Abstract
Without ribosome biogenesis, translation of mRNA into protein ceases and cellular growth stops. We asked whether ribosome biogenesis is cell cycle regulated in the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe, and we determined that it is not regulated in the same manner as in metazoan cells. We therefore turned our attention to cellular sensors that relay cell size information via ribosome biogenesis. Our results indicate that the small subunit (SSU) processome, a complex consisting of 40 proteins and the U3 small nucleolar RNA necessary for ribosome biogenesis, is not mitotically regulated. Furthermore, Nan1/Utp17, an SSU processome protein, does not provide a link between ribosome biogenesis and cell growth. However, when individual SSU processome proteins are depleted, cells arrest in the G1 phase of the cell cycle. This arrest was further supported by the lack of staining for proteins expressed in post-G1. Similarly, synchronized cells depleted of SSU processome proteins did not enter G2. This suggests that when ribosomes are no longer made, the cells stall in the G1. Therefore, yeast cells must grow to a critical size, which is dependent upon having a sufficient number of ribosomes during the G1 phase of the cell cycle, before cell division can occur.
INTRODUCTION
Ribosomes are essential for the translation of mRNA into protein and are necessary for cell growth. Synthesis of the small and large ribosomal subunits begins with the transcription of rDNA into a single 35S pre-rRNA transcript within the cell nucleolus. In yeast, the 35S pre-rRNA is processed into the 5.8S, 18S, and 25S rRNAs. Pre-rRNA processing occurs through distinct pathways at 10 known processing sites by many trans-acting factors, including small nucleolar ribonucleoproteins (snoRNPs; Lafontaine and Tollervey, 1995; Kressler et al., 1999; Gerbi et al., 2001, 2003). The 18S rRNA is incorporated into the small ribosomal subunit, whereas the 5.8S and 25S rRNAs are incorporated into the large ribosomal subunit. The two ribosomal subunits are transported from the nucleolus through the nucleoplasm to the cytoplasm where they are assembled into a fully functional ribosome.
The cellular sensors that coordinate ribosome biogenesis and cell division are largely unknown (Jorgensen et al., 2002). Cell cycle regulation of ribosome biogenesis was first observed in metazoan cells, where several laboratories reported a lack of RNA synthesis in mitotic cells (Taylor, 1960; Prescott and Bender, 1962). Since then, others have shown that both pre-rRNA transcription and processing cease during mitosis in mammalian cells (Gautier et al., 1994; Dundr and Olson, 1998; Sirri et al., 2000). When mitotic transcription of the pre-rRNA is forced by inhibition of cdc2-cyclin B kinase, defects in processing of the pre-rRNA are observed (Sirri et al., 2000). These results indicate that both transcription and processing of the pre-rRNA are inhibited during mitosis in mammalian cells.
Recently, ribosome biogenesis has been linked to the cell cycle through studies of tumorigenesis (Ruggero and Pandolfi, 2003). Changing the dynamics of protein synthesis can promote cell transformation and cancer progression. In mammalian cells, there is evidence that links the tumor suppressor gene p53 to proteins required for ribosome biogenesis. For example, cell cycle arrest caused by deletion of Bop1, a protein required for large subunit biogenesis and cell cycle progression at G1, can be restored by inactivation of p53 (Strezoska et al., 2000; Pestov et al., 2001a,b; Strezoska et al., 2002). Links between ribosome biogenesis and the p53 pathway also have been observed by others (David-Pfeuty et al., 2001). Many of the proteins required for ribosome biogenesis have conserved orthologues, suggesting that these processes occur through similar mechanisms in all eukaryotes.
During the mid-to-late 1970s, several studies aimed to determine whether ribosome biogenesis in yeast is cell cycle regulated in the same manner as in metazoan cells (Shulman et al., 1973; Wain and Staatz, 1973; Sogin et al., 1974; Elliot and McLaughlin, 1979). Collectively, these studies revealed that the rate of synthesis of the mature rRNAs remained constant throughout the cell cycle, suggesting that ribosome biogenesis is not cell cycle regulated in yeast. It remains unknown whether pre-rRNA processing defects occur during mitosis.
Contrary to earlier findings, a few examples of proteins linking ribosome biogenesis and the cell cycle in the yeast Saccharomyces cerevisiae have recently been identified. Depletion of the nucleolar yeast protein Nop15 leads to defects in both cytokinesis and processing of the 5.8S and 25S prerRNAs (Oeffinger and Tollervey, 2003). Similarly, when Sda1, a protein required for large subunit biogenesis, and Swe1, a protein required for cell cycle progression at G2/M, are both depleted, cells are unable to reenter the cell cycle at G1 (Saracino et al., 2004). This result is not unexpected because Sda1 protein has previously been shown to be required for cell cycle progression during G1 (Buscemi et al., 2000; Zimmerman and Kellogg, 2001). In addition, a protein required for large ribosomal subunit biogenesis, Nop7/Yph1, is required for normal S-phase progression as part of the origin of replication complex (Du and Stillman, 2002). To date, the role of proteins required for small subunit biogenesis in cell cycle regulation has not been determined. In addition, it is unclear how ribosome biogenesis and the cell cycle are coordinately regulated.
Our laboratory has recently identified a new, large ribonucleoprotein that contains the U3 snoRNA and at least 40 proteins, which is required for 18S rRNA biogenesis (Dragon et al., 2002). This complex has been named the small subunit (SSU) processome because it is required for processing of RNA precursors of 18S, the small subunit rRNA. A similar complex was isolated by Grandi et al. (2002) and termed the 90S pre-ribosome. We sought to determine whether the SSU processome might provide a link between ribosome biogenesis and the cell cycle. One of the SSU processome proteins, Net1-associated nucleolar protein 1 or U3-associated protein 17 (Nan1/Utp17), had originally been suggested to be associated with Net1, a protein required for mitotic exit at G2/M (Shou et al., 1999). Therefore, Nan1/Utp17 was hypothesized to be involved in mitotic exit through this association.
We sought to determine whether ribosome biogenesis is cell cycle regulated in yeast in the same way as metazoan cells. Ribosome biogenesis is a highly regulated process and, in metazoan cells, ceases at the G2/M phase of the cell cycle. Our results indicate that ribosome biogenesis is not cell cycle regulated in either S. cerevisiae or Schizosaccharomyces pombe in the same manner as in mammalian cells. These differences can be explained by the different types of mitoses that mammalian and yeast cells undergo. Mammalian cells undergo an open mitosis, where their nuclear envelope breaks down and nucleolus fragments. Yeast undergo a closed mitosis, where their nuclear envelope remains intact throughout the cell cycle. In addition, the SSU processome protein Nan1/Utp17 did not provide a link between these two processes. However, growth in size and ribosome biogenesis are tightly linked in all eukaryotic cells. Recent genomic studies have supported these observations by implicating the depletion of many ribosomal proteins and proteins required for ribosome biogenesis lead to growth in size defects (Jorgensen et al., 2002), whereas overexpression or deletion of these proteins cause cell cycle defects (Kondoh et al., 2000; Pestov et al., 2001a; Tallada et al., 2002). Our results support these observations, because depletion of SSU processome proteins leads to G1 arrest. This suggests that if rRNA cannot be made at G1, then progression through the cell cycle is blocked.
MATERIALS AND METHODS
Yeast Strains and Media
All yeast strains were derived from YPH499 (Mata, ura3-52, lys2-80, ade2-101, trp1-D 63, his3-D 200, leu2-D 1). S. cerevisiae yeast strains were grown in rich medium YPD (1% yeast extract, 2% peptone, 2% glucose) or YPG/R (1% yeast extract, 2% peptone, 2% galactose, 2% raffinose) where specified. S. pombe was grown in YEPD (1% yeast extract, 2.5 mg/l phloxin B [P 4030; Sigma-Aldrich, St. Louis, MO], 2% glucose) and EMM (4110-012; Bio 101, Vista, CA) where specified. Utp1, Utp2, Utp3, Utp4, Utp5, Utp6, Utp7, Utp8, Utp9, Utp10, Utp11, Utp12, Utp13, Utp14, Utp15, Utp16, and Utp17 triple HA-tagged strains and GAL-3xHA-tagged strains were from Dragon et al. (2002). The NET1-3xHA strain was a gift from Danesh Moazed (Straight et al., 1999). Integration of the TAP tag at NAN1 and 3xHA at NET1 in YPH499 were created as described previously (Rigaut et al., 1999). For cell synchronization studies, expression of the relevant proteins was placed under the control of the GAL promoter (GAL::RPS14A, GAL::UTP1, GAL::UTP2, and GAL::UTP4; Longtine et al., 1998). In these strains, deletion of BAR1 (Δbar1), integration of the TAP tag at CLN2 (Rigaut et al., 1999) and integration of the 3xHA tag at CLB2 (Knop et al., 1999) were carried out.
Metabolic Labeling of RNA
S. cerevisiae were grown to early log phase in YPD and shifted to medium minus methionine for 48 h. Nocodazole-treated cultures were incubated with 12.5 μg/ml nocodazole after 44 h of growth in medium lacking methionine for 4 h. Cells were examined by light microscopy to verify cell cycle arrest. Ten milliliters of cells at OD600 0.4–0.5 were then pulsed for 10 min with 50 μCi of [3H]methyl-methionine. RNA was extracted and 10,000 cpm of each sample was analyzed as described previously (Dunbar et al., 1997). S. pombe were grown to early log phase in YEPD and then depleted of methionine for 48 h by growth in EMM medium. Nocodazole-treated cultures were incubated with 12.5 μg/ml nocodazole after 44 h of growth for 4 h in EMM medium. Cells were examined by light microscopy to verify cell cycle arrest. Ten milliliters of cells at OD600 0.4–0.5 were pulsed for 10 min with 50 μCi of [3H]methyl-methionine. RNA was extracted and 10,000 counts per sample were analyzed as described previously (Dunbar et al., 1997).
Protein-Protein Coimmunoprecipitation
Nan1/Net1 coimmunoprecipitation experiments were carried out with Net1-3xHA, Nan1-TAP, YPH499 (parent strain), and Net1-3xHA/Nan1-TAP-tagged strains. Net1 immunoprecipitations were carried out with 200 μl each of anti-hemagglutinin (HA) (12CA5 culture supernatant) with glass bead extracts (Lee and Baserga, 1999) and blotted with anti-PAP antibodies (1:6666) as described previously (Rigaut et al., 1999). Immunoprecipitation of Nan1-TAP was carried out with 20 μl of IgG-Sepharose beads (Amersham Biosciences, Uppsala, Sweden) and blotted with anti-HA (12CA5 culture supernatant) at a dilution of 1:666.
Coimmunoprecipitation of 3xHA-tagged SSU processome proteins (Utp1, Utp2, Utp3, Utp4, Utp5, Utp6, Utp7, Utp8, Utp9, Utp10, Utp11, Utp13, Utp14, Utp15, Utp16, and Utp17) was carried out with 200 μl each of anti-HA (12CA5 culture supernatant) and glass bead extracts. Immunoprecipitated proteins were run on 10% SDS-PAGE gels and Western blotted with anti-Mpp10 antibodies (Lee and Baserga, 1999). The NET1-3xHA strain was obtained from Danesh Moazed (Straight et al., 1999).
Protein Depletion
Twenty milliliters of cells from YPH499, GAL::3xHA-UTP1, GAL::3xHA-UTP2, GAL::3xHA-UTP3, GAL::3xHA-UTP4, GAL::3xHA-UTP5, GAL::3xHA-UTP6, GAL::3xHA-UTP7, GAL::3xHA-UTP8, GAL::3xHA-UTP9, GAL::3xHA-UTP10, GAL::3xHA-UTP11, GAL::3xHA-UTP12, GAL::3xHA-UTP13, GAL::3xHA-UTP14, GAL::3xHA-UTP15, GAL::3xHA-UTP16, and GAL::3xHA-UTP17 were grown to early log phase in YPG/R. Two milliliters of cells at OD600 0.5 were collected and washed with water. Cells were shifted into YPD media for 24 h. At 3, 6, 12, and 24 h of growth in YPD, 2 ml of cells at OD600 0.5 were collected and washed with water. Protein extracts were prepared as described previously (Kushnirov, 2000). Protein was run on 10% SDS-PAGE gels and Western blotted with anti-HA antibodies (Dragon et al., 2002).
Flow Cytometry
Two milliliters of cells from YPH499, GAL::3xHA-RPS14A, GAL::3xHA-UTP1, GAL::3xHA-UTP2, GAL::3xHA-UTP3, GAL::3xHA-UTP4, GAL::3xHA-UTP5, GAL::3xHA-UTP6, GAL::3xHA-UTP7, GAL::3xHA-UTP8, GAL::3xHA-UTP9, GAL::3xHA-UTP10, GAL::3xHA-UTP11, GAL::3xHA-UTP12, GAL::3xHA-UTP13, GAL::3xHA-UTP14, GAL::3xHA-UTP15, GAL::3xHA-UTP16, and GAL::3xHA-UTP17 were collected from early log phase cultures of yeast strains grown in YPG/R and fluorescence-activated cell sorting (FACS) analyzed as described previously (Burton and Solomon, 2000). These cultures were then shifted into YPD for 24 h, and 2 ml of culture was collected from early log phase (Burton and Solomon, 2000). GAL::RPS14A, GAL::UTP1, GAL::UTP2, GAL::UTP4 yeast strains bearing Δbar1, Cln2-TAP, Clb2-3xHA were grown to early log phase in YPG/R liquid media and then shifted into YPD liquid media for 21 h. Cells were synchronized with α-factor (2 μg/ml) for 2.5 h in YPD liquid media. Two milliliters of cells at OD600 0.5 were analyzed by FACS after synchronization with α-factor and after 105 min of release by washing out α-factor (Burton and Solomon, 2000).
Immunofluorescence
The GAL::3xHA-UTP and GAL::3xHA-NET1 strains were used for indirect immunofluorescence as described previously (Dunbar et al., 2000). Rat antitubulin (YOL1/34) (diluted 1:100; Kilmartin et al., 1982) and rabbit anti-Mpp10 polyclonal antibodies (diluted 1:2000; Lee and Baserga, 1997) were detected by tetramethylrhodamine B isothiocyanate (TRITC)-conjugated donkey anti-rat IgG (diluted 1:200) and FITC-conjugated donkey anti-rabbit IgG (diluted 1:200) secondary antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA). Immunofluorescence was carried out on strains where the gene was expressed from a conditional promoter, including UTP18, and NET1 from 4 ml of cells grown to early log phase in YPG/R (undepleted) and after 23 h in YPD (depleted).
α-Factor Synchronization
GAL::RPS14A, GAL::UTP1, GAL::UTP2, GAL::UTP4 yeast strains bearing Δbar1, Cln2-TAP, Clb2–3xHA were grown to early log phase in YPG/R liquid media and then shifted into YPD liquid media for 21 h. Cells were synchronized with α-factor (2 μg/ml) for 2.5 h in YPD liquid media. Cells were pelleted, washed with water, and resuspended into media containing glucose. One milliliter of cells at OD600 0.4–0.5 was pelleted, washed with water, and protein was extracted every 15 min after α-factor release for 105 min. Proteins were run on 10% SDS-PAGE gel for 50 min at 200 V, transferred onto nitrocellulose membrane, and Western blotted for the TAP tag to detect Cln2 (anti-PAP antibodies) or Clb2 (anti-HA antibodies) and exposed to film.
RESULTS
Because it is well known that ribosome biogenesis ceases during mitosis in mammalian cells, we sought to determine whether rRNA is made and processed during mitosis in yeasts. To do this, we pulsed interphase (untreated cells) or mitotically arrested yeast cells with [3H]methyl-methionine and looked for defects in transcription and processing of pre-rRNA (Figure 1). Cell cycle arrest was verified by light microscopy that showed that >90% of the cells were arrested as large-budded cells, indicating a block at G2/M (our unpublished data). Equal amounts of tritiated RNA were resolved on denaturing agarose gels. These results show that mitotically arrested S. cerevisiae cells transcribe and process the nascent 35S pre-rRNA equally well during interphase and mitosis (Figure 1). We also conducted similar experiments in S. pombe, because they are more similar to mammalian cells with respect to their cell cycle. As described above, equal amounts of tritiated RNA were resolved on denaturing agarose gels. We found that they, too, continue to make and process pre-RNA during mitosis (Figure 1). Cell cycle arrest was again verified by light microscopy where 90% of the cells were arrested as large, elongated cells, indicating a block at G2/M (our unpublished data). Together, these results suggest that ribosome biogenesis continues during mitosis in the yeasts S. cerevisiae and S. pombe, unlike in metazoan cells.
Figure 1.
rRNA transcription and processing occurs during mitosis in S. cerevisiae and S. pombe. Cells were grown in media lacking methionine for 48 h. Cells were arrested in mitosis with the drug nocodazole. Both interphase and mitotic cells were pulsed for 10 min with [3H]methyl-methionine, rRNA extracted, and analyzed by gel electrophoresis and blotting.
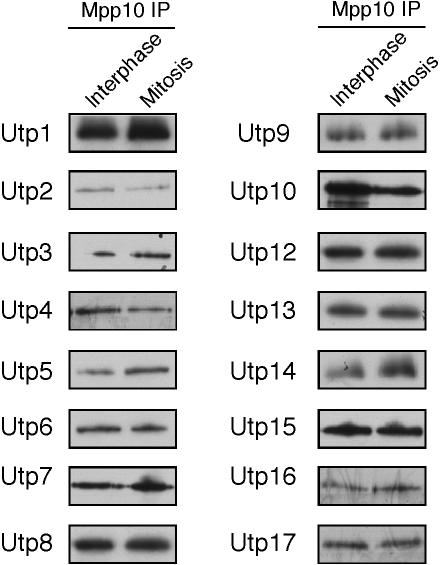
Next, we determined whether synthesis of the SSU processome is itself mitotically regulated in S. cerevisiae by testing whether it was intact during mitosis. This was done by assessing the ability of SSU processome proteins to associate with each other. To assess whether the SSU processome was intact, coimmunoprecipitation of 3xHA-tagged SSU processome proteins with Mpp10 was tested in interphase and mitotically arrested cells. Utp1, Utp2, Utp3, Utp4, Utp5, Utp6, Utp7, Utp8, Utp9, Utp10, Utp12, Utp13, Utp14, Utp15, Utp16, and Utp17 were all able to coimmunoprecipitate Mpp10 during interphase as well as during mitosis (Figure 2). These results suggest that the SSU processome remains intact during mitosis.
Figure 2.
The SSU processome remains intact in mitotic cells. 3xHA-tagged SSU processome proteins Utp1-10, 12-17 were immmunoprecipitated from glass bead extracts made from interphase or mitotically arrested cells, by using beads conjugated with anti-HA antibodies. SSU processome proteins were tested for their ability to coimmunoprecipitate Mpp10 by Western blot with anti-Mpp10 antibody.
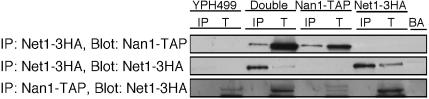
Previous results indicated that one SSU processome component, Utp17/Nan1, might link ribosome biogenesis with the exit from mitosis (Shou et al., 1999). Utp17 was originally named Nan1 for Net1-associated nucleolar protein 1. Utp17/Nan1 was originally purified as a Net1-associated protein through affinity chromatography (Shou et al., 1999). Net1 is a component of the regulator of nucleolar silencing and telophase (RENT) complex, which is important for mitotic exit through its association with Cdc14 and silencing through its association with Sir2 (Shou et al., 1999). Utp17/Nan1 was subsequently purified as a component of the SSU processome, the results were extensively validated, and it was shown to be required for generation of the 18S rRNA (Dragon et al., 2002). YPH499, the untagged parent strain, is used as a control for this and subsequent experiments. We asked whether Utp17/Nan1 was also a true component of the RENT complex by examining whether it could coimmunoprecipitate Net1. We constructed double-tagged strains where Nan1 was tagged with the TAP tag (Rigaut et al., 1999) and Net1 with 3xHA. When Net1–3xHA was immunoprecipitated and then Western blotted for Nan1, the same amount of Nan1 coimmunoprecipitated in the double-tagged strain as in the single Nan1-TAP strain (Figure 3), thus representing background. We have confirmed that Net1–3xHA was enriched by coimmunoprecipitation by blotting with anti-HA antibodies (Figure 3). We obtained the same result by coimmunoprecipitating Nan1-TAP first and then Western blotting for Net1–3xHA (Figure 3). This suggests that Nan1 and Net1 do not coimmunoprecipitate. These results suggest that Nan1 is likely not Net1-associated during interphase and is therefore not a component of the RENT complex, contrary to what has been previously reported.
Figure 3.
Net1 and Nan1 do not coimmunoprecipitate. Nan1/Net1 coimmunoprecipitation experiments were carried out with Net1-3xHA-, Nan1-TAP–, YPH499-, and Net1-3xHA/Nan1-TAP–tagged yeast strains. YPH499 (untagged parent strain), Net1-3xHA, and Nan1-TAP strains were used as controls. Net1-3xHA was immunoprecipitated with beads conjugated to anti-HA antibodies (IP, immunoprecipitation lane), and totals (T, total, representing 5% of the total protein extracted) were Western blotted with PAP antibodies, recognizing protein A. This blot was stripped and reprobed with anti-HA antibodies. Nan1-TAP was immunoprecipitated with IgG beads and Western blotted with anti-HA antibodies. Beads alone (BA) was used as a control.
We determined whether ribosome biogenesis was required for cell cycle progression by asking whether depletion of individual SSU processome proteins would lead to arrest at a particular stage of the cell cycle. Because all but one of the SSU processome proteins are essential, their depletion ultimately affects cell growth. In these strains, the SSU processome proteins are under the control of a galactose-inducible promoter and tagged with a 3xHA tag. In the presence of galactose, the protein is made; however, in glucose, protein levels are depleted. Growth of cells depleted of SSU processome proteins begins to decline after 12 h of depletion (Figure 4A). Growth of cells depleted of Utp16, a nonessential SSU processome component, was only slightly slowed in comparison with the parent strain YPH499, as expected. After 24 h of protein depletion, the SSU processome components were not detectable by Western blot (Figure 4B).
Figure 4.
Depletion of the essential SSU processome proteins slows growth. Strains expressing Utp1-17 from a galactose-inducible/glucose-repressible promoter with a 3xHA tag were grown to early log phase in galactose/raffinose media (undepleted) and then shifted into glucose media (depleted) for 24 h. YPH499 (the parent untagged strain) was used as a control. After 0, 3, 6, 12, and 24 h of depletion, cells were analyzed for growth by OD600 (A) and for protein expression by Western blotting with anti-HA antibodies (B).
The cell cycle profiles of SSU processome-depleted strains were compared with those obtained from control strains YPH499 (parent strain) and GAL::3xHA-RPS14A, after growth in galactose and glucose. Rps14A is a redundant ribosomal protein; that is, depletion of one of the Rps14 proteins (Rps14A or Rps14B) does not appreciably affect cell growth. When the normally expressed Rps14A is depleted, protein levels of Rps14B are increased 10-fold (Paulovich et al., 1993). Up-regulation of Rps14B protein levels occur through posttranslational modification of RPS14B (Li et al., 1995; Fewell and Woolford, 1999). The growth of YPH499 and GAL::3xHA-RPS14A in galactose yields a cell cycle profile indicative of cells primarily in the G1 phase of the cell cycle upon FACS sorting (Figure 5, Undepleted). This is because galactose is a poor carbon source and the cells grow slowly. However, when these strains are shifted to glucose and analyzed by FACS sorting, the cell cycle profile for these two strains shifts to nearly equal G1 and G2 peaks (Figure 5, Depleted). Thus, this is the expected cell cycle distribution result after the switch from galactose to glucose if cells are cycling normally. When SSU processome proteins are not depleted (Utp1-17 grown in galactose) the yeast strains yield cell cycle profiles identical to those obtained by FACS sorting of YPH499 and GAL::3xHA-RPS14A, with a prominent G1 peak (Figure 5, Undepleted). Cells with integrated galactose promoters grew slower in galactose media and had a more prominent G1 peak than YPH499. However, when these cells are shifted into glucose to deplete the indicated proteins and analyzed by FACS sorting, the cell cycle profiles are different from those obtained from cells that are cycling normally. In yeast depleted of SSU processome proteins Utp1-15, and Utp17 by growth in glucose (Figure 5, Depleted) the G1 peak continues to be much larger than the G2 peak, with little change upon shift to glucose (compare Depleted YPH499 and Rps14A to the Utp proteins). This indicates arrest in the G1 phase of the cell cycle. In contrast, yeast depleted of Utp16, a nonessential SSU processome protein, yielded a cell cycle profile similar to the YPH499 and GAL::3xHA-RPS14A strains, with equal G1 and G2 peaks (Figure 5). Thus, as expected, depletion of the nonessential Utp16 protein does not cause arrest at G1.
Figure 5.
Depletion of SSU processome proteins leads to G1 arrest. Strains expressing Utp1-17 and Rps14A from galactose-inducible/glucose-repressible promoters were grown in early log phase in galactose/raffinose media (undepleted) and then shifted into glucose (depleted) for 24 h. YPH499 (the parent untagged strain) and Rps14A (a redundant ribosomal protein) were used as controls. Cells were stained with propidium iodide and FACS sorted.
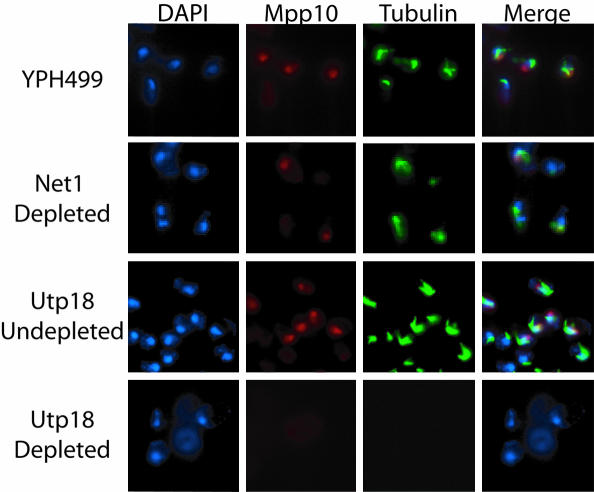
The G1 arrest upon essential Utp protein depletion was further supported by lack of immunofluorescence of a marker for the G2 phase of the cell cycle (Figure 6). Tubulin staining is indicative of progression to G2 phase of the cell cycle, because tubulin is only detectable in G2 when the mitotic spindle is present. YPH499, GAL::3xHA-NET1, and GAL::3xHA-UTP18 were grown to early log phase in galactose-containing medium and then shifted to glucose-containing medium for 24 h. Cells were analyzed by immunoflouresence for the presence of cellular DNA (blue); Mpp10, an SSU processome component (red); and tubulin (green). As expected, both Mpp10 and tubulin were expressed in the parent YPH499 strains (Figure 6). Similarly, cells depleted of Net1, a protein required for cell cycle progression at G2/M (Straight et al., 1999) resulted in expression of Mpp10 and tubulin (Figure 6). However, when Utp18 was depleted, Mpp10 and tubulin expression were no longer observed (Figure 6). This suggests that these cells no longer form mitotic spindles and is consistent with an arrest in G1. Because Mpp10 is a component of the SSU processome, the lack of Mpp10 staining in Utp18-depleted cells suggests that the SSU processome is no longer intact. Therefore, the lack of ribosomes after depletion of SSU processome proteins leads to stalling in the G1 phase of the cell cycle. Yeast depleted of SSU processome proteins are able to form small buds, suggesting that these yeast may be stalled in late G1 (our unpublished data). Therefore, this suggests that the SSU processome is required for cell cycle progression at G1.
Figure 6.
Immunofluorescence of depleted SSU processome proteins is consistent with G1 arrest. YPH499, GAL::3xHA-NET1, and GAL::3xHA-UTP18 yeast strains were grown to early log phase in galactose/raffinose media (undepleted) and then shifted into glucose (depleted) for 23 h. YPH499 (the parent untagged strain) and GAL::3xHA-NET1 (a protein required for mitotic exit) were used as controls. Immunofluorescence was carried out using antibodies to tubulin and detected with TRITC (green), and Mpp10 was detected with FITC (red). DAPI (blue) was used to stain the cellular DNA.
To further verify this arrest, we created yeast strains that could be synchronized and monitored for the expression of additional cell cycle markers. Yeast strains were created by the disruption of BAR1 that could be easily synchronized in G1 by the addition of low concentrations of the mating-type pheromone α-factor. In the Δbar1 strain, the cell cycle cyclins Cln2 and Clb2 were chromosomally tagged with TAP and 3xHA, respectively. Cln2 is a cell cyclin that is expressed during G1, whereas Clb2 is a cyclin that is expressed at G2. In normally cycling cells, after they are released from the G1 arrest, the cells will first express the cell cyclin Cln2-TAP and then express Clb2-3xHA as the cells progress into G2. The cells remain synchronized for one cell cycle (∼100 min) after being released from α-factor. The necessary strains were constructed for GAL::UTP1, GAL::UTP2, GAL::UTP4, and GAL::RPS14A by deletion of Bar1 (Δbar1) and the integration of TAP tag and the HA tag at CLN2 (Cln2-TAP) and CLB2 (Clb2–3xHA) in each strain.
SSU processome-depleted cells were synchronized with α-factor and then released to determined whether cells depleted of SSU processome proteins were then able to enter G2. The strains (construction described in the above paragraph) were grown in medium containing galactose (Undepleted) until OD600 0.2–0.6. The cells were shifted into glucose media (Depleted) for 21 h and then arrested in G1 with α-factor for 2.5 h. Cells were released from the G1 arrest (by washing α-factor out of the media), and the extracts were analyzed by Western blot or the cells sorted by FACS. The same number of cells were collected every 15 min after the G1 arrest, protein extracted, and Western blotted for the G1 cyclin Cln2-TAP or the G2 cyclin Clb2-3xHA via their respective tags (Figure 7A). After release from the G1 arrest, cells depleted of Rps14A, a redundant nonessential ribosomal protein, expressed Cln2 beginning at ∼30 min after release, and the levels of this cyclin declined over time. In this same strain, protein levels of Clb2 were visualized at ∼75 min after release and remained constant after this time point. Expression of this protein demonstrated that these cells were cycling from G1 into the G2 stage of the cell cycle. In contrast, cells depleted of Utp1, Utp2, and Utp4 only expressed the G1 cyclin Cln2, although expression of this protein was variable from strain to strain (Figure 7A). Expression of the G2 cyclin could not be detected at all when these SSU processome components were depleted.
Figure 7.
Synchronized depleted SSU processome proteins arrest in G1 upon cell cycle release. GAL::RPS14A, GAL::UTP1, GAL::UTP2, GAL::UTP4 yeast strains bearing Δbar1, Cln2-TAP, Clb2-3xHA were grown to early log phase in galactose/raffinose media (undepleted) and then shifted to glucose (depleted) for 21 h. a-Factor (2 μg/ml) was added to cells for 2.5 h and then washed out of the media. (A) Western blot on synchronized and released yeast. Protein was extracted from equal amounts of cells every fifteen minutes after the α-factor release and Western blotted with PAP antibodies (to visualize Cln2-TAP) and anti-HA antibodies (to visualize Clb2-3xHA). (B) FACS analysis of synchronized and released yeast. Cells were arrested with α-factor for 2.5 h and analyzed for DNA content (1C, 1 DNA content; 2C, 2 DNA content) by FACS. After 105 min of release from α-factor, yeast were again analyzed for DNA content by FACS sorting.
The cell cycle arrest resulting from depletion of SSU processome components was further analyzed by FACS analysis of synchronized cells (Figure 7B). Cells arrested with α-factor for 2.5 h or released from this arrest for 105 min were collected and analyzed for one or two DNA contents (1C or 2C, respectively). After incubation with α-factor, Rps14A-, Utp1-, Utp2-, and Utp4-depleted cells had predominantly arrested in G1 (1C DNA at 0 min). When released from this arrest for 105 min, Rps14A-depleted cells were predominantly in G2 with a 2C DNA content at 105 min, whereas Utp1-, Utp2-, and Utp4-depleted cells were still arrested in G1 (1C at 105 min). Together, these results suggest that the SSU processome is required for cell cycle progression at G1.
DISCUSSION
Ribosomes are the protein factories of the cell and without them cellular growth stops. Due to the highly regulated nature of ribosome biogenesis, we hypothesized that the activity of the SSU processome must be coordinately regulated with other cellular processes such as mitosis and growth in cell size. Although ribosome biogenesis is mitotically regulated in metazoan cells, our results suggest that ribosome biogenesis continues during mitosis in the yeasts S. cerevisiae and S. pombe. Even though ribosome biogenesis is not mitotically regulated in yeast, our data support the observations that ribosome biogenesis is critical for cell division to occur. Genetic depletion of SSU processome proteins leads to a G1 arrest as observed by FACS sorting, immunofluorescence, and Western blotting of G1- and G2-specific factors.
In mammalian cells, ribosome biogenesis is mitotically regulated. In previously described experiments, mammalian cells were mitotically arrested with colchicine, a drug that inhibits microtubule assembly (Prescott and Bender, 1962). We arrested yeast with the analogous drug nocodazole to arrest the cells at the same point during the cell cycle when transcription of rRNA was previously observed to cease. However, we did not observe any difference in transcription or processing of the rRNA. Therefore, yeast and mammalian cells regulate ribosome biogenesis differently. One possible reason for this lies in the biology of cell division; yeast have a “closed” mitosis, whereas mammalian cells have an “open” mitosis. Yeast cells in mitosis maintain their nuclear envelope, whereas the nuclear envelope dissolves in mammalian cells. Because the structure of the nucleus and nucleolus is completely disrupted during mitosis in mammalian cells, they cease making ribosomes. In contrast, the yeast nuclear envelope remains intact throughout the cell cycle, thereby enabling ribosome biogenesis to continue.
These results are consistent with Elliot and McLaughlin where they pulsed S. cerevisiae throughout the cell cycle (obtained by cell elutriation) with [3H]methionine and [3H]uracil and examined the rate of synthesis of 18S, 25S, 5.8S, and 5S rRNAs (Elliot and McLaughlin, 1979). They similarly concluded that rRNA is transcribed during mitosis. However, they did not analyze the pre-rRNAs, as we have done here. Similarly, our results are consistent with those of Sogin et al. (1974) who also looked at synthesis of rRNAs during mitosis in S. cerevisiae by the incorporation of 14C and 32P. Both studies concluded that overall levels of rRNA increased during the cell cycle but could not differentiate this from the doubling rate. In another study, the levels of the SSU processome protein Mpp10 remained constant throughout the cell cycle (Burton and Solomon, 2000).
We also investigated whether Nan1/Utp17 provides a link between ribosome biogenesis and mitotic exit. Nan1/Utp17 was originally purified through its association with Net1, a protein required for mitotic exit and silencing. However, our coimmunoprecipitation experiments suggest that Nan1/Utp17 and Net1 do not interact. In addition, when proteins required for mitotic exit are depleted, these cells arrest in G2/M. When Nan1 is depleted, these cells arrest in G1, suggesting that Nan1 is not required for G2/M. Although it is clear that the majority of Nan1/Utp17 is associated with the SSU processome, it is possible that small, substoichiometric amounts of Nan1/Utp17 do associate with Net1. In addition, Nan1 may have been found to be associated with Net1 in the affinity purification because it resides in the nucleolus. Because Nan1 is not primarily associated with Net1, we suggest Nan1 should be renamed and referred to as Utp17.
Cell division is highly coordinated with cellular growth and size (Jorgensen et al., 2004). Progression through START is regulated through growth in size where cells must reach a minimum cell size before S phase can begin (Johnston and Singer, 1978; Moore, 1988). Many of the cellular sensors that regulate cell progression through START have remained elusive. Recently, Jorgensen et al. (2002) conducted a genome-wide approach to identify proteins that regulate growth in size. This group suggested that deletion of genes involved in ribosome biogenesis lead to decreased cell size. For example, Jorgensen et al. (2002) found using microarrays that inducing Sfp1 expression, a gene implicated in cell size control, may target mRNA expression of Utp4, Utp6, Utp10, Utp17, and Utp20. This indirectly suggests that expression of a subset of SSU processome proteins is necessary for cells to grow in size. Recent work by Mnaimneh et al. (2004) also used a genome-wide approach to find essential genes that when depleted lead to decreased cell size. This group also found a link between proteins required for ribosome biogenesis and cell size. Our data support the findings that the SSU processome proteins are required for cell cycle progression at G1. This suggests that if a cell cannot make ribosomes and ultimately synthesize proteins, these cells cannot reach the critical cell size required for division to occur (Johnston and Singer, 1978). Because ribosome biogenesis and protein synthesis are intertwined, we cannot rule out the possibility that the G1 arrest caused by depletion of SSU processome proteins may be due to decreased protein synthesis as well as to the deficits in ribosome biogenesis.
Reaching the minimum cell size necessary for cell division is regulated through the target of rapamycin (TOR) pathway (Fingar and Blenis, 2004). It is clear that cell growth and ribosome biogenesis are linked somehow through the TOR signaling pathway (Thomas, 2000). In higher eukaryotes, the 40S ribosomal protein S6 seems to be critical for TOR signaling and cell cycle progression (Thomas, 2000; Fingar et al., 2004). For example, when transgenic mice have ribosomal protein S6 deleted in their liver cells, these cells arrest in late G1 phase of the cell cycle and cannot proceed into S phase (Volarevic et al., 2000).
What is the minimal number of ribosomes required for cell cycle progression in yeast? Previous observations suggest that there may be a threshold for ribosome number below which yeast cannot grow. Two different mutations in the SSU processome protein Mpp10 both affect levels of the 18S rRNA at 30°C. However, the mpp10-2 mutant results in a greater decrease in 18S rRNA levels than the mpp10-1 mutant (Lee and Baserga, 1997). Surprisingly, the mpp10-1 strain grows normally whereas the mpp10-2 strain grows very slowly. This suggests that the number of 40S ribosomes in the mpp10-1 strain is sufficient for cell cycle progression, even though it is less than normal. In contrast, in the mpp10-2 mutant, the number of 40S ribosomes is below the threshold for rRNA production and cell growth. We believe that future analysis of these mutants will begin to elucidate the relationship between ribosome number, cell growth, and cell division.
Although ribosome biogenesis is not cell cycle regulated in yeasts, SSU processome proteins are required for cell cycle progression at G1. These results support the observation that cell size is critical for cell division to occur, because ribosome biogenesis proteins have been shown to be important for cellular growth in size. Our results suggest that the SSU processome, a complex required for biogenesis of the small ribosomal subunit, may be critical for regulating cell growth and division. In the future, it will be important to understand what specific signals convey to a yeast cell that it has sufficiently grown in size to progress from G1 to S.
Acknowledgments
We thank Danesh Moazed for the Net1-3xHA-tagged strain and Sandra Wolin for the S. pombe. We thank Rocco Carbone from the Yale Cancer Center Flow Cytometry Shared Resource at Yale University School of Medicine. This facility is supported by a grant from National Institute of Health (CA-16359). We also thank Janet Burton, Erica Champion, Marisa Dolled-Filhart, Jennifer Kalish, Rebecca Lackman, and Catherine Sterling for proofreading the manuscript. K.A.B. is currently supported by a predoctoral fellowship from the National Institute of Health (GM-67564) and previously supported by the Research Service Award (GM-07499) from the National Institutes of Health National Institute of General Medical Sciences. This work was supported by the National Institute of Health grant GM-52581 to S.J.B.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04–06–0515. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–06–0515.
References
- Burton, J.L., and Solomon, M.J. (2000). Hsl1p, a Swe1p inhibitor, is degraded via the anaphase-promoting complex. Mol. Biol. Cell 20, 4614-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscemi, G., Saracino, F., Masnada, D., Carbone, M.L. (2000). The Saccharomyces cerevisiae SDA1 gene is required for actin cytoskeleton organization and cell cycle progression. J. Cell Sci. 113, 1199-1211. [DOI] [PubMed] [Google Scholar]
- David-Pfeuty, T., Nouvian-Dooghe, Y., Sirri, V., Roussel, P., and Hernandez-Verdun, D. (2001). Common and reversible regulation of wild-type p53 function and of ribosomal biogenesis by protein kinases in human cells. Oncogene 20, 5951-5963. [DOI] [PubMed] [Google Scholar]
- Dragon, F., et al. (2002). A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417, 967-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, Y.C., and Stillman, B. (2002). Yph1p, an ORC-interacting protein: potential links between cell proliferation control, DNA replication, and ribosome biogenesis. Cell 109, 835-848. [DOI] [PubMed] [Google Scholar]
- Dunbar, D.A., Dragon, F., Lee, S.J., and Baserga, S.J. (2000). A nucleolar protein related to ribosomal protein L7 is required for an early step in large ribosomal subunit biogenesis. Proc. Natl. Acad. Sci. USA 97, 13027-13032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar, D.A., Wormsley, S., Agentis, T.M., and Baserga, S.J. (1997). Mpp10p, a U3 small nucleolar ribonucleoprotein component required for pre-18S rRNA processing in yeast. Mol. Cell. Biol. 17, 5803-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr, M., and Olson, M.O.J. (1998). Partially processed pre-rRNA is preserved in association with processing components in nucleolus-derived foci during mitosis. Mol. Biol. Cell 9, 2407-2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliot, S., and McLaughlin, C. (1979). Regulation of RNA synthesis in yeast III. Mol. Gen. Genet. 169, 237-243. [DOI] [PubMed] [Google Scholar]
- Fewell, S.W., and Woolford, J.L., Jr. (1999). Ribosomal protein S14 of Saccharomyces cerevisiae regulates its expression by binding to RPS14B pre-mRNA and to 18S rRNA. Mol. Cell. Biol. 19, 826-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingar, D.C., and Blenis, J. (2004). Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene 23, 3151-3171. [DOI] [PubMed] [Google Scholar]
- Fingar, D.C., Richardson, C.J., Tee, A.R., Cheatham, L., Tsou, C., and Blenis, J. (2004). mTOR controls cell cycle progression through its cell growth effectors S6K1 and 4E-BP1/eukaryotic translation initiation factor 4E. Mol. Cell. Biol. 24, 200-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier, T., Fomproix, N., Masson, C., Azum-Gelade, M.C., Gas, N., and Hernandez-Verdun, D. (1994). Fate of specific nucleolar perichromosomal proteins during mitosis: cellular distribution and association with U3 snoRNA. Biol. Cell 82, 81-93. [DOI] [PubMed] [Google Scholar]
- Gerbi, S.A., Borovjagin, A.V., Ezrokhi, M., and Lange, T.S. (2001). Ribosome biogenesis: role of small nucleolar RNA in maturation of eukaryotic rRNA. Cold Spring Harb. Symp. Quant. Biol. 66, 575-590. [DOI] [PubMed] [Google Scholar]
- Gerbi, S.A., Borovjagin, A.V., and Lange, T.S. (2003). The nucleolus: a site of ribonucleoprotein maturation. Curr. Opin. Cell Biol. 15, 318-325. [DOI] [PubMed] [Google Scholar]
- Grandi, P., et al. (2002). 90S Pre-Ribosome Include the 35S Pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell 10, 105-115. [DOI] [PubMed] [Google Scholar]
- Johnston, G.C., and Singer, R.A. (1978). RNA synthesis and control of cell division in the yeast S. cerevisiae. Cell 14, 951-958. [DOI] [PubMed] [Google Scholar]
- Jorgensen, P., Nishikawa, J.L., Breitkreutz, B.J., and Tyers, M. (2002). Systematic identification of pathways that couple cell growth and division in yeast. Science 297, 395-400. [DOI] [PubMed] [Google Scholar]
- Kilmartin, J.V., Wright, B., and Milstein, C. (1982). Rat monoclonal antitubulin antibodies derived by using a new nonsecreting rat cell line. J. Cell Biol. 93, 576-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop, M., Siegers, K., Pereira, G., Zachiariae, W., Winsor, B., Nasmyth, K., and Schiebel, E. (1999). Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15, 963-972. [DOI] [PubMed] [Google Scholar]
- Kondoh, H., Yuasa, T., and Yanagida, M. (2000). Mis3 with a conserved RNA binding motif is essential for ribosome biogenesis and implicated in the start of cell growth and S phase checkpoint. Genes Cells 5, 525-541. [DOI] [PubMed] [Google Scholar]
- Kressler, D., Linder, P., and de la Cruz, J. (1999). Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 7897-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnirov, V.V. (2000). Rapid and reliable protein extraction from yeast. Yeast 16, 857-860. [DOI] [PubMed] [Google Scholar]
- Lafontaine, D., and Tollervey, D. (1995). Trans-acting factors in yeast prerRNA and pre-snoRNA processing. Biochem. Cell Biol. 73, 803-812. [DOI] [PubMed] [Google Scholar]
- Lee, S.J., and Baserga, S.J. (1997). Functional separation of pre-rRNA processing steps revealed by truncation of the U3 small nucleolar ribonucleoprotein component, Mpp10. Proc. Natl. Acad. Sci. USA 94, 13536-13541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S.J., and Baserga, S.J. (1999). Imp3p and Imp4p: two specific components of the U3 small nucleolar ribonucleoprotein that are essential for pre-18S rRNA processing. Mol. Cell. Biol. 19, 5441-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z., Paulovich, A.G., and Woolford, J.L., Jr. (1995). Feedback inhibition of the yeast ribosomal protein gene CRY2 is mediated by the nucleotide sequence and secondary structure of CRY2 pre-mRNA. Mol. Cell. Biol. 15, 6454-6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., McKenzie, A.R., Demarini, D.J., Shah, N.G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J.R. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953-961. [DOI] [PubMed] [Google Scholar]
- Mnaimneh, S., et al. (2004). Exploration of essential gene functions via titratable promoter alleles. Cell 118, 31-44. [DOI] [PubMed] [Google Scholar]
- Moore, S.A. (1988). Kinetic evidence for a critical rate of protein synthesis in the Saccharomyces cerevisiae yeast cell cycle. J. Biol. Chem. 263, 9674-9681. [PubMed] [Google Scholar]
- Oeffinger, M., and Tollervey, D. (2003). Yeast Nop15p is an RNA-binding protein required for pre-rRNA processing and cytokinesis. EMBO J. 22, 6573-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulovich, A.G., Thompson, J.R., Larkin, J.C., Li, Z., and Woolford, J.L., Jr. (1993). Molecular genetics of cryptopleurine resistance in Saccharomyces cerevisiae: expression of a ribosomal protein gene family. Genetics 135, 719-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestov, D.G., Stockelman, M.G., Strezoska, Z., and Lau, L.F. (2001a). ERB1, the yeast homolog of mammalian Bop1, is an essential gene required for maturation of the 25S and 5.8S ribosomal RNAs. Nucleic Acids Res. 29, 3621-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestov, D.G., Strezoska, Z., and Lau, L.F. (2001b). Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: effects of nucleolar protein Bop1 on G(1)/S transition. Mol. Cell. Biol. 21, 4246-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott, D.M., and Bender, M.A. (1962). Synthesis of RNA and protein during mitosis in mammalian tissue culture cells. Exp. Cell Res. 26, 260-268. [DOI] [PubMed] [Google Scholar]
- Rigaut, G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M., and Séraphin, B. (1999). A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotech. 17, 1030-1032. [DOI] [PubMed] [Google Scholar]
- Ruggero, D., and Pandolfi, P.P. (2003). Does the ribosome translate cancer? Nat. Rev. Cancer 3, 179-192. [DOI] [PubMed] [Google Scholar]
- Saracino, F., Bassler, J., Muzzini, D., Hurt, E., and Carbone, M.L.A. (2004). The yeast kinase Swe1 is required for proper entry into cell cycle after arrest due to ribosome biogenesis and protein synthesis defects. Cell Cycle 3, 648-654. [PubMed] [Google Scholar]
- Shou, W., Seol, J.H., Shevchenko, A., Baskerville, C., Moazed, D., Chen, Z.W.S., Jang, J., Shevchenko, A., Charbonneau, H., and Deshaies, R.J. (1999). Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 97, 233-244. [DOI] [PubMed] [Google Scholar]
- Shulman, R.W., Hartwell, L.H., and Warner, J.R. (1973). Synthesis of ribosomal proteins during the yeast cell cycle. J. Mol. Biol. 73, 513-525. [DOI] [PubMed] [Google Scholar]
- Sirri, V., Roussel, P., and Hernandez-Verdun, D. (2000). In vivo release of mitotic silencing of ribosomal gene transcription does not give rise to precursor ribosomal RNA processing. J. Cell Biol. 148, 259-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogin, S.J., Carter, B.L.A., and Halvorson, H.O. (1974). Changes in the rate of ribosomal RNA synthesis during the cell cycle in Saccharomyces cerevisiae. Exp. Cell Res. 89, 127-138. [DOI] [PubMed] [Google Scholar]
- Straight, A.F., Shou, W., Dowd, G.J., Turck, C.W., Deshaies, R.J., Johnson, A.D., and Moazed, D. (1999). Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell 97, 245-256. [DOI] [PubMed] [Google Scholar]
- Strezoska, Z., Pestov, D.G., and Lau, L.F. (2000). Bop1 is a mouse WD40 repeat nucleolar protein involved in 28S and 5. 8S RRNA processing and 60S ribosome biogenesis. Mol. Cell. Biol. 20, 5516-5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strezoska, Z., Pestov, D.G., and Lau, L.F. (2002). Functional inactivation of the mouse nucleolar protein Bop1 inhibits multiple steps in pre-rRNA processing and blocks cell cycle progression. J. Biol. Chem. 277, 29617-29625. [DOI] [PubMed] [Google Scholar]
- Tallada, V.A., Daga, R.R., Palomeque, C., Garzon, A., and Jimenez. (2002). Genome-wide search of Schizosaccharomyces pombe genes causing overexpression-mediated cell cycle defects. Yeast 19, 1139-1151. [DOI] [PubMed] [Google Scholar]
- Taylor, J.H. (1960). Nucleic acid synthesis in relation to the cell division cycle. Ann. N.Y. Acad. Sci. 90, 409-421. [DOI] [PubMed] [Google Scholar]
- Thomas, G. (2000). An encore for ribosome biogenesis in the control of cell proliferation. Nat. Cell Biol. 2, E71-E72. [DOI] [PubMed] [Google Scholar]
- Volarevic, S., Stewart, M.J., Ledermann, B., Zilberman, F., Terracciano, L., Montini, E., Grompe, M., Kozma, S.C., and Thomas, G. (2000). Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science 288, 2045-2047. [DOI] [PubMed] [Google Scholar]
- Wain, W.H., and Staatz, W.D. (1973). Rates of synthesis of ribosomal protein and total ribonucleic acid through the cell cycle of the fission yeast Schizosaccharomyces pombe. Exp. Cell Res. 81, 269-278. [DOI] [PubMed] [Google Scholar]
- Zimmerman, Z.A., and Kellogg, D.R. (2001). The Sda1 protein is required for passage through start. Mol. Biol. Cell 12, 201-219. [DOI] [PMC free article] [PubMed] [Google Scholar]