Abstract
Lipids are important nutrients that proliferating cells require to maintain energy homeostasis as well as to build plasma membranes for newly synthesized cells. Previously, we identified nutrient-sensing checkpoints that exist in the latter part of the G1 phase of the cell cycle that are dependent upon essential amino acids, Gln, and finally, a checkpoint mediated by mammalian target of rapamycin (mTOR), which integrates signals from both nutrients and growth factors. In this study, we have identified and temporally mapped a lipid-mediated G1 checkpoint. This checkpoint is located after the Gln checkpoint and before the mTOR-mediated cell cycle checkpoint. Intriguingly, clear cell renal cell carcinoma cells (ccRCC) have a dysregulated lipid-mediated checkpoint due in part to defective phosphatase and tensin homologue (PTEN). When deprived of lipids, instead of arresting in G1, these cells continue to cycle and utilize lipid droplets as a source of lipids. Lipid droplets have been known to maintain endoplasmic reticulum homeostasis and prevent cytotoxic endoplasmic reticulum stress in ccRCC. Dysregulation of the lipid-mediated checkpoint forces these cells to utilize lipid droplets, which could potentially lead to therapeutic opportunities that exploit this property of ccRCC.
Keywords: amino acid, cell cycle, checkpoint control, glutamine, lipid, mTOR complex (mTORC), START, renal cancer, lipids, checkpoints
Introduction
The key decisions regarding whether a mitotic cell will continue to divide are made during the G1 phase of the cell cycle. There are series of checkpoints that monitor first for the presence of growth factors that indicate that it is appropriate to undergo another cell division, and then whether there are sufficient nutrients for the cell to double its mass and divide into two daughter cells (1). Growth factors need to be present for a dividing cell to avoid cell cycle exit into a state of quiescence commonly called G0. This checkpoint is commonly referred to as the restriction point originally described by Arthur Pardee and colleagues (2, 3). This site was carefully mapped to a site about 3.5 h after mitosis for virtually all mammalian cells in culture (4, 5). After passing the restriction point, the cell is committed to replicating its genome and dividing into two daughter cells. However, there are series of additional checkpoints that monitor nutrient sufficiency to determine whether enough raw material is available to the cell to make two daughter cells. We previously reported that we could distinguish checkpoints for essential amino acids (EAA)2 and for the conditionally EAA Gln (6). These metabolic checkpoints mapped downstream from the restriction point and upstream from another checkpoint mediated by mTOR, the mammalian target of rapamycin (6). This site very late in G1 has also been referred to as the restriction point in that it is also dependent upon growth factors (7). It was proposed that the metabolic checkpoints represent the evolutionary equivalent of START in the yeast cell cycle (1) where nutrient sufficiency is determined prior to entering S-phase and the replication of the genome (8). In this study, we identify another distinguishable metabolic checkpoint that is sensitive to the presence of exogenous lipids. This site can be temporally mapped downstream of the EAA and Gln checkpoints and upstream from the mTOR checkpoint. We propose that this checkpoint is part of the START-like checkpoints that evaluate nutrient sufficiency prior to committing to replication of the genome. This checkpoint was found to be dysregulated in renal cancer cell lines lacking phosphatase and tensin homologue (PTEN) and could possibly create therapeutic opportunities for cancer cells where this checkpoint is dysregulated.
Results
Depriving Cells of Lipids Arrests Cells in G1
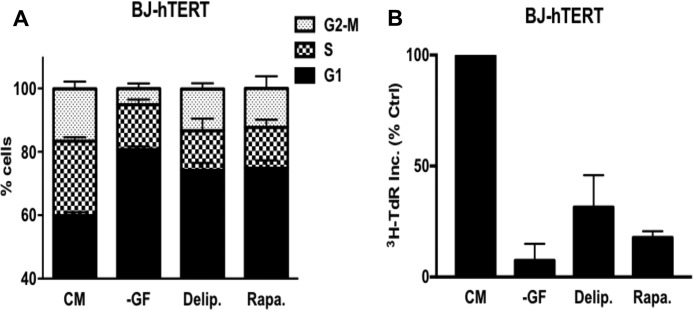
It was recently reported that Ras-driven cancer cells have an acute need for exogenously supplied lipids (9, 10). During the course of this study, we noticed that non-Ras-driven cancers were arrested in G1 (10). We extended this study to the immortalized human fibroblast cell line BJ-hTERT. We subjected these cells to serum deprivation and rapamycin for 48 h, which we demonstrated previously arrested the BJ-hTERT cells in G1 (6). As shown in Fig. 1A, these treatments caused a G1 cell cycle arrest as determined by flow cytometry, which measures the DNA content per cell. There was an increase in cells with G1 DNA content and a reduction in the amount of S-phase and G2/M DNA content, indicating a G1 arrest. We also treated the BJ-hTERT cells with delipidated serum and observed a similar increase in G1 DNA content and reduction in S-phase and G2/M DNA content cells (Fig. 1A), indicating a G1 arrest. We also measured the effect of lipid deprivation on DNA synthesis as measured by the uptake of [3H]thymidine deoxyribose (TdR). The BJ-hTERT cells were treated as in Fig. 1A except that [3H]TdR was added for the last 24 h of treatment. As shown in Fig. 1B, [3H]TdR incorporation was dramatically reduced with all three treatments. Although the reduction in [3H]TdR with delipidated serum was not quite as strong, this was likely due to the inability to remove all lipids from serum. These data demonstrate that in response to lipid deprivation, BJ-hTERT fibroblasts arrest in G1.
FIGURE 1.
Depriving cells of lipids arrests cells in G1. A, BJ-hTERT cells were plated at 30% confluence in DMEM containing 10% FBS. After 24 h, cells were shifted to complete medium (CM), no growth factors (−GF), medium containing 10% delipidated serum (Delip.), or CM containing rapamycin (Rapa.) for 48 h, after which the cells were harvested and analyzed for cell cycle distribution by measuring DNA content/cell. The CM contained 10% dialyzed FBS instead of 10% FBS. Error bars represent the standard deviation from independent experiments repeated 3 times. B, BJ-hTERT cells were plated and shifted to the conditions explained above. Cells were labeled with [3H]TdR for the final 24 h of treatment, after which the cells were collected and the incorporated label (3H-TdR Inc.) was determined by scintillation counting. Ctrl, control. Values were normalized to the cpm for CM, which was given a value of 100%. Error bars represent the standard deviation for experiments repeated 3 times.
Lipid Deprivation Arrests Cells Downstream of the Gln Checkpoint and Upstream of the mTOR Checkpoint
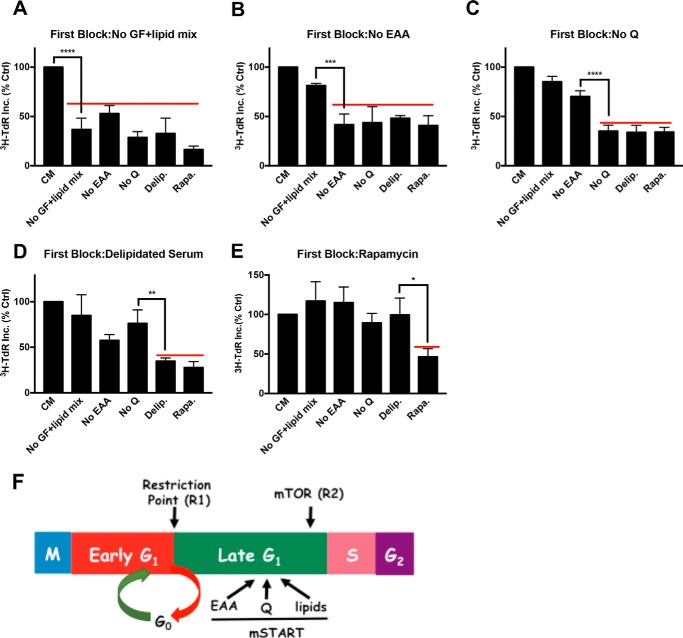
To temporally distinguish the lipid G1 cell cycle arrest from the arrest caused by serum withdrawal, EAA and Gln deprivation, and rapamycin, we performed a series of sequential blocking experiments. In brief, cells were exposed to various blocking conditions for 48 h to cause complete cell cycle arrest. At this point, the first block was removed and a second block was applied along with [3H]TdR for 24 h. If the second block applied is either at the same point or downstream of the first block, then [3H]TdR incorporation should not occur. However, if the second block site is upstream of the first block, then the cells should progress into S-phase and incorporate the label. The level of [3H]TdR incorporated by cells released into complete medium after a first block was considered to be 100%. We first examined cell cycle progression after a first block of serum withdrawal (Fig. 2A). To better distinguish the effect of growth factors from that of lipids in serum, the serum withdrawal block included a mixture of lipids (no growth factors plus lipid mix) (see “Experimental Procedures”). As reported previously (6), EAA and Gln deprivation, as well as rapamycin treatment, prevented significant [3H]TdR incorporation. If we used delipidated serum (i.e. growth factors without lipids) as a second block, there was still very little [3H]TdR (Fig. 2A). These data indicate that the lipid checkpoint is downstream from the growth factor-dependent restriction point.
FIGURE 2.
Lipid deprivation arrests cells downstream of the Gln checkpoint and upstream from the mTOR checkpoint. BJ-hTERT cells were plated and shifted to various first blocking conditions for 48 h as in Fig. 1A. The cells were subsequently shifted to CM or different second block conditions containing [3H]TdR for 24 h, after which the cells were collected and the incorporated label (3H-TdR Inc.) was determined. The first blocks were: A, no serum (no growth factors) plus lipid mix (No GF+lipid mix) (see “Experimental Procedures”); B, no EAA; C, no Gln (Q); D, delipidated serum; and E, rapamycin (20 μm). Ctrl, control; No Q, no Gln; Delip., delipidated serum; Rapa., rapamycin. F, schematic summarizing results from the double block mapping experiments for metabolic checkpoints for EAA, Gln, and lipids that are hypothesized to represent a mammalian START (mSTART). Also shown are the relative positions of the two restriction points (R1 and R2) that respond to growth factors that have been described (5, 7). Error bars represent the standard deviation for experiments repeated at least 3 times. One-way analysis of variance was used to generate p values for establishing the significance between the primary block condition and the immediate upstream block condition (*, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001).
We next examined the effects of EAA (Fig. 2B) and Gln (Fig. 2C) deprivation as a first block. In both cases, a second block with delipidated serum still prevented [3H]TdR incorporation, indicating that the lipid checkpoint was downstream from both the EAA and Gln checkpoints. As reported previously (6), the Gln site was downstream from the EAA site, as indicated by the ability of Gln deprivation to prevent [3H]TdR incorporation when EAA were restored (Fig. 2B), whereas deprivation of EAA as a second block did not suppress [3H]TdR incorporation if the first block was Gln deprivation (Fig. 2C). If the first block was delipidated serum, the readdition of non-delipidated serum and a second block with EAA, Gln, or growth factor deprivation did not suppress [3H]TdR incorporation (Fig. 2D). In contrast, rapamycin treatment did prevent [3H]TdR incorporation observed when non-delipidated serum was provided (Fig. 2D). Lastly, if the first block was rapamycin, a second block with delipidated serum did not prevent [3H]TdR incorporation (Fig. 2E), indicating that the lipid checkpoint is upstream from the mTOR-dependent checkpoint. The second blocks after rapamycin treatment consistently led to elevated [3H]TdR incorporation relative to the control (Fig. 2E). This was likely because rapamycin blocks cell cycle progression better than nutrient deficiency (Fig. 2A) (6). Nutrient deficiency is leaky due to autophagy and other scavenging mechanisms, and therefore the cells are less synchronous and do not enter S-phase as uniformly as do the rapamycin-treated cells. Collectively, the data in Fig. 2 demonstrate that the site in G1 where cells arrest in response to lipid deprivation is between the Gln checkpoint and the mTOR checkpoint (Fig. 2F).
Effect of Lipid Deprivation on Cancer Cell Proliferation and Cell Cycle Progression
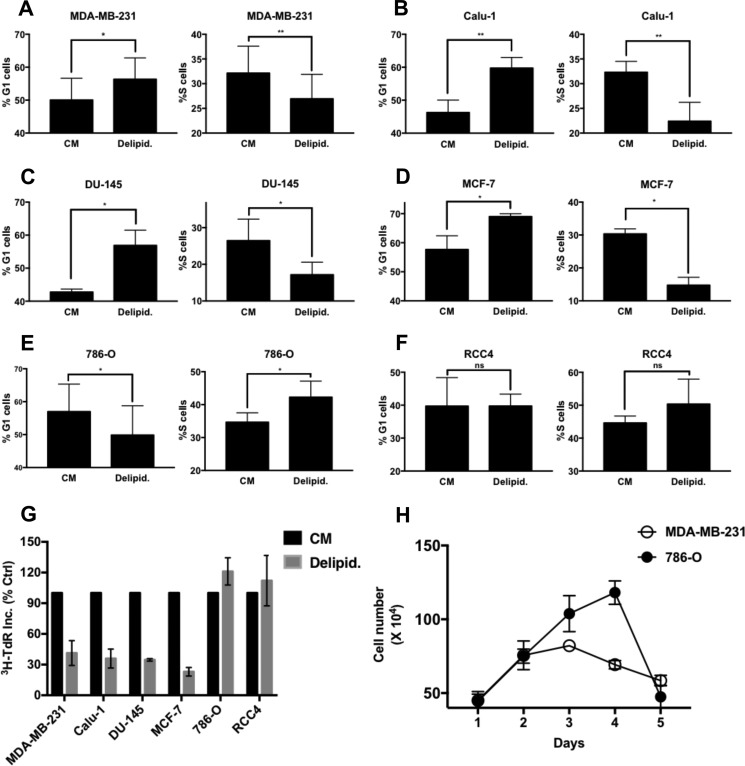
We previously reported that cancer cells harboring Ras mutations bypassed the Gln checkpoint and arrested in S-phase instead (6, 11). We therefore examined the impact of lipid deprivation on several human cancer cell lines to determine whether there were similar bypasses of the G1 lipid checkpoint. We examined the impact of lipid deprivation on MDA-MB-231 and MCF7 breast, Calu1 lung, DU-145 prostate, and 786-O and RCC4 renal cancer cell lines. As shown in Fig. 3, MDA-MB-231 breast (Fig. 3A), Calu1 lung (Fig. 3B), DU-145 prostate (Fig. 3C), and MCF7 breast (Fig. 3D) cancer cells all displayed an increase in G1 DNA content and a reduction in S-phase DNA content in response to lipid deprivation, indicating G1 cell cycle arrest. However, both 786-O (Fig. 3E) and RCC4 (Fig. 3F) renal cancer cell lines did not arrest in G1, as indicated by reduced levels of cells with G1 DNA content. Although there tended to be somewhat higher levels of cells with S-phase content with lipid deprivation in the renal cancer cell lines (Fig. 3, E and F), the cells were not arrested in S-phase, as indicated by continued DNA synthesis and uptake of [3H]TdR in the 786-O and RCC4 cells. In addition, 786-O cells continued proliferation, leading to increased cell number in delipidated serum, whereas MDA-MB-231 cells ceased proliferation in delipidated serum (Fig. 3H). However, after 4 days, the 786-O cells started to die. Thus, unlike the S-phase arrest observed with Gln deprivation in Ras-driven cancer cell lines (6, 11), the renal cancer cells did not arrest in S-phase in response to lipid deprivation.
FIGURE 3.
Effect of lipid deprivation on cancer cell proliferation and cell cycle progression. A–F, MDA-MB-231, Calu-1, DU-145, MCF-7, 786-O, and RCC4 cells were plated at 30% confluence in CM in 10-cm plates. After 24 h, cells were shifted to CM or medium containing delipidated serum (Delipid.) for 48 h, at which time the cells were harvested, fixed, stained with propidium iodide, and analyzed for distribution in G1 and S-phase of the cell cycle by measuring DNA content/cell as described under “Experimental Procedures.” The percentage of G1 and S-phase cells in CM relative to that in delipidated serum is presented. Error bars represent the standard deviation from experiments repeated 3 times. G, the cells were plated in CM in 12-well plates and treated with conditions as explained in A. Cells were labeled with [3H]TdR for the final 24 h of treatment, after which the cells were collected and the incorporated label (3H-TdR Inc.) was determined by scintillation counting. Values were normalized to the cpm for CM, which was given a value of 100%. Ctrl, control; Error bars represent the standard deviation for experiments repeated 2 times. H, the proliferation of 786-O and MDA-MB-231 cells in delipidated serum was determined by counting the number cells over 5 days. Error bars represent the standard deviation for experiments repeated 2 times. Where indicated, paired t tests were performed using GraphPad Prism software (not significant (ns), p > 0.05; *, p < 0.05; **, p < 0.01).
Effect of Prolonged Lipid Deprivation on Clear Cell Renal Cancer Cells
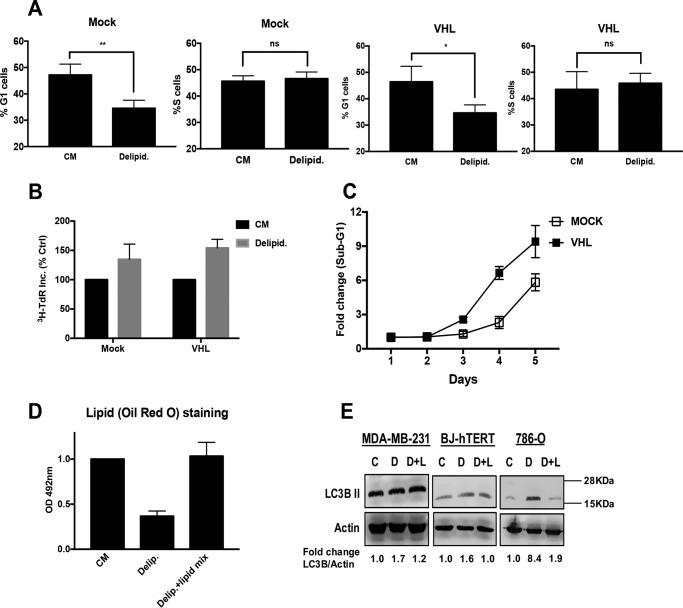
Both of the renal cancer cell lines used have a defect in the von Hippel-Lindau (VHL) tumor suppressor protein, which leads to the stabilization of hypoxia-inducible factor (HIF)1α and HIF2α (RCC4) or just HIF2α (786-O). We therefore examined the impact of lipid deprivation on cell cycle progression in 786-O with restored VHL expression. We found that 786-O cells with restored VHL expression were not arrested in G1 (Fig. 4A). Consistent with this observation, 786-O cells with restored VHL expression still incorporated [3H]TdR under conditions of lipid deprivation (Fig. 4B). Thus, the mechanism of bypass apparently does not depend on loss of VHL and stabilized HIF2α. However, we did notice a difference between the 786-O and the 786-O cells with restored VHL expression in response to lipid deprivation. As was shown in Fig. 3H, 786-O cells began to die after 4 days in delipidated serum. However, the cells with restored VHL died sooner than the parental 786-O cells, as indicated by an increase in sub-G1 DNA content in the 786-O-VHL cells earlier than in the paren-tal 786-O cells (Fig. 4C). A hallmark of clear cell renal carcinoma cells is a high concentration of lipid droplets that is dependent upon the loss of VHL that gives these cells the clear cell phenotype (12). Thus, it is possible that the basis for the lack of arrest is the utilization of the high lipid content of the HIF2α-dependent accumulation of lipid droplets in 786-O cells. To examine the effect of proliferation in delipidated serum on the level of lipid droplets, we quantified the level of lipids by staining the 786-O cells with Oil Red O stain followed by extraction and then testing for absorbance at 492 nm to quantify the level of lipids in cells maintained for 48 h in complete medium, medium with delipidated serum, and delipidated serum and the lipid mix. As shown in Fig. 4D, the cells maintained in delipidated serum for 48 h had a 65% reduction in lipid content. Because lipid metabolism is regulated by autophagy (13), we examined the ability of lipid deprivation to induce expression of the autophagy marker LC3B II. MDA-MB-231 cells, BJ-hTERT, and 786-O cells were maintained in complete medium, delipidated medium, or delipidated medium plus the lipid mix for 48 h, at which time the levels of LC3B II were determined by Western blotting analysis. As shown in Fig. 4E, only the 786-O cells showed an increase in LC3B II expression upon lipid deprivation. Collectively, the data in Fig. 4 demonstrate that restoring VHL in 786-O cells does not reestablish a G1 arrest upon lipid deprivation. In addition, treatment of BJ-hTERT cells with CoCl2, which mimics hypoxia and elevates HIFα levels (14), had no effect on the ability of these cells to arrest in G1 upon lipid deprivation (data not shown). Although these data indicate that VHL loss is not responsible for overriding the late G1 lipid checkpoint, the data do suggest that the high lipid content in the 786-O cells prolongs survival of cells that continue to proliferate when deprived of lipids.
FIGURE 4.
Renal cancer cells utilize lipid droplets to progress through the cell cycle. A, 786-O-VHL and 786-O-Mock cells were plated in DMEM containing 10% FBS (CM). After 24 h, cells were shifted to CM or medium containing 10% delipidated serum (Delipid.) for 48 h. The cells were then harvested, fixed, stained with propidium iodide, and analyzed for distribution in G1 and S-phase of the cell cycle by measuring DNA content/cell as described in the legend for Fig. 1. Error bars represent the standard deviation from experiments performed 2 times (not significant (ns), p > 0.05; *, p < 0.05; **, p < 0.01). B, 786-O Mock and VHL cells were plated in 12-well plates at 30% confluence in CM. After 24 h, cells were shifted to CM or medium containing delipidated serum for 48 h. Cells were labeled with [3H]TdR for the final 24 h of treatment, after which the cells were collected and the incorporated label (3H-TdR Inc.) was determined by scintillation counting. Values were normalized to the cpm for CM, which was given a value of 100%. Error bars represent the standard deviation for experiments repeated 3 times. Ctrl, control. C, 786-O-Mock and 786-O-VHL cells were plated in CM as above. After 24 h, cells were shifted to medium containing 10% delipidated serum. At the indicated time points, cells were harvested, fixed, stained with propidium iodide, and analyzed for sub-genomic DNA content/cell as described under “Experimental Procedures” at the indicated times. Error bars represent the standard deviation from experiments performed 2 times. D, 786-O cells were plated at 40% confluence in RPMI 1640 medium containing 10% FBS for 24 h. Cells were then shifted to CM, medium containing delipidated serum, or medium containing delipidated serum with lipid mix added. After 48 h, cells were stained with Oil Red O as described under “Experimental Procedures.” OD, optical density. Error bars represent the standard deviation from experiments performed 3 times. Paired t tests were performed as in Fig. 3. E, MDA-MB-231, BJ-hTERT and 786-O cells were plated and treated as explained earlier. Cells were harvested after 48 h of treatment. The levels of LC3B II and actin were determined by Western blotting analysis. The data shown are representative of experiments repeated at least 2 times (C = Complete medium, D = delipidated serum, D+L = delipidated serum with lipid mix). -Fold change of LC3B II and actin was measured using Image Studio Lite software.
Loss of PTEN Promotes Passage of the Late G1 Lipid Checkpoint
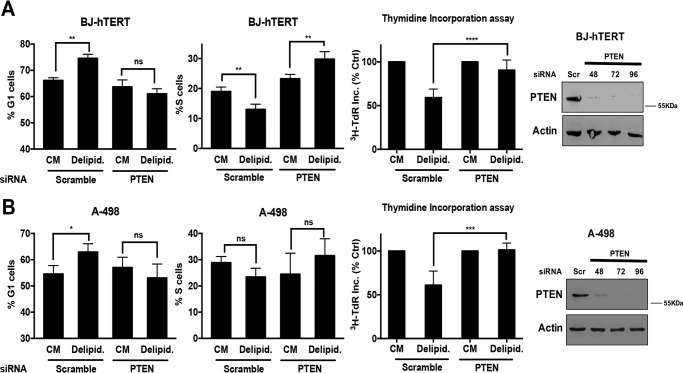
In addition to being defective for VHL, the 786-O renal cancer cells are also defective for PTEN (15), which is common in renal cancer (16). We therefore investigated the effect of PTEN suppression on the impact of lipid deprivation on G1 cell cycle progression. We first tried to restore PTEN expression in the 786-O cells; however, restoring PTEN expression in these cells resulted in substantial loss of cell viability (data not shown). This was not surprising in that PTEN loss leads to activated mTORC1, which has been widely associated with cancer cell survival (17). We next examined the effect of suppressing PTEN expression on G1 cell cycle progression when BJ-hTERT cells were deprived of lipids. BJ-hTERT cells were treated with either scrambled or PTEN siRNA, and then cell cycle status was evaluated in the presence of serum and delipidated serum as in Fig. 1. As shown in Fig. 5A, delipidated serum increased the number of cells in G1 in cells transfected with scrambled siRNA in G1, but not in cells transfected with PTEN siRNA. The BJ-hTERT cells with suppressed PTEN expression now failed to arrest in G1 in response to lipid deprivation. Lipid deprivation also suppressed [3H]TdR incorporation in the BJ-hTERT cells, but not in cells with suppressed PTEN expression (Fig. 5A). We also examined the effect of lipid deprivation on A-498 renal cancer cells that express PTEN (18). As shown in Fig. 5B, lipid deprivation led to an increase in G1 DNA content cells and a decrease in S-phase content cells. However, if PTEN expression was suppressed, the population of G1 cells dropped, whereas the population of S-phase cells increased. Similarly, lipid deprivation suppressed [3H]TdR incorporation in the A-498 cells, but not in the A-498 cells, with suppressed PTEN expression. These data demonstrate that loss of PTEN contributes to bypass of the lipid checkpoint in renal cancer cells.
FIGURE 5.
Loss of PTEN promotes passage of the late G1 lipid checkpoint. A, BJ-hTERT cells were plated at 60% confluence in 6-well plates in CM. After 24 h, cells were transfected with either scrambled or PTEN siRNA. 24 h later, cells were transferred to CM or medium containing 10% delipidated serum (Delipid.) for 48 h. Cells were then collected and cell cycle distribution was determined as in Figs. 3 and 4. Error bars represent the standard deviation for experiments repeated 3 times. For thymidine incorporation assays, cells were labeled with [3H]TdR for the final 24 h of treatment, after which the cells were collected and the incorporated label (3H-TdR Inc.) was determined by scintillation counting. Ctrl, control. Values were normalized to the cpm for CM, which was given a value of 100%. Error bars represent the standard deviation for experiments repeated 4 times. Western blotting was also performed to check the levels of PTEN. The data shown are representative of experiments repeated 2 times. B, A-498 cells were plated at 60% confluence in 6-well plates in CM. After 24 h, cells were transfected with either scrambled or PTEN siRNA. 24 h later, cells were transferred to CM or medium containing 10% delipidated serum for 48 h. Cells were then collected, and cell cycle distribution was determined as in A. Error bars represent the standard deviation for experiments repeated 4 times. Thymidine incorporation assays were performed as in A. Error bars represent the standard deviation for experiments repeated 3 times. Paired t tests were performed as in Figs. 3 and 4. Western blotting analysis of PTEN levels was performed as in A. The data shown are representative of experiments repeated 2 times (not significant (ns), p > 0.05; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001).
Discussion
In this study, we have characterized and mapped a distinct G1 cell cycle checkpoint that is dependent on lipids. This checkpoint is in late G1 and appears to be grouped with two other metabolic checkpoints that monitor the presence of EAA and Gln. As shown in Fig. 2F, the lipid checkpoint maps between the Gln checkpoint and the checkpoint mediated by mTOR. We are proposing that this collection of distinguishable late G1 metabolic checkpoints represents the evolutionary equivalent of START of the yeast cell cycle (8, 19).
The metabolic checkpoints are flanked by growth factor-dependent checkpoints: the restriction point (2, 5) and the mTOR-dependent checkpoint (7, 20, 21). Both of these growth factor-dependent checkpoints have been referred to as a restriction point (1, 5, 7), but they are clearly distinguishable (6). A significant difference between the two growth factor-dependent checkpoints is the point from which cells enter the cell cycle. If cells enter G1 phase after transiting through mitosis, growth factors are required to get past the first restriction point, which is about 3.5 h after mitosis (4). However, if the cells start from quiescence/G0, then in addition to growth factors, such as platelet-derived growth factor, to exit G0, additional growth factors such as insulin-like growth factor are required for progression through late G1 (22). For actively dividing cells entering G1 from mitosis, there is no apparent need for growth factors in late G1. However, rapamycin will arrest cells that are coming from mitosis (6, 20), indicating that there is a mechanism for activating mTOR that does not need growth factors present.
mTOR has been referred to as an integrator of both growth factor and nutrient signals and a controller of cell growth (21, 23). mTOR has for a long time been known as a sensor of amino acids (24), and much has been learned recently about the mechanism for amino acid sensing by mTOR (25). It has also been proposed that mTOR can respond to both glucose and fatty acids via the de novo biosynthesis of phosphatidic acid (26, 27), a metabolite that is critical for the stability of the mTOR complexes mTORC1 and mTORC2 (28). It has also been reported that mTOR can be activated in a manner that is dependent on lysophosphatidic acid acyltransferase (29), a key enzyme in the de novo biosynthesis of phosphatidic acid. Phosphatidic acid is also a product of glycolysis and therefore can be an indicator of glucose sufficiency. Gln is commonly converted into fatty acids that can be incorporated into phosphatidic acid (30), and thus, phosphatidic acid can also be an indicator of sufficient Gln. Thus, there are several mechanisms by which mTOR can respond to the same nutrients that are recognized as additional metabolic checkpoints that are distinguishable from the mTOR checkpoint, which also needs growth factor input as well. The question that emerges from these observations is: why is there an apparent redundancy of nutrient-sensing mechanisms? The finding reported here that there is a distinct mechanism for sensing lipids compliments the ability to sense EAA and Gln, all of which are critical sources of carbon and nitrogen needed for cell growth. Clearly, the cell needs to monitor the presence of nutrients carefully before committing to replicating the genome and doubling its mass. The redundant mechanisms for nutrient sensing underscore the importance of this cellular function.
In our previous studies characterizing the Gln checkpoint in cancer cells, we made the discovery that Ras-driven cancer cells bypassed this late G1 checkpoint and instead arrested in S-phase (6) due to a need for Gln in nucleotide biosynthesis (31). Of significance, this created a synthetic lethality for rapamycin (11), which preferentially induces apoptosis in cells arrested in S-phase (32, 33). In this study, we identified two renal cancer cell lines that bypass the lipid checkpoint and continue to divide. Importantly, the ability of a class of cancer cells that bypass the lipid checkpoint helps validate the existence of the late G1 lipid checkpoint; it is the exception that validates that the lipid checkpoint is real.
Two common genetic alterations in renal cancers are the loss of both VHL and PTEN expression (16). The renal cancer cells used in this study lacked expression of VHL, a ubiquitin ligase for HIFα (34). HIFα is a transcription factor that promotes a metabolic transformation that causes lipid droplet accumulation in renal cancer cells that gives them a clear cell phenotype (35). Restoration of the VHL gene did not restore sensitivity to lipid deprivation. However, it diminished the amount of lipid in the cells and shortened the number of cell divisions before cell death in response to lipid deprivation. In contrast, suppression of PTEN in either BJ-hTERT fibroblasts or A-498 renal cancer cells with wild type PTEN became insensitive to lipid deprivation. Both 786-O and RCC4 renal cancer cells, which bypass the lipid checkpoint, have suppressed levels of PTEN expression (18). Thus, it appears that it is the loss of PTEN that is mostly responsible for the override of the lipid checkpoint we have identified in this work. The override of this checkpoint in renal cancer cells may also provide new opportunities for therapeutic intervention in clear cell renal carcinoma, in that these cells continue to proliferate under lipid-deprived conditions until they die.
Experimental Procedures
Cells and Cell Culture Conditions
The human cancer cell lines BJ-hTERT, MDA-MB-231, MCF-7, Calu-1, DU-145, 786-O, RCC4, and A-498 cells were obtained from the American Tissue Type Culture Collection. The 786-O-VHL and 786-O-Mock cells were generated as described previously (36). All the cells mentioned above except 786-O and RCC4 were maintained in DMEM (Sigma D6429) supplemented with 10% fetal bovine serum (Sigma F4135). 786-O cells were maintained in RPMI 1640 (Sigma R8758) supplemented with 10% FBS.
Materials
Reagents were obtained from the following sources. Antibodies against LC-3B (2775) and PTEN (9188) were obtained from Cell Signaling Technology; the antibody against actin (60008-1) was obtained from Proteintech; anti-mouse- and anti-rabbit HRP-conjugated secondary antibodies were obtained from Promega. For the EAA deprivation, DMEM lacking Arg, Leu, and Lys (D9443) and dialyzed FBS (F0392) were obtained from Sigma. For the Gln deprivation, DMEM lacking Gln (D5546) and dialyzed FBS were obtained from Sigma. For the lipid deprivation, delipidated fetal bovine serum (900-123) was obtained from Gemini Bio Products, fatty acid mixture (11905) was obtained from Invitrogen, and fatty acid-free bovine serum albumin (BP9704) was obtained from Fisher Scientific. Rapamycin (R-5000) was obtained from LC Laboratories. Ultima Gold scintillation fluid (6013681) and [3H]TdR (20 Ci/mmol, 1 mCi/ml) (NET-027E) were obtained from PerkinElmer. Negative control scrambled siRNA (D-001206-13-05), siRNAs targeted against PTEN (M-003023-02-0005), were obtained from Dharmacon.
Flow Cytometric Analysis
Cell cycle distribution was determined by flow cytometry as described previously (6). Briefly, cells were fixed in 70% ethanol, stained using propidium iodide, and passed through 70-μm meshes to remove cell aggregates. Fluorescence intensity corresponding to DNA content in different phase of cell cycle was measured by flow cytometry (FACSCalibur; Becton Dickinson), and analyzed using WinCycle software (Phoenix Flow Systems).
Western Blotting Analysis
Proteins were extracted from cultured cells in M-PER (Thermo Scientific 78501). Equal amounts of proteins were subjected to SDS-PAGE on polyacrylamide separating gels. Electrophoresed proteins were then transferred to nitrocellulose membrane. After transfer, membranes were blocked in an isotonic solution containing 5% nonfat dry milk in phosphate-buffered saline. Membranes were then incubated with primary antibodies as described in the text. The dilutions were used as per vendor's instructions. Depending on the origin of the primary antibody, either anti-mouse- or anti-rabbit HRP-conjugated IgG was used for detection using the ECL system (Thermo Scientific 34080).
Thymidine Incorporation Assay
Cells were labeled with 1 μCi/ml [3H]TdR. At the indicated times, cells were washed twice with 1 ml of phosphate-buffered saline, and then precipitated twice with 1 ml of 10% trichloroacetic acid. The precipitates were solubilized in 0.5 ml of 0.5% SDS, 0.5 M NaOH solution, and the extent of TdR incorporation was quantified using 75 μl of sample and 3 ml of scintillation fluid. Each experiment was performed two times.
Lipid (Oil Red O) Staining
Lipid (Oil Red O) staining kit (MAK194) was purchased from Sigma. The staining was conducted as per the vendor's protocol. To measure quantitatively, stained cells were washed three times with 60% isopropyl alcohol for 5 min each time with gentle rocking. Oil Red O stain was extracted with 100% isopropyl alcohol for 5 min with gentle rocking. Absorbance at 492 nm was measured for the collected samples, and 100% isopropyl alcohol was used as background to subtract the background signal.
Transient Transfections
Cells were plated in 6-well plates in medium containing 10% FBS overnight. Transfections with siRNAs (100 nm) in Lipofectamine RNAiMAX were then performed according to vendor instructions. After 6 h, reagents were replaced with fresh 10% FBS and cells were allowed to incubate for the indicated times as described under “Results.”
Lipid Mix Supplementation
Fatty acid mix was obtained from Invitrogen (11905) and supplied to cells as a 1:200 dilution complexed with 10% BSA (Sigma) in a 2:1 ratio for the final concentration of lipids in the medium of 0.375 mg/liter. The exact composition of the fatty acid mixture is provided in supplemental Table S1.
Author Contributions
D. P., D. S., A. C., M. S., and D. A. F. conceptualized the study; D. P., M. S., and D. A. F. developed the methodology; D. P. and V. M. performed the investigations; and D. P., M. O., and D. A. F. wrote the manuscript.
Supplementary Material
Acknowledgments
Research Centers in Minority Institutions Award RR-03039 from the National Center for Research Resources of the National Institutes of Health, which supports infrastructure and instrumentation in the Biological Sciences Department at Hunter College, is acknowledged.
This work was supported by NCI, National Institutes of Health Grants R01-CA046677 and R01-CA179542 (to D. A. F). This work was also supported by Canadian Institutes of Health Research Grant MOP-77718 (to M. O.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Table S1.
- EAA
- essential amino acid(s)
- HIF
- hypoxia-inducible factor
- mTOR
- mammalian target of rapamycin
- TdR
- thymine deoxyribose
- VHL
- von Hippel-Lindau
- PTEN
- phosphatase and tensin homologue
- CM
- complete medium.
References
- 1. Foster D. A., Yellen P., Xu L., and Saqcena M. (2010) Regulation of G1 cell cycle progression: distinguishing the restriction point from a nutrient-sensing cell growth checkpoint(s). Genes Cancer 1, 1124–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pardee A. B. (1974) A restriction point for control of normal animal cell proliferation. Proc. Natl. Acad. Sci. U.S.A. 71, 1286–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blagosklonny M. V., and Pardee A. B. (2002) The restriction point of the cell cycle. Cell Cycle 1, 103–110 [PubMed] [Google Scholar]
- 4. Zetterberg A., and Larsson O. (1985) Kinetic analysis of regulatory events in G1 leading to proliferation or quiescence of Swiss 3T3 cells. Proc. Natl. Acad. Sci. U.S.A. 82, 5365–5369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zetterberg A., Larsson O., and Wiman K. G. (1995) What is the restriction point? Curr. Opin. Cell Biol. 7, 835–842 [DOI] [PubMed] [Google Scholar]
- 6. Saqcena M., Menon D., Patel D., Mukhopadhyay S., Chow V., and Foster D. A. (2013) Amino acids and mTOR mediate distinct metabolic checkpoints in mammalian G1 cell cycle. PLoS ONE 8, e74157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Planas-Silva M. D., and Weinberg R. A. (1997) The restriction point and control of cell proliferation. Curr. Opin. Cell Biol. 9, 768–772 [DOI] [PubMed] [Google Scholar]
- 8. Hartwell L. H., Culotti J., Pringle J. R., and Reid B. J. (1974) Genetic control of the cell division cycle in yeast. Science 183, 46–51 [DOI] [PubMed] [Google Scholar]
- 9. Kamphorst J. J., Cross J. R., Fan J., de Stanchina E., Mathew R., White E. P., Thompson C. B., and Rabinowitz J. D. (2013) Hypoxic and Ras-transformed cells support growth by scavenging unsaturated fatty acids from lysophospholipids. Proc. Natl. Acad. Sci. U.S.A. 110, 8882–8887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Salloum D., Mukhopadhyay S., Tung K., Polonetskaya A., and Foster D. A. (2014) Mutant Ras elevates dependence on serum lipids and creates a synthetic lethality for rapamycin. Mol. Cancer Ther. 13, 733–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Saqcena M., Mukhopadhyay S., Hosny C., Alhamed A., Chatterjee A., and Foster D. A. (2015) Blocking anaplerotic entry of glutamine into the TCA cycle sensitizes K-Ras mutant cancer cells to cytotoxic drugs. Oncogene 34, 2672–2680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Qiu B., Ackerman D., Sanchez D. J., Li B., Ochocki J. D., Grazioli A., Bobrovnikova-Marjon E., Diehl J. A., Keith B., and Simon M. C. (2015) HIF2α-dependent lipid storage promotes endoplasmic reticulum homeostasis in clear-cell renal cell carcinoma. Cancer Discov. 5, 652–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M., Tanaka K., Cuervo A. M., and Czaja M. J. (2009) Autophagy regulates lipid metabolism. Nature 458, 1131–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Epstein A. C., Gleadle J. M., McNeill L. A., Hewitson K. S., O'Rourke J., Mole D. R., Mukherji M., Metzen E., Wilson M. I., Dhanda A., Tian Y. M., Masson N., Hamilton D. L., Jaakkola P., Barstead R., et al. (2001) C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107, 43–54 [DOI] [PubMed] [Google Scholar]
- 15. Schneider E., Keppler R., Prawitt D., Steinwender C., Roos F. C., Thüroff J. W., Lausch E., and Brenner W. (2011) Migration of renal tumor cells depends on dephosphorylation of Shc by PTEN. Int. J. Oncol. 38, 823–831 [DOI] [PubMed] [Google Scholar]
- 16. Brenner W., Färber G., Herget T., Lehr H. A., Hengstler J. G., and Thüroff J. W. (2002) Loss of tumor suppressor protein PTEN during renal carcinogenesis. Int. J. Cancer 99, 53–57 [DOI] [PubMed] [Google Scholar]
- 17. Song M. S., Salmena L., and Pandolfi P. P. (2012) The functions and regulation of the PTEN tumour suppressor. Nat. Rev. Mol. Cell Biol. 13, 283–296 [DOI] [PubMed] [Google Scholar]
- 18. Kearney A. Y., Fan Y. H., Giri U., Saigal B., Gandhi V., Heymach J. V., and Zurita A. J. (2015) 8-Chloroadenosine sensitivity in renal cell carcinoma is associated with AMPK activation and mTOR pathway inhibition. PLoS ONE 10, e0135962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Johnson A., and Skotheim J. M. (2013) Start and the restriction point. Curr. Opin. Cell Biol. 25, 717–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chatterjee A., Mukhopadhyay S., Tung K., Patel D., and Foster D. A. (2015) Rapamycin-induced G1 cell cycle arrest employs both TGF-β and Rb pathways. Cancer Lett. 360, 134–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fingar D. C., and Blenis J. (2004) Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene 23, 3151–3171 [DOI] [PubMed] [Google Scholar]
- 22. Stiles C. D., Capone G. T., Scher C. D., Antoniades H. N., Van Wyk J. J., and Pledger W. J. (1979) Dual control of cell growth by somatomedins and platelet-derived growth factor. Proc. Natl. Acad. Sci. U.S.A. 76, 1279–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Laplante M., and Sabatini D. M. (2012) mTOR signaling in growth control and disease. Cell 149, 274–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Avruch J., Long X., Ortiz-Vega S., Rapley J., Papageorgiou A., and Dai N. (2009) Amino acid regulation of TOR complex 1. Am. J. Physiol. Endocrinol. Metab. 296, E592–E602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Efeyan A., Zoncu R., and Sabatini D. M. (2012) Amino acids and mTORC1: from lysosomes to disease. Trends Mol. Med. 18, 524–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Foster D. A. (2013) Phosphatidic acid and lipid-sensing by mTOR. Trends Endocrinol. Metab. 24, 272–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Foster D. A., Salloum D., Menon D., and Frias M. A. (2014) Phospholipase D and the maintenance of phosphatidic acid levels for regulation of mammalian target of rapamycin (mTOR). J. Biol. Chem. 289, 22583–22588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Toschi A., Lee E., Xu L., Garcia A., Gadir N., and Foster D. A. (2009) Regulation of mTORC1 and mTORC2 complex assembly by phosphatidic acid: competition with rapamycin. Mol. Cell. Biol. 29, 1411–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Blaskovich M. A., Yendluri V., Lawrence H. R., Lawrence N. J., Sebti S. M., and Springett G. M. (2013) Lysophosphatidic acid acyltransferase β regulates mTOR signaling. PLoS ONE 8, e78632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. DeBerardinis R. J., Mancuso A., Daikhin E., Nissim I., Yudkoff M., Wehrli S., and Thompson C. B. (2007) Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl. Acad. Sci. U.S.A. 104, 19345–19350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Patel D., Menon D., Bernfeld E., Mroz V., Kalan S., Loayza D., and Foster D. A. (2016) Aspartate rescues S-phase arrest caused by suppression of glutamine utilization in KRas-driven cancer cells. J. Biol. Chem. 291, 9322–9329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Saqcena M., Patel D., Menon D., Mukhopadhyay S., and Foster D. A. (2015) Apoptotic effects of high-dose rapamycin occur in S-phase of the cell cycle. Cell Cycle 14, 2285–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gan W., Liu P., and Wei W. (2015) Cell cycle status dictates effectiveness of rapamycin. Cell Cycle 1–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Robinson C. M., and Ohh M. (2014) The multifaceted von Hippel-Lindau tumour suppressor protein. FEBS Lett. 588, 2704–2711 [DOI] [PubMed] [Google Scholar]
- 35. Schönenberger D., Harlander S., Rajski M., Jacobs R. A., Lundby A. K., Adlesic M., Hejhal T., Wild P. J., Lundby C., and Frew I. J. (2016) Formation of renal cysts and tumors in Vhl/Trp53-deficient mice requires HIF1α and HIF2α. Cancer Res. 76, 2025–2036 [DOI] [PubMed] [Google Scholar]
- 36. Stickle N. H., Chung J., Klco J. M., Hill R. P., Kaelin W. G. Jr, and Ohh M. (2004) pVHL modification by NEDD8 is required for fibronectin matrix assembly and suppression of tumor development. Mol. Cell. Biol. 24, 3251–3261 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.