds, and this was the only relative condition that differed among the three VHH-RTA complexes.
TABLE 2.
Summary of RTA-VHH structures
| E1a | V1C7a | V5E1a | |
|---|---|---|---|
| dmin (Å) | 3.1 | 1.8 | 1.7 |
| Space group | F222 | P3221 | P21 |
| Rb/Rfreec (%) | 22.4/27.5 | 17.3/20.9 | 20.0/24.8 |
| PDB code | 5BOZ | 5J56 | 5J57 |
a All complexes were formed by co-crystallization.
b R = Σ‖Fo| − |Fc‖/Σ‖Fo‖, where Fo and Fc denote observed and calculated structure factors, respectively.
c Rfree was calculated using 5% of data excluded from refinement.
FIGURE 6.
X-ray crystal structures of RTA-VHH complexes. Structures of RTA in complex with VHHs E1 (A), V1C7 (B), and V5E1 (C). RTA (green) is presented in a similar orientation for each panel with α-helices D–G indicated, as necessary. VHHs are shaded in cyan with CDR1–3 colored blue, yellow, and red, respectively.
FIGURE 7.
X-ray crystal structures of VHHs E1, V1C7, and V5E1. The structures of VHHs E1 (A), V1C7 (B), and V5E1 (C) are drawn as ribbon diagrams colored cyan with CDR1–3 colored blue, yellow, and red, respectively. The VHHs are oriented with CDR3 projecting out front.
V5E1, however, has two intramolecular disulfide bridges: the canonical disulfide bond between Cys-22 and Cys-96 and the less frequently observed disulfide bond between Cys-50 and Cys-109 that links CDR2 and CDR3 (Fig. 1). Two other potent ricin toxin-neutralizing VHHs, E5 and F5, exhibit this same secondary disulfide bond (17, 18), which makes it tempting to speculate that stabilizing CDR2-CDR3 through a covalent linkage positively impacts toxin neutralizing activity. Finally, we noted that RTA assumed a very similar conformation in all three RTA-VHH complexes, as evidenced by root mean square deviation values of 0.5 to 0.6 Å when Cα atoms of RTA (PDB code 1RTC) were superimposed onto each of the RTA-VHH crystal structures. Thus, RTA does not undergo any significant conformational changes upon complex formation with E1, V1C7, or V5E1. As an aside, shape complementarity is not a strong predictor of either relative binding affinity or toxin neutralizing activity, as evidenced by the fact that V5E1 had a shape complementarity score of 0.66, as compared with E1 and V1C7, which had higher shape complementarity scores of 0.76 and 0.75, respectively (data not shown). Thus, other factors must account for differences in toxin neutralizing activity observed among the three different VHHs.
Molecular Interactions between RTA and VHHs E1 and V1C7
The region of RTA that contains SyH7's linear epitope, residues 187–198, is in close proximity to secondary structural elements that are likely to constitute the epitopes of other cluster II mAbs like PA1, PH12, and TB12, as well as E1, V1C7, and V5E1 (Fig. 3). These structural elements include six α-helices (A and C–G), three loop regions between α-helices C-D, D-E, and F-G, a single 310-helix between α-helices C and D (referred to as 310-helix C-D), the loops between α-helix A and β-strand a, and β-strands e and d (referred to as loops A-a and e-d) (6, 21, 22).
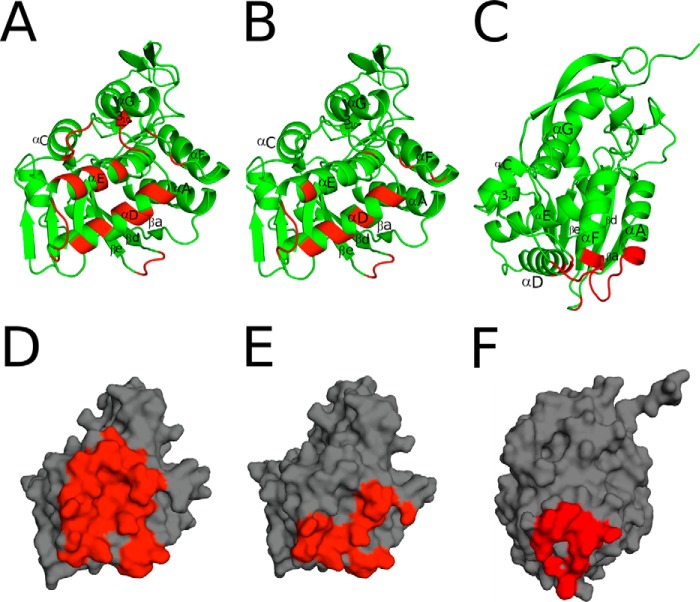
Analysis of the VHH-RTA crystal structures revealed that E1 and V1C7 recognize overlapping but distinct epitopes on RTA. E1 makes contact with α-helix C (residue 130), α-helix D (residues 142 and 145–147), and α-helix E (residues 166, 167, and 170). E1 also interacts with loops e-d (residues 65–67), C-D (residues 131–132), D-E (residues 156,158), F-G (residues 197,199), as well as the 310-helix C-D (residues 135–136) (Fig. 8A). V1C7's interface with RTA is limited to α-helix D (residues 141–142, 145–146, 149, and 153) and α-helix E (residue 163), along with several additional interactions with loops e-d (residues 65–68), C-D (residue 138), D-E (residues 157–160), and F-G (residues 195, 197) (Fig. 8B). As compared with V1C7, E1 forms as many as 10 additional hydrogen bonds with RTA (12 versus 22), depending on which of the six RTA-E1 complexes within the asymmetric unit is used to analyze the RTA-E1 interface (Table 1; supplemental Fig. S1). E1 establishes a buried surface area (bsa) with RTA of ∼2,000 Å2, composed of 63% non-polar and 37% polar atoms. This is in contrast to V1C7, which has a bsa of 1,440 Å2 with RTA, consisting of 59% non-polar and 41% polar atoms (Fig. 8, D and E). Altogether, the larger E1-RTA interface with more H-bonds likely contributes significantly to the 5-fold stronger binding affinity of E1 for ricin toxin, as compared with V1C7 (Table 1). E1 and V1C7 each form one salt-bridge with RTA as follows: E1 residue Lys-114 interacts with Arg-166 of RTA, and V1C7 residue Asp-59 binds Arg-197 in RTA. It is unclear at this time whether the differences in toxin neutralizing activity between E1 and V1C7 are associated with E1's greater degree of interaction with RTA, different contact residues on RTA, and/or its enhanced affinity for ricin holotoxin.
FIGURE 8.
Interfaces of E1, V1C7, and V5E1 with RTA. A–C, RTA (green) is drawn as a ribbon diagram with E1- (A), V1C7- (B), and V5E1 (C)-interacting regions colored red. RTA secondary structural elements are labeled as follows: α-helices A and C–G, β-strands a, d, e, and the 310-helix between α-helices C-D. D–F, surface representations of RTA (gray) with E1 (D), V1C7 (E), and V5E1 (F) contact points highlighted in red.
Molecular Interactions between RTA and V5E1
There is little overlap between the structural epitope recognized by V5E1 and the E1 and V1C7 epitopes, even through all three antibodies are, by definition, situated within cluster II. The distinctiveness of V5E1's epitope, as compared with V1C7 and E1, is most apparent when the RTA subunit in the V5E1-RTA crystal structure is independently superpositioned onto the RTA subunits within the corresponding structures of RTA-V1C7 and RTA-E1 (Fig. 4, D and E). V5E1 makes contact with loops A-a (residues 14–20) and e-d (residue 65), α-helix D (residue 141,145), and loop F-G (residues 192–196) (Fig. 8C). The interface between V5E1 and RTA contains 55% non-polar and 45% polar atoms and is relatively small (bsa = 1,133 Å2), considering its tight binding affinity for ricin (Kd = 0.02 nm) and its potent toxin-neutralizing capacity (IC50 = 0.5 nm) (Table 1). V5E1 forms a total of 10 hydrogen bonds and two salt bridges with RTA. The salt bridges occur between CDR1 residue Arg-28 and RTA residue Glu-145 and CDR3 residue Glu-101 with RTA residue Arg-193.
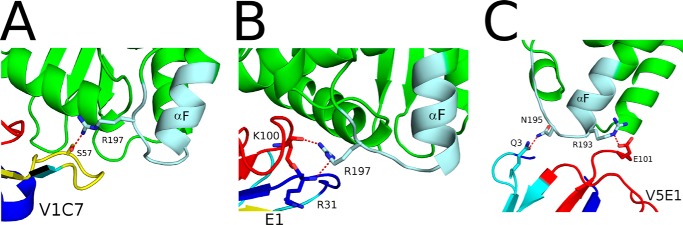
A closer examination of V5E1's interaction with RTA residues 187–198 is warranted considering that these residues are proposed to constitute the core of SyH7's epitope (15). V5E1 buries 481 Å2 of this region of RTA and establishes three H-bonds between framework residue Gln-3 and RTA residues Tyr-194 and Asn-195, as well as the salt bridge between CDR3 residue Glu-101 and RTA residue Arg-193, as noted above (Fig. 9C). By comparison, V1C7 forms only a single H-bond with this region (i.e. CDR2 residue Ser-57 interacts with RTA residue Arg-197) and buries just 241 Å2 (Fig. 9A). E1 forms two hydrogen bonds with this area of RTA (i.e. CDR1 residue Arg-31 with RTA residue Arg-197 and CDR3 residue Lys-100 with RTA residue Arg-197) and buries 260 Å2 (Fig. 9B). We speculate that contact with RTA residues 193–195 may be critical for toxin neutralizing activity for reasons that will be described under the “Discussion.”
FIGURE 9.
Close-up of the VHH interactions with the SyH7 epitope on RTA. RTA (green), the SyH7 epitope (colored pale cyan), and VHHs (cyan), V1C7 (A), E1 (B), and V5E1 (C) are drawn as ribbon diagrams. CDR1–3 are colored blue, yellow, and red, respectively. Key side chains are drawn as sticks and color-coordinated to the main chain. Hydrogen bonds are represented as red dashes.
Another striking feature of the RTA-V5E1 complex is that the entire central region of the CDR3 (residues 105–111) is positioned away from the surface of RTA, making it accessible for potential interactions with other molecules, notably RTB. Indeed, superpositioning the RTA subunit within the RTA-V5E1 complex onto RTA within ricin holotoxin (PDB code 2AAI) reveals that V5E1 could contact RTB residues 147–149, 237–239, and 254, resulting in a potential gain of 237 Å2 of bsa, as compared with the V5E1-RTA complex (Fig. 10). The association with RTB would also create an additional H-bond between V5E1 CDR3 residue Arg-110 and RTB residue Ala-237, which may be consequential in terms of V5E1's toxin neutralizing activity. Arg-110 is ∼4.0 Å from RTB residue Ser-238 of RTB, which, in turn, forms an H-bond with Asn-255 (Fig. 10, inset). Asn-255 is involved in direct interaction with Gal/GalNAc ligands (23). Thus, perturbation of the position of Asn-255 by Arg-110 in V5E1 would be expected to interfere with RTB's ability to adhere to Gal/GalNAc substrates.
FIGURE 10.
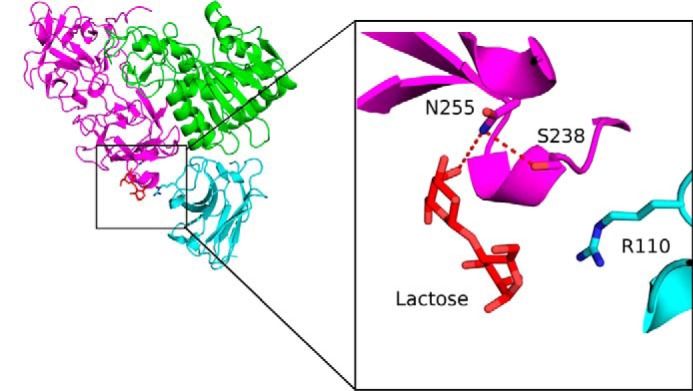
Potential interactions of V5E1 with ricin holotoxin. RTB from ricin holotoxin (PDB code 2AAI) was superpositioned onto RTA-V5E1. RTA is colored green, V5E1 in cyan, and RTB in magenta. Lactose bound to RTB is shown as red sticks. The inset illustrates the proximity of V5E1 residue Arg-110 with RTB residues Asn-255 and Ser-238 involved in lactose recognition. Key residues forming interactions are drawn as sticks and color-coordinated to their respective main chain color. Hydrogen bonds are represented as red dashes.
To address this experimentally, we performed a solid phase binding assay in which microtiter plates coated with ASF were probed with biotin-ricin in the absence or presence of V5E1. V5E1 reduced ricin-ASF interactions in a dose-dependent manner with maximal inhibition being ∼60% when antibody concentrations were equal to or in excess of 30 nm (Fig. 11A). In contrast, V1C7 (Fig. 11A, squares) had no effect on the ricin-ASF interaction, even at 330 nm. We also performed toxin binding assays in which fluorescently labeled ricin was mixed with V5E1, applied to THP-1 monocyte cells on ice (to enable binding but not internalization), and then subjected to flow cytometry. Similar to the ASF assay, V5E1 demonstrated a dose-dependent reduction (∼60%) in ricin binding to cell surfaces (Fig. 11B).
FIGURE 11.
V5E1 partially interferes with ricin attachment to terminal galactosides. A, dilutions of V5E1 (triangles) and V1C7 (squares) were mixed with biotinylated ricin and then applied to microtiter plates coated with ASF, as described under the “Experimental Procedures.” The % inhibition (y axis) values are based on ricin binding in the absence of antibodies. B, V5E1 at indicated concentrations was mixed with FITC-ricin and then applied to THP-1 cells at 4 °C. After 30 min, the THP-1 cells were washed, and the amount of bound ricin was determined by flow cytometry. The % inhibition (y axis) values are based on ricin binding in the absence of antibodies. Each panel is a representative experiment conducted in triplicate.
Finally, we reasoned that if V5E1 has an increased binding affinity for ricin holotoxin (as compared with RTA) then soluble ricin would be more effective than soluble RTA at competitively inhibiting V5E1 attachment to surface-immobilized ricin. V5E1 was mixed with equimolar amounts of RTA or ricin in solution and then subjected to SPR using a ricin-coated sensor. In the absence of soluble ricin or RTA, V5E1 elicited an RU value of 140 (supplemental Fig. S2). When V5E1 was mixed with 60 nm RTA or 60 nm ricin, the RU values dropped to 100 and 40, respectively, indicating that on a molar basis ricin holotoxin is more effective than RTA at competitively inhibiting V5E1 binding. The RU values were reduced to 40 and <5 when V5E1 was mixed with 120 nm RTA or ricin, further indicating that V5E1 preferentially associates with soluble ricin over RTA. In contrast, soluble ricin and RTA were equally effective at inhibiting VHH E1 from binding to plate-bound ricin (supplemental Fig. S3). We conclude from these studies that V5E1 is able to functionally interfere with ricin attachment to cellular receptors, possibly through an interaction with residues within domain 2γ of RTB.
Discussion
We have previously proposed that ricin toxin-neutralizing antibodies are directed against four distinct “hot spots” on the surface of RTA, which we refer to as epitope clusters I–IV. Cluster II is defined by SyH7, a mAb that recognizes a linear epitope within RTA's α-helix F (residues 187–198). Three additional mAbs, PA1, PH12, and TB12, were grouped within cluster II based on their abilities to interfere with SyH7 binding to ricin toxin, as determined by ELISA and SPR (Fig. 3) (14). TB12, PA1, and PH12 are some of the most potent toxin-neutralizing mAbs in our collection (Fig. 3C), and each has been shown to passively protect mice against ricin challenge. Thus, so-called epitope cluster II appears to represent a particularly vulnerable region on ricin toxin.
In this study, we provide the first structural insights into epitope cluster II, as well as clues to contact points on RTA that are associated with toxin neutralizing activity. We solved the X-ray crystal structures of three different cluster II-specific VHHs, each in complex with RTA. The two VHHs with weak toxin neutralizing activities, E1 and V1C7, have overlapping structural epitopes on RTA that primarily encompass residues within α-helix D (residues 141–152) and α-helix E (residues 161–180). E1's slightly lower dissociation constant may explain its marginally better toxin neutralizing activity as compared with V1C7. In contrast, V5E1's structural epitope is focused on the N terminus of α-helix A (residues 18–32) and the C terminus of α-helix F (182–194), as well as residues in the F-G loop. Thus, we tentatively conclude that differences in the structural epitopes recognized by E1, V1C7, and V5E1 are primarily responsible for the observed differences in toxin neutralizing activities among the three antibodies.
V5E1 ranks as one of the most potent toxin-neutralizing single chain antibodies in our collection (16, 24, 25). V5E1 was isolated from the so-called HobJo phage-displayed single domain camelid antibody library, which was generated from two alpacas (Vicugna paco) that had been repeatedly immunized with RiVax, RTB, and ricin toxoid in the presence of aluminum salts and CpG as adjuvants (16). Although it is estimated that the HobJo library consists of >107 unique VHH clones,7 exploiting the diversity of the antibody repertoire has not been trivial. The first ricin-specific VHHs isolated from the HobJo libraries were based on affinity enrichments using RTA or RTB bound directly to polystryrene surfaces (16). These initial screens were biased toward epitopes in cluster I, as evidenced by the fact that 12 out of first 18 anti-RTA VHHs that we characterized were competitively inhibited from binding to RTA by PB10 (16–18). To circumvent this issue, we devised five different screening/masking/capture strategies, which will be detailed in a separate manuscript.6 V5E1 was isolated from a screen in which ricin holotoxin was captured with a cluster I-specific antibody (VHH F5) and masked with a cluster III antibody (VHH F6) (16, 17). It is unlikely that V5E1 would have been isolated without masking cluster I and, possibly, cluster III epitopes.
The X-ray crystal structure of the V5E1-RTA complex offers several clues as to the possible mechanism(s) underlying V5E1's toxin neutralizing activity. First, V5E1 contacts loop F-G (residues 192–196), a region located within an electrostatically positive patch of RTA involved in mediating contact with ribosomes (21, 26). In a yeast model, site-directed mutagenesis has implicated Arg-193 and Arg-196 as being important in ribosome binding (27). It is therefore notable that V5E1 forms a salt bridge with Arg-193, as well as H-bonds with Tyr-194 and Asn-195. α-Helix F and loop F-G also constitutes the core of SyH7's epitope, as initially suggested by pepscan analysis (15) and confirmed by hydrogen deuterium exchange/mass spectroscopy.8 In an in vitro translation assay, SyH7 reduces RTA's ability to inactivate ribosomes by >1,000-fold, presumably because the mAb buries RTA's electrostatic positive patch and/or physically occludes RTA-ribosome contact (15). Preliminary evidence indicates that V5E1 also partially interferes with the ribosome-inactivating properties of RTA in vitro.9
Second, superpositioning of the X-ray crystal structure of the V5E1-RTA complex onto ricin holotoxin revealed that V5E1 would be able to contact RTB's subdomain 2γ, near one of two galactose-binding pockets involved in ricin uptake into host cells (23). RTB's two galactose-binding pockets are located on opposite ends of the molecule (28). Although the two elements are homologous to each other, they have slightly different substrate specificities and affinities; subdomain 1α is specific for galactose (Gal) and is of low affinity, whereas subdomain 2γ recognizes Gal and N-acetylgalactosamine (GalNAc) and is of higher affinity (29, 30). Our experimental results are consistent with V5E1 affecting the lectin activity of subdomain 2γ, as the antibody reduced the attachment of ricin to plate-bound ASF and cell surface receptors by ∼60%. It is also possible that V5E1, by virtue of its potential interaction with subdomain 2γ, alters the dynamics of ricin uptake and/or intracellular transport (29, 31).
The V5E1-RTA crystal structure reveals a potential shortcoming of two candidate ricin toxin subunit vaccines, RiVax and RVEc (13). As noted above, V5E1 appears to recognize a quaternary epitope in which CDR1 makes contact with RTA, whereas CDR3 makes contact with RTA and RTB. From an immunological perspective, V5E1 most certainly arose as a consequence of affinity maturation and somatic hypermutation in response to ricin toxoid vaccination and not in response to either of the individual subunits. The implications for vaccine design are significant considering that the two candidate vaccines being tested are derivatives of RTA, without RTB. RiVax is a recombinant full-length version of RTA with point mutations at residues Tyr-80 and Val-76 to abolish RTA's RNA N-glycosidase activity and to eliminate the protein's propensity to induce vascular leak syndrome, respectively (10, 32). RVEc is a truncated derivative of RTA that lacks a small hydrophobic loop in the N terminus (residues 34–43), as well as C-terminal residues 199–267 (33–35). Thus, neither RiVax nor RVEc would be expected to elicit V5E1-like antibodies. In an effort to overcome this issue, we reconstituted RiVax with RTB in vitro and used this novel toxoid immunogen to vaccinate mice, as well as two additional alpacas.10 Although these studies are still ongoing, preliminary analysis of the antibody responses in mice suggests that RiVax toxoid does indeed boost toxin-neutralizing titers as compared with RiVax alone.
Although the crystal structure of the V5E1-RTA complex offers clues as to the mechanisms by which V5E1 neutralizes ricin, it is not obvious from the E1-RTA and V1C7-RTA structures why E1 and V1C7 have only weak (not strong) neutralizing activity. They each recognize structural epitopes nested within cluster II, as defined by competition ELISAs with SyH7, PA1, TB12, and PH12 (Fig. 2). In the case of V1C7, one possible explanation relates to its relatively low binding affinity for ricin, as compared with V5E1 (3.2 nm versus 0.02 nm). It is tempting to speculate, for example, that a 10-fold improvement in V1C7's binding affinity would result in a corresponding increase in toxin neutralizing activity. In a separate study, we have identified three V1C7-like family members from the HobJo VHH library that differ in their relative affinities and toxin neutralizing activities. The three new family members were subjected to homology modeling by Rosetta using the V1C7-RTA complex as a template to identify residues in CDR1–3 that account for differences in affinity.11 Our long term goal is to engineer higher affinity variants of V1C7 as a means to parse out the relative contributions of affinity and epitope specificity in ricin toxin neutralization.
Experimental Procedures
Materials
Ricin toxin (RCA-II), biotinylated ricin (b-ricin), and RTA were obtained from Vector Laboratories (Burlingame, CA). HRP-anti-E-tag antibodies (Bethyl, Montgomery, TX) and HRP-anti-M13 antibodies were purchased from GE Healthcare. All other chemicals and reagents were purchased from Sigma unless noted otherwise. Epitope-tagged VHHs were produced and purified as described (24).
ELISA, Surface Plasmon Resonance, and Flow Cytometry
The basic ELISA protocol has been described (16). Nunc-Immuno plates (ThermoScientific, Swedesboro, NJ) were coated overnight at 4 °C with target antigen (1 μg/ml), blocked for 2 h with 2% goat serum in 0.1% PBST, and then incubated for 1 h with 5-fold serial dilutions of VHHs and then anti-E-HRP antibodies diluted 1:10,000. For competition assays, murine IgGs (1 μg/ml) were coated onto wells overnight and then blocked for 2 h. In a separate dilution plate, VHHs were diluted into biotinylated ricin at the fixed EC90 concentration for each individual coated mAb. These mixtures were then transferred into the mAb-coated plates and allowed to bind for 1 h. After washing, bound biotinylated ricin was detected with streptavidin-HRP (1:1000) (Thermo Fisher) and developed with SureBlue Peroxidase Substrate (KPL). The reaction was quenched with 1 m phosphoric acid, and absorbance was read at 450 nm using a VersaMax Microplate Reader (Molecular Devices, Sunnyvale, CA). VHH affinity for ricin was determined by SPR using the ProteOn XPR36 (Bio-Rad), as described previously (18). Ricin binding assays to THP-1 cells were described previously (36).
Identification of Cluster II VHHs
The RTA-specific VHHs were identified using the basic panning strategy described previously with some modifications (16). JIY-E1 (referred to as E1) was identified in a pan against RTA-coated immunotubes, essentially as described (16). V1C7 was identified using a screen in which ricin was captured by surrogate receptor ASF to retain proper conformation of ricin. V5E1 was identified in a pan against ricin captured by the cluster I-specific VHH JIV-F5 (16), to mask that immunodominant region. Cluster III was also masked with an excess of VHH JIV-F6. The details of the screens are described elsewhere.12
Vero Cell Cytotoxicity Assays
The Vero cell cytotoxicity assay has been described in detail elsewhere (37). Vero cells grown in DMEM containing 10% FBS were seeded (5 × 104 per ml) in 96-well cell culture plates and incubated at 37 °C overnight. The cells were then overlaid with ricin (10 ng/ml, 150 pm) in the absence or presence of 5-fold serial dilutions of VHHs and incubated at 37 °C for 2 h. The cells were then washed, and fresh medium was applied. Cell viability was assessed 45–48 h later using CellTiter-Glo (Promega, Madison, WI). In cytotoxicity assays in which VHH was competed against mAb, VHH concentration was held steady, although the mAb concentration was diluted 2-fold.
Cloning, Expression, and Purification of Untagged VHHs for Crystallization
PCR amplicons corresponding to E1 (residues 1–134), V1C7 (residues 1–136), V5E1 (residues 1–134), and RTA (residues 1–267) were subcloned into the N-terminally deca-histidine maltose-binding protein-tagged MCSG9 expression vector using a standard ligase-independent cloning protocol. All four proteins were expressed in Escherichia coli strain BL21(DE3)-pRARE. The transformed bacteria were grown at 37 °C in TB medium and induced at 20 °C with 0.1 mm isopropyl 1-thio-β-d-galactopyranoside at an A600 of 0.6 for ∼16 h. After induction, cells were harvested and resuspended in 20 mm Tris-Cl, pH 7.5, and 150 mm NaCl. The cell suspension was sonicated and centrifuged at 30,000 × g for 30 min. After centrifugation, the protein-containing supernatant was purified by nickel-affinity and size-exclusion chromatography on an AKTAxpress system (GE Healthcare), which consisted of a 1-ml nickel affinity column followed by a Superdex 200 16/60 gel filtration column. The elution buffer consisted of 0.5 m imidazole in binding buffer, and the gel filtration buffer consisted of 20 mm HEPES, pH 7.6, 150 mm NaCl, and 20 mm imidazole. Fractions containing protein were pooled and subjected to tobacco etch virus protease cleavage (1:20 weight ratio) for 3 h at room temperature to remove the deca-histidine maltose-binding protein tag. The cleaved protein was passed over a 1-ml nickel-nitrilotriacetic acid-agarose (Qiagen) and a 1-ml amylose-agarose gravity column to remove the added tobacco etch virus protease, cleaved residues, and uncleaved fusion protein. To generate RTA-VHH protein complexes, after purification RTA was mixed in a 1:1 stoichiometry with the purified VHH and incubated on ice for 1 h. Purified RTA-VHH complex was concentrated to a final total concentration of 10 mg/ml for crystallization experiments.
Crystallization and Data Collection
All three RTA-VHH complex crystals were grown by sitting drop vapor diffusion at 20 °C using a protein to reservoir volume ratio of 1:1 with total drop volumes of 0.4 μl. Crystals of the RTA-E1 complex were grown against crystallization buffer containing 1.5 m ammonium sulfate and 25% glycerol. Crystals of the RTA-E1 complex nucleated within 24 h and grew slowly to a full size of ∼60 μm over a period of 10–14 days. Crystals of the RTA-V1C7 complex were grown with a crystallization buffer containing 100 mm sodium acetate, pH 4.6, and 8% PEG 4000. Crystals of the RTA-V1C7 complex nucleated within 24 h and grew to full size of ∼100 μm within 5 days. Crystals of the RTA-V5E1 complex were grown using a crystallization buffer containing 100 mm Tris, pH 7.0, and 20% PEG 1000. Crystals of the RTA-V5E1 complex took 8 days to nucleate and continued growing very slowly for another 7 days until they reached a full size of ∼60 μm. All crystals were flash-frozen in liquid nitrogen after a short soak in the appropriate crystallization buffers supplemented with 20–25% ethylene glycol. Data were collected at the 24-ID-C and 24-ID-E beamlines at the Advanced Photon Source, Argonne National Laboratory.
Structure Determination and Refinement
The structures of each RTA-VHH complex was solved by molecular replacement using the program Phaser (38). Molecular replacement calculations were performed using the coordinates of the ricin A chain as a search model for RTA (PDB code 1RTC) in all three RTA-VHH complexes. The VHH coordinates used as a search model for all three RTA-VHH complexes was D10 (PDB code 4LGR) with all three of the CDRs removed from the search model. For the RTA-V1C7 and RTA-V5E1 structures, the resulting phase information was used to autobuild most of the model for the both RTA-VHH structures using the program ARP (39). Some additional manual building of each model was performed with COOT (40). For the RTA-E1 structure, the resulting phase information was used insert the E1 sequence and manually build the mode in COOT. All structural refinement was done employing the PHENIX package (41). Twinned refinement was performed for the RTA-E1 complex using the twin operator -k,-h, -l with a twinning fraction of 0.015. During refinement a cross-validation test set was created from a random 5% of the reflections. Data collection and refinement statistics are listed in supplemental Table S1. Molecular graphics were prepared using PyMOL (Schrödinger, LLC, New York).
Antibody-Antigen Shape Correlation
We examined the shape correlation parameter Sc (42) to see whether there was a correlation between toxin neutralizing capacity, binding affinity (Kd), and shape complementarity within the RTA-VHH interfaces described here.
Accession Numbers
The structures generated in this study were deposited in the Protein Data Bank under accession numbers 5BOZ (E1), 5J56 (V1C7), and 5J57 (V5E1) as described in Table 1.
Author Contributions
M. J. R., D. J. V., M. S. C., and Y. R. conducted the experiments described in paper; M. J. R., D. J. V., and N. J. M. analyzed the results and wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We gratefully acknowledge the 24-ID-C and 24-ID-E beamline staff at the Advanced Photon Source for their assistance in data collection and Dr. Jon Robertus (University of Texas, Austin) for providing us with a plasmid encoding RTA. We thank the Wadsworth Center's Immunology Core for assistance with flow cytometry and SPR. We thank Amanda Poon for assistance with the ricin binding assays to THP-1 cells.
This work was supported by NIAID Contract No. HHSN272201400021C from the National Institutes of Health. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Figs. S1–S3 and Table S1.
The atomic coordinates and structure factors (codes 5BOZ, 5J56, and 5J57) have been deposited in the Protein Data Bank (http://wwpdb.org/).
R. Toth, D. Weis, D. Volkin, and N. Mantis, manuscript in preparation.
D. Vance, J. Tremblay, C. Shoemaker, and N. Mantis, manuscript in preparation.
C. Shoemaker, unpublished results.
R. Toth, N. Mantis, D. Weis, and D. Volkin, manuscript in preparation.
T. Czjaka, D. Vance, M. Rudolph, and N. Mantis, unpublished results.
D. Vance, C. Shoemaker, and N. Mantis, unpublished results.
A. Bazzoli, D. Vance, M. Rudolph, J. Karanicolas, and N. Mantis, manuscript in preparation.
D. Vance, J. Tremblay, C. Shoemaker, and N. Mantis, manuscript in preparation.
- RTA
- ricin toxin enzymatic subunit A
- RTB
- ricin toxin-binding subunit B
- VHH
- single domain camelid antibody
- bsa
- buried surface area
- CDR
- complementarity determining region
- PDB
- Protein Data Bank
- SPR
- surface plasmon resonance
- ER
- endoplasmic reticulum
- ASF
- asialofetuin.
References
- 1. Endo Y., Mitsui K., Motizuki M., and Tsurugi K. (1987) The mechanism of action of ricin and related toxins on eukaryotic ribosomes. J. Biol. Chem. 262, 5908–5912 [PubMed] [Google Scholar]
- 2. Endo Y., and Tsurugi K. (1987) RNA N-glycosidase activity of ricin A-chain. Mechanism of action of the toxic lectin ricin on eukaryotic ribosomes. J. Biol. Chem. 262, 8128–8130 [PubMed] [Google Scholar]
- 3. Sandvig K., Olsnes S., and Pihl A. (1976) Kinetics of binding of the toxic lectins abrin and ricin to surface receptors of human cells. J. Biol. Chem. 251, 3977–3984 [PubMed] [Google Scholar]
- 4. Spooner R. A., and Lord J. M. (2015) Ricin trafficking in cells. Toxins 7, 49–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jandhyala D. M., Thorpe C. M., and Magun B. (2012) Ricin and Shiga toxins: effects on host cell signal transduction. Curr. Top. Microbiol. Immunol. 357, 41–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Montfort W., Villafranca J. E., Monzingo A. F., Ernst S. R., Katzin B., Rutenber E., Xuong N. H., Hamlin R., and Robertus J. D. (1987) The three-dimensional structure of ricin at 2.8 A. J. Biol. Chem. 262, 5398–5403 [PubMed] [Google Scholar]
- 7. Ready M. P., Kim Y., and Robertus J. D. (1991) Site-directed mutagenesis of ricin A-chain and implications for the mechanism of action. Proteins 10, 270–278 [DOI] [PubMed] [Google Scholar]
- 8. Castelletti D., Fracasso G., Righetti S., Tridente G., Schnell R., Engert A., and Colombatti M. (2004) A dominant linear B-cell epitope of ricin A-chain is the target of a neutralizing antibody response in Hodgkin's lymphoma patients treated with an anti-CD25 immunotoxin. Clin. Exp. Immunol. 136, 365–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pincus S. H., Smallshaw J. E., Song K., Berry J., and Vitetta E. S. (2011) Passive and active vaccination strategies to prevent ricin poisoning. Toxins 3, 1163–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Smallshaw J. E., Ghetie V., Rizo J., Fulmer J. R., Trahan L. L., Ghetie M. A., and Vitetta E. S. (2003) Genetic engineering of an immunotoxin to eliminate pulmonary vascular leak in mice. Nat. Biotechnol. 21, 387–391 [DOI] [PubMed] [Google Scholar]
- 11. Reisler R. B., and Smith L. A. (2012) The need for continued development of ricin countermeasures. Adv. Prev. Med. 2012, 149737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Roy C. J., Brey R. N., Mantis N. J., Mapes K., Pop I. V., Pop L. M., Ruback S., Killeen S. Z., Doyle-Meyers L., Vinet-Oliphant H. S., Didier P. J., and Vitetta E. S. (2015) Thermostable ricin vaccine protects rhesus macaques against aerosolized ricin: epitope-specific neutralizing antibodies correlate with protection. Proc. Natl. Acad. Sci. U.S.A. 112, 3782–3787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vance D. J., and Mantis N. J. (2016) Progress and challenges associated with the development of ricin toxin subunit vaccines. Expert Rev. Vaccines 15, 1213–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. O'Hara J. M., Kasten-Jolly J. C., Reynolds C. E., and Mantis N. J. (2014) Localization of non-linear neutralizing B cell epitopes on ricin toxin's enzymatic subunit (RTA). Immunol. Lett. 158, 7–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. O'Hara J. M., Neal L. M., McCarthy E. A., Kasten-Jolly J. A., Brey R. N. 3rd, and Mantis N. J. (2010) Folding domains within the ricin toxin A subunit as targets of protective antibodies. Vaccine 28, 7035–7046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vance D. J., Tremblay J. M., Mantis N. J., and Shoemaker C. B. (2013) Stepwise engineering of heterodimeric single domain camelid VHH antibodies that passively protect mice from ricin toxin. J. Biol. Chem. 288, 36538–36547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rudolph M. J., Vance D. J., Cassidy M. S., Rong Y., Shoemaker C. B., and Mantis N. J. (2016) Structural analysis of nested neutralizing and non-neutralizing B cell epitopes on ricin toxin's enzymatic subunit. Proteins 84, 1162–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rudolph M. J., Vance D. J., Cheung J., Franklin M. C., Burshteyn F., Cassidy M. S., Gary E. N., Herrera C., Shoemaker C. B., and Mantis N. J. (2014) Crystal structures of ricin toxin's enzymatic subunit (RTA) in complex with neutralizing and non-neutralizing single-chain antibodies. J. Mol. Biol. 426, 3057–3068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhu Y., Dai J., Zhang T., Li X., Fang P., Wang H., Jiang Y., Yu X., Xia T., Niu L., Guo Y., and Teng M. (2013) Structural insights into the neutralization mechanism of monoclonal antibody 6C2 against ricin. J. Biol. Chem. 288, 25165–25172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Muyldermans S., Cambillau C., and Wyns L. (2001) Recognition of antigens by single-domain antibody fragments: the superfluous luxury of paired domains. Trends Biochem. Sci. 26, 230–235 [DOI] [PubMed] [Google Scholar]
- 21. Katzin B. J., Collins E. J., and Robertus J. D. (1991) Structure of ricin A-chain at 2.5 A. Proteins 10, 251–259 [DOI] [PubMed] [Google Scholar]
- 22. Rutenber E., Katzin B. J., Ernst S., Collins E. J., Mlsna D., Ready M. P., and Robertus J. D. (1991) Crystallographic refinement of ricin to 2.5 A. Proteins 10, 240–250 [DOI] [PubMed] [Google Scholar]
- 23. Rutenber E., Ready M., and Robertus J. D. (1987) Structure and evolution of ricin B chain. Nature 326, 624–626 [DOI] [PubMed] [Google Scholar]
- 24. Herrera C., Tremblay J. M., Shoemaker C. B., and Mantis N. J. (2015) Mechanisms of ricin toxin neutralization revealed through engineered homodimeric and heterodimeric camelid antibodies. J. Biol. Chem. 290, 27880–27889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Herrera C., Vance D. J., Eisele L. E., Shoemaker C. B., and Mantis N. J. (2014) Differential neutralizing activities of a single domain camelid antibody (VHH) specific for ricin toxin's binding subunit (RTB). PLoS ONE 9, e99788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li X. P., Chiou J. C., Remacha M., Ballesta J. P., and Tumer N. E. (2009) A two-step binding model proposed for the electrostatic interactions of ricin a chain with ribosomes. Biochemistry 48, 3853–3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li X. P., Kahn P. C., Kahn J. N., Grela P., and Tumer N. E. (2013) Arginine residues on the opposite side of the active site stimulate the catalysis of ribosome depurination by ricin A chain by interacting with the P-protein stalk. J. Biol. Chem. 288, 30270–30284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wales R., Richardson P. T., Roberts L. M., Woodland H. R., and Lord J. M. (1991) Mutational analysis of the galactose binding ability of recombinant ricin B chain. J. Biol. Chem. 266, 19172–19179 [PubMed] [Google Scholar]
- 29. Newton D. L., Wales R., Richardson P. T., Walbridge S., Saxena S. K., Ackerman E. J., Roberts L. M., Lord J. M., and Youle R. J. (1992) Cell surface and intracellular functions for ricin galactose binding. J. Biol. Chem. 267, 11917–11922 [PubMed] [Google Scholar]
- 30. Zentz C., Frénoy J. P., and Bourrillon R. (1978) Binding of galactose and lactose to ricin. Equilibrium studies. Biochim. Biophys. Acta 536, 18–26 [DOI] [PubMed] [Google Scholar]
- 31. Yermakova A., Klokk T. I., Cole R., Sandvig K., and Mantis N. J. (2014) Antibody-mediated inhibition of ricin toxin retrograde transport. MBio 5, e00995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Smallshaw J. E., Firan A., Fulmer J. R., Ruback S. L., Ghetie V., and Vitetta E. S. (2002) A novel recombinant vaccine which protects mice against ricin intoxication. Vaccine 20, 3422–3427 [DOI] [PubMed] [Google Scholar]
- 33. Carra J. H., Wannemacher R. W., Tammariello R. F., Lindsey C. Y., Dinterman R. E., Schokman R. D., and Smith L. A. (2007) Improved formulation of a recombinant ricin A-chain vaccine increases its stability and effective antigenicity. Vaccine 25, 4149–4158 [DOI] [PubMed] [Google Scholar]
- 34. McHugh C. A., Tammariello R. F., Millard C. B., and Carra J. H. (2004) Improved stability of a protein vaccine through elimination of a partially unfolded state. Protein Sci. 13, 2736–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Olson M. A., Carra J. H., Roxas-Duncan V., Wannemacher R. W., Smith L. A., and Millard C. B. (2004) Finding a new vaccine in the ricin protein fold. Protein Eng. Des. Sel. 17, 391–397 [DOI] [PubMed] [Google Scholar]
- 36. Yermakova A., and Mantis N. J. (2011) Protective immunity to ricin toxin conferred by antibodies against the toxin's binding subunit (RTB). Vaccine 29, 7925–7935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wahome P. G., and Mantis N. J. (2013) High-throughput, cell-based screens to identify small-molecule inhibitors of ricin toxin and related category B ribosome inactivating proteins (RIPs). Curr. Protoc. Toxicol. 2013, Chapter 2, Unit 2.23 [DOI] [PubMed] [Google Scholar]
- 38. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Morris R. J., Perrakis A., and Lamzin V. S. (2003) ARP/wARP and automatic interpretation of protein electron density maps. Methods Enzymol. 374, 229–244 [DOI] [PubMed] [Google Scholar]
- 40. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lawrence M. C., and Colman P. M. (1993) Shape complementarity at protein/protein interfaces. J Mol. Biol. 234, 946–950 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.