Abstract
Plant lipid transfer proteins (LTPs) are small, cysteine-rich proteins secreted into the extracellular space. They belong to the pathogenesis-related proteins (PR-14) family and are believed to be involved in several physiological processes including plant disease resistance, although their precise biological function is still unknown. Here, we show that a recombinant tobacco LTP1 is able to load fatty acids and jasmonic acid. This LTP1 binds to specific plasma membrane sites, previously characterized as elicitin receptors, and is shown to be involved in the activation of plant defense. The biological properties of this LTP1 were compared with those of LTP1-linolenic and LTP1-jasmonic acid complexes. The binding curve of the LTP1-linolenic acid complex to purified tobacco plasma membranes is comparable to the curve obtained with LTP1. In contrast, the LTP1-jasmonic acid complex shows a strongly increased interaction with the plasma membrane receptors. Treatment of tobacco plants with LTP1-jasmonic acid resulted in an enhancement of resistance toward Phytophthora parasitica. These effects were absent upon treatment with LTP1 or jasmonic acid alone. This work presents the first evidence for a biological activity of a LTP1 and points out the crucial role of protein-specific lipophilic ligand interaction in the modulation of the protein activity.
INTRODUCTION
Plant nonspecific lipid transfer proteins (LTPs) are low molecular mass, generally basic proteins that are ubiquitous in the plant kingdom. Two multigenic families have been identified: type I LTP (9 kDa, LTP1) and type II LTP (7 kDa, LTP2). They share several structural features, the most striking being a consensus motif of eight cysteine residues that are engaged in four disulfide bridges with identical connectivity (Kader, 1996; Douliez et al., 2000). In both type I and type II LTPs, these disulfide bonds stabilize an a-helix fold in which a hydrophobic cavity or tunnel can bind different types of lipids and hydrophobic molecules except sterols (Douliez et al., 2000; Marion et al., 2004).
Although numerous structural and physiological data are available, the precise function of these proteins in the physiology of plants is still debated. Historically, the in vitro lipid transfer ability demonstrated by Kader (1975) led to the hypothesis that these proteins could be involved in the intracellular trafficking of membrane lipids. However, such a role has been ruled out when it was shown that these proteins have an extracellular location, generally at the periphery of plant organs and are synthesized as preproteins with an N-terminal peptide signal (Coutos-Thévenot et al., 1993; Thoma et al., 1993; Arondel et al., 2000). LTPs are massively secreted during somatic embryogenesis of grape or carrot cells and might be involved in plant development mechanisms (Sterk et al., 1991; Fleming et al., 1992; Coutos-Thévenot et al., 1993). For these reasons, and based on the binding properties of fatty acid derivatives, it was suggested that LTP could be involved in the transport of cutin monomers during cuticle layer formation (Sterk et al., 1991; Meijer et al., 1993). However, no clear experimental evidence of such a role has been obtained yet. More recently, results from several groups indicated a role for LTPs in plant defense mechanisms. Transcripts that encode LTPs accumulated in grape cell suspension culture treated by a proteinaceous elicitor from the phytopathogenic fungus Botrytis cinerea (Gomès et al., 2003). Different 9-kDa basic polypeptides sharing high homology with LTPs are produced during plant-microorganism interactions (Blilou et al., 2000). Moreover, some of them are active against several pathogens, and expression of barley LTP in transgenic tobacco and Arabidopsis thaliana enhances tolerance to bacterial pathogens (Molina et al., 1993; Cammue et al., 1995; Molina and Garcia-Olmedo, 1997; Carvalho et al., 2001). This led to classify LTPs in the pathogenesis-related proteins, under the denomination PR-14 (Van Loon and Van Strien, 1999). Moreover, recent lines of evidences indicate that LTPs could have a role in plant defense signaling. An A. thaliana T-DNA tagged mutant, dir1-1 (defective in induced resistance 1-1) is unaffected in the local resistance against virulent or avirulent Pseudomonas syringae, but is unable to develop SAR (systemic acquired resistance) against virulent Pseudomonas (Maldonado et al., 2002). The wild-type DIR1 gene encodes a 102 amino acid putative nonspecific LTP. Expression of the PR-1 gene, a SAR marker, is lost in leaves distant to the infection sites in dir 1-1 plants, indicating that DIR1 is required for long-distance signaling during SAR, either by generating or transporting an essential mobile signal. However, transgenic plants overexpressing the DIR1 gene did not appear to exhibit constitutive SAR. This implies that DIR1 is probably not the mobile signal itself, but rather acts in cooperation with a mobile cosignal.
Besides this, it has been shown that a LTP1 from wheat endosperm binds to high affinity sites of tobacco plasmalemma (Buhot et al., 2001), previously identified as elicitin receptors (Wendehenne et al., 1995; Bourque et al., 1999). Elicitins are low molecular mass (10 kDa) proteins, secreted by the phytopathogenic Oomycetes Phytophthora or Pythium, which induce a hypersensitive reaction and SAR in tobacco plants (Ponchet et al., 1999). These proteinaceous elicitors of resistance display an internal cavity that can accommodate and transfer hydrophobic compounds such as sterols (Vauthrin et al., 1999) and fatty acids (Osman et al., 2001a). Prerequisite steps to the induction of plant responses are the formation of a sterol-elicitin complex before its recognition by receptors located in plant plasma membranes (Osman et al., 2001b). This prompted us to investigate whether plant LTPs, which share some common features with elicitins at both the function (extracellular lipid-binding proteins) and the structural three-dimensional level (Buhot et al., 2001), could also have their biological activity modulated by lipid ligands. To test this hypothesis, we chose the tobacco LTP1 (GenBank no. X62395), which has a constitutive, low-level expression throughout the plant, with a higher expression in epidermis and shoot apex (Fleming et al., 1992; Canevascini et al., 1996). This protein has been heterologously overexpressed in Pichia pastoris and its solution structure has just been solved by heteronuclear multidimensional NMR. It shows that its fold is comparable to those previously reported for other plant LTPs (Da Silva et al., unpublished results). In this work, after having checked that the recombinant tobacco LTP1 was functional and could load various fatty acids, we focused on two potential ligands. The first one is the oxylipin jasmonic acid (JA), a well known signaling molecule in plant response to wounding and to pathogen attack (Wasternack and Parthier, 1997; Blée, 2002; Stratmann, 2003) and the second one is its precursor, linolenic acid (LA), a polyunsaturated fatty acid widely represented in plant lipids. To test the hypothesis that LA and/or JA could modulate the biological activity of tobacco LTP1, we studied the ability of the LTP1-JA and LTP1-LA complexes to promote tobacco resistance against Phytophthora parasitica, in relation with their ability to interact with elicitin plasmalemma receptors.
MATERIALS AND METHODS
Materials
Nicotiana tabacum var. xanthi. Seeds were generously given by the Institut du Tabac (Bergerac, France). Plants were grown under controlled conditions (24 ± 2°C, 16 h light, 100 μEm-2 s-1). P. parasitica strain 329 from IPMSV collection (INRA Antibes) was grown in 10-cm-diameter Petri dish on maltagar (1%, wt/vol) for 7 d, at 25°C in the dark (Bonnet et al., 1996).
Chemicals. All fatty acids, JA, and 2-p-toluidinonaphtalene-6-sulfonate (TNS; potassium salt) were purchased from Sigma (Saint Quentin-en-Yvelines, France). Lipids and TNS were dissolved in ethanol and stored at -20°C. Cryptogein was obtained as previously described (Bonnet et al., 1996). LTP1 or LTP1-lipid complexes were used in this study. For binding experiments on purified plasma membrane, complexes were obtained by vortexing (1 min, room temperature) an aqueous solution of LTP1 and an ethanolic solution of the lipid in a 1/2 stoichiometry, with a final ethanol concentration of 1%. For plant protection experiments, the solutions were premixed such a way that the applied amounts were the same as those used for protein or lipid treatment alone, i.e., 5 μg (0.5 nmol) for LTP1 and 1 nmol for the lipids. Control treatments were cryptogein 1 μg (0.1 nmol) and 1% ethanol in water.
Overexpression of the Tobacco LTP1. The tobacco LTP1 (GenBank Accession Number X62395) is constitutively expressed in Nicotiana tabacum young aerial organs and especially in the shoot apex (Fleming et al., 1992). The tobacco LTP1 cDNA was first amplified by PCR with oligo 1 (5′-CTAGCTAGATACTTCATT-3′) and 2 (5′-ATAGACGAACACATCATATT-3′) using tobacco leaves cDNA as template. The PCR fragment was then cloned into pGEM-T easy (Promega, Madison, WI) and sequenced to verify its complete identity to X62395. The coding sequence was then reamplified using two oligos, oligo 3 (5′-GGGTATCTCTCGAGAAAAGAGAGGCTGAAGCTGCCATAACCTGTGGCC-3′) and 4 (5′-AGCCTCTCTTTTCTCGAGATTCTTATTTCATCAGCCTTACTGG-3′) as forward and reverse primers, respectively. The amplified fragment was purified and cloned into pPIC9. The correct orientation of the encoding sequence was checked by PCR screening and confirmed by DNA sequencing. P. pastoris transformation (strain GS115, obtained from Invitrogen, Carlsbad, CA) was performed as previously described (Osman et al., 2001b). The yeast transformant strains were cultivated on a derived YNB medium. All cultures were achieved at 29°C, in the dark, on rotary shakers. After 5 d of culture, the medium was recovered after centrifugation (10,000 × g, 10 min, 4°C). After pH adjustment to 2 by addition of TFA, the supernatant was loaded onto Amberlite XAD7 (Fluka, Buchs, Switzerland), preequilibrated with 0.1% aqueous TFA. After washing with 0.1% aqueous TFA and 10% CH3CN, 0.1% TFA, the LTP1 was eluted by 40% CH3CN, 0.1% TFA. After CH3CN evaporation under vacuum, the fraction was adjusted to pH 7.0 with 1 M Tris and loaded onto a Trisacryl SP ion exchange column (M grade, Biosepra, Malboro, MA), preequilibrated with Tris-HCl (20 mM, pH 7.0) buffer. A step gradient of NaCl in the same buffer was applied to the column: 0.1, 0.25, 0.5 M. The protein was recovered in the 0.25 M fraction. This fraction was adjusted to pH 2.0 with TFA, and CH3CN was added to reach a 10% (vol/vol) final concentration; then it was settled on a reversed-phase column (Uptisphere C4, 300 Å, 20 μm, Interchim, Lyon, France). After washing with CH3CN 10%, 0.1% TFA, the protein was eluted with 20% CH3CN, 0.1% TFA. The fraction containing the tobacco LTP1 was dialyzed extensively against ultrapure water and freeze-dried. The protein content and purity were checked throughout the purification by SDS-PAGE and HPLC.
Protein Compliance Tests. The recombinant tobacco LTP1 was analyzed in LC/MS (electrospray mode). The N-terminus was sequenced and circular dichroism spectrum was recorded as previously described (Elmorjani et al., 2004).
Methods
Lipid Binding. The ability of the tobacco LTP1 to bind lipids was assayed by monitoring the displacement of the fluorescent probe TNS. Fluorescence experiments were performed at 25°C in a Shimadzu RF 5301 PC spectrofluorimeter (Kyoto, Japan), as previously described (Mikès et al., 1998; Osman et al., 2001a), with minor modifications. The excitation and emission wavelengths were set at 320 and 437 nm, respectively. TNS, with or without fatty acids or JA, were incubated for 1 min in a stirred cuvette containing 2 ml of measurement buffer (175 mM mannitol, 0.5 mM K2SO4 0.5 mM CaCl2, and 5 mM Mes, pH 7.0) before the fluorescence was recorded (F0). Then, the tobacco LTP1 was added and after 2 min, fluorescence was recorded at equilibrium (F). Results are expressed either as percentage of LTP1-TNS complex fluorescence according to ((F - F0)/FC) × 100, where FC is the fluorescence of the LTP1-TNS complex in absence of FA or JA; or as fluorescence (F - F0, ΔF), in arbitrary units.
Binding Experiments Using 125I-labeled Proteins. Purified plasma membranes were obtained as previously described by Vauthrin et al. (1999). Iodination of the tobacco LTP1 was performed as previously described (Blein et al., 1991). Binding experiments and replacement experiments were carried out as previously described (Bourque et al., 1998). Mathematical analysis was done using the binding theoretical curves described in Buhot et al., (2001).
Plant Treatment and Determination of Necrosis Intensity. Fifty-day-old tobacco plants were used. Protein or lipid-protein mix in 1% ethanolic solutions were applied on the stem section of freshly decapitated plants (five plants for each treatment). Ethanol (1%) was added on control plants. Twenty-four hours later, a mycelial plug was dropped onto the cut petiole of a leaf (the third fully expanded leaf from the apex) and covered with a piece of aluminum foil. Seven days after the inoculation, the length of the necroses was measured on the stem (internal necroses) corresponding to the brown or desiccated zones (Bonnet et al., 1996). Results are expressed as percentage of protection ±SD.
RESULTS
Recombinant Tobacco LTP1 Compliance Tests
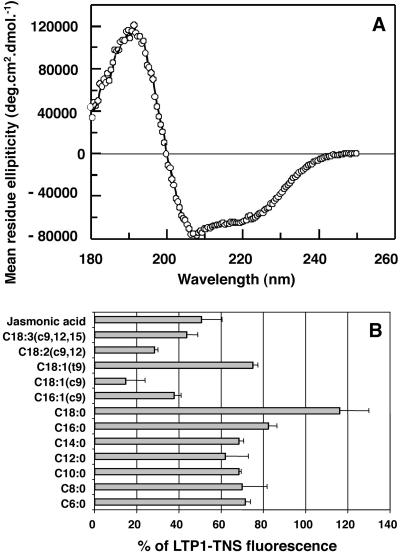
About 15 mg of LTP1 at homogeneity were recovered per liter of yeast culture. N-terminal sequencing of the protein yielded EAEAAIT instead of AIT, showing that a part of the yeast signal peptide was integrated into the recombinant LTP1. Mass measurement gave 9565.76 ± 0.81 Da for a calculated mass of 9572.69 Da with the EAEA extension. The difference between measured and calculated masses (about -7 MU) is probably due to the formation of four disulfide bridges necessary for protein folding. Analysis of the circular dichroism (Figure 1A) using the CONTIN software based on the method developed by Provencher and Glockner (1981) returned a helix content of 53%, in agreement with the previous high helicity found for these proteins (Douliez et al., 2000). This result and the fact that the molecular mass is in agreement with the presence of four disulfide bonds show that the recombinant tobacco LTP1 has a fold similar to that of the other plant type I LTPs. This has been recently confirmed by determination of the structure of this recombinant protein by multidimensional heteronuclear NMR (Da Silva et al., unpublished results).
Figure 1.
Biochemical properties of the recombinant tobacco LTP1. (A) Circular dichroism spectrum. (B) Effect of fatty acids on the fluorescence level of the LTP1-TNS complex. FAs or JA (16 μM) and TNS (3 μM) were incubated together for 1 min and then LTP1 (250 nM) was added. Results are expressed as the percentage of the fluorescence of the LTP1-TNS (control, no FA or JA added, fluorescence level; 100 ± 6.01%). Experiments were performed in triplicate and results are expressed as the mean values ± SD.
Tobacco LTP1-Fatty Acid Binding
To check whether the recombinant LTP1 was able to bind fatty acids, the protein was incubated with TNS, a fluorescent probe previously used to study the interaction between various lipids and elicitins. Elicitins show some structural homologies with LTPs at the three-dimensional level, including a hydrophobic pocket that can accommodate hydrophobic ligands (Mikès et al., 1998). From the initial level of fluorescence, a quenching is recorded when a lipid is able to compete with the bound probe inside the protein. Several groups of lipids were used: saturated FAs with C6 to C18 chain length, unsaturated FAs C16 or C18 harboring one to three double bonds, and JA.
When LTP1 was added to a mixture of TNS and FAs, the fluorescence of TNS was lower as compared with a control performed by adding LTP1 to TNS alone (Figure 1B). This indicates that fatty acids are able to compete with the fluorescent probe for binding to the protein. No significant interaction between FAs and TNS was detected (our unpublished results). Saturated FAs with a carbon number between 6 and 14 displayed identical behavior and were moderately effective in competing with TNS (71–62% of the control fluorescence). When the carbon number increased to 16 or 18, the displacement of the TNS was less efficient, the C18:0 fatty acid leading to a fluorescence level identical to that of the TNS control. In contrast, unsaturated fatty acids with the same chain length more efficiently displace the TNS probe. Three groups of ligands can be distinguished according to TNS displacement efficiency: 1) high: linoleic acid (C18:2, all cis) and oleic acid (C18:1, cis) leading to 28 and 15% of the control fluorescence, respectively; 2) medium: C16:1 cis, C18:3 (all cis), and jasmonic acid leading to 38, 44 and 50% of fluorescence, respectively; 3) low: C18:1 trans and saturated FAs (with the exception of C18:0, which does not displace TNS at all) with a fluorescence level of ca. 75% of the control.
Dixon's Representation of LTP1-JA Interactions
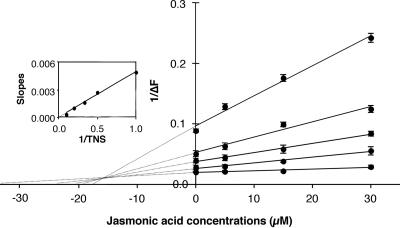
Interestingly, JA, a key component involved in plant response to injury and pathogen attack, was rather effective in competing with TNS. To better characterize the interaction between LTP1 and JA and to rule out nonspecific binding to the protein, a Dixon plot was drawn (Figure 2). All straight lines, obtained by linear regression, cross at a single point left of the ordinate axis and above the abscissa axis, indicating a pure competitive inhibition phenomenon. Moreover, the replot of the slopes of the Dixon plot vs. 1/TNS concentration is a straight line crossing the origin (Figure 2, inset). The apparent Kd for TNS is 8.28 ± 0.82 μM and the Ki for JA is 15.55 ± 1.09 μM. The existence of the LTP1-JA complex has been confirmed by Quadrupole-Time of Flight (Q-TOF) mass spectrometry (our unpublished results).
Figure 2.
Dixon's plot of the binding competition between JA and TNS to LTP1. Various amount of JA (0–30 μM) and TNS (1–10 μM) were incubated together for 1 min and then LTP1 (250 nM) was added. From top to bottom, linear regressions lead to the following cr2 values: 0.989, 0.972, 0.993, 0.981, and 0.915 for TNS concentrations of 1, 2, 3, 5, or 10 μM, respectively. Inset: replot of the slopes of the Dixon plots and linear regression (cr2 = 0.993). Experiments were performed in triplicate and results are expressed as the mean values ± SD.
Binding of LTP1 to Tobacco Plasma Membranes
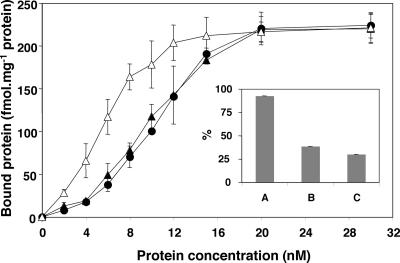
LTP1, LTP1-LA, and LTP1-JA complexes bind to plasma membranes, the saturability level being similar (Figure 3 and Table 1). However, 50% saturability was obtained at ∼6.5, 10.5, and 11 nM for LTP1-JA complex, LTP1-LA complex and LTP1, respectively. The shapes of the binding curves are sigmoidal and the mathematical treatment previously used for a wheat LTP1 binding experiment (Buhot et al., 2001) was applied to determine the binding parameters. The three ligands exhibit a high-affinity for the binding sites (KB is ∼2 nM), but the allosteric constant parameter (T) is much lower in the case of LTP1-JA complex (Table 1).
Figure 3.
Specific interactions between tobacco LTP1 and high-affinity sites located on tobacco plasma membranes. Plasma membrane preparations were incubated with various concentrations of 125I-labelled LTP1. Specific binding was determined by subtracting nonspecific binding from total binding. Experiments were repeated at least five times, and results are expressed as the mean values ± SD (fmol bound LTP1/mg plasmalemma proteins). •, LTP1; ▴, LTP1-LA; and ▵, LTP1-JA. Inset: displacement of 125I-labelled LTP1 specifically bound to plasma membrane by lysozyme (A), unlabeled LTP1 (B), or unlabeled cryptogein (C), 30 min after the addition of the unlabeled protein. The experiments were repeated three times, and results are the mean values ± SD.
Table 1.
Thermodynamic parameters of the binding of tobacco LTP1 to plasma membranes
| N (fmol/mg protein) | KB (nM) | T | E | |
|---|---|---|---|---|
| Tobacco LTP1 | 253 | 1.9 | 1393 | 7×10–3 |
| Tobacco LTP1 + linolenic acid | 263 | 1.9 | 1045 | 8×10–3 |
| Tobacco LTP1 + jasmonic acid | 248 | 2.5 | 56 | 7×10–3 |
| Wheat LTP1a | 248 | 1.6 | 1252 | 3×10–3 |
| Cryptogeina | 246 | 2.0 | 30 | 7×10–3 |
Values were obtained by fitting experimental data from Figure 3 with the allosteric binding model defined by Monod, Wyman, and Changeux (Monod et al., 1965) and hypothesizing a four-subunit receptor (Buhot et al., 2001). N, KB, T, and E represent the values of: the number of sites, the apparent dissociation constant, the allosteric constant, and the error function, respectively.
Data from Buhot et al. (2001)
Displacement Experiments
Displacement experiments were performed in order to determine if LTP1 and cryptogein share the same binding sites. Tobacco plasma membranes were preincubated with 125I-labeled LTP1, before the addition of unlabeled LTP1, cryptogein, or lyzozyme as a negative control. Both LTP1 and cryptogein were able to displace labeled LTP1, whereas lyzozyme was not (Figure 3, inset). The displacements were similar, with radioactivity associated with the membranes decreasing from 100 to 38 and 31% within 30 min after the addition of unlabeled LTP1 or cryptogein, respectively (Figure 3, inset).
Effect of LTP1-JA Complex on Tobacco Plants Inoculated with P. parasitica
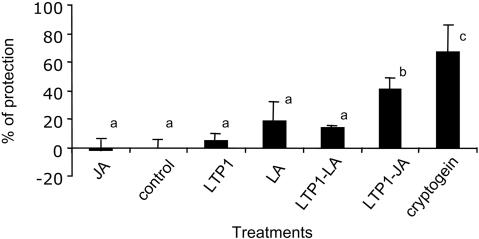
Twenty-four hours before inoculation, tobacco plants were decapitated and a drop 1% ethanolic solution containing either LTP1, LA, JA, or the complexes LTP1-LA or LTP1-JA was laid on the cut stem. Cryptogein, an elicitin from P. cryptogea, known to elicit a high level of protection in tobacco (Bonnet et al., 1996), was used as positive control. Then, the pathogen was inoculated on a cut petiole of a distal leaf. After 7 d, P. parasitica has invaded the stem, resulting in typical symptoms with tissue browning followed by a collapse. The classical quantitation of disease development via the measurement of the length of necrotic tissue has been performed. The obtained results show that compared with the protection induced by cryptogein (67%), LTP1, LTP1-LA, or LA induces a poor protection (5, 14, and 20%, respectively; Figure 4). JA treatment has almost no effect (-2%), but the complex LTP1-JA (42%) induces a better protection than LTP1 or JA alone (Figure 4). The Student (α = 0.05) test revealed that cryptogein and LTP1-JA unambiguously differ from the ethanol control, whereas LTP1-LA, LTP1, LA, and JA do not.
Figure 4.
Level of tobacco resistance to P. parasitica induced by cryptogein and tobacco LTP1. Ethanolic elicitor solutions were applied on the stem section of freshly decapitated plants (cryptogein 0.1 nmol/plant, LA or JA 1 nmol/plant, LTP1 or LTP1-LA or LTP1-JA 0.5 nmol/plant). Ethanol (1%) was added on control plants. Twenty-four hours later, a mycelial plug was dropped on the cut petiole of the third fully expanded leaf from the apex. Seven days after inoculation, stems were split in two, and the internal necroses, corresponding to the brown or desiccated zones, measured. Results are expressed as the means of percentage of protection ± SD from three replicate experiments and five plants for each treatment. Small letters represent the result of a Student-Newman-Keuls test at 0.05.
DISCUSSION
Tobacco LTP1 Binds Several Lipids, Including the Signaling Molecule JA
It has long been known that LTPs, from various plant and animal species, are able to bind FAs and to transport phospholipids between membrane systems in vitro (Kader, 1975; Bourgis and Kader, 1997), They show either narrow (Dansen et al., 1999; Balendiran et al., 2000) or broad (Zachowski et al., 1998; Tassin-Moidrot et al., 2000) specificity toward the nature of the transferred lipids. Despite a high degree of sequence identity and similar global folding (Poznanski et al., 1999), LTPs can exhibit structural differences leading to different affinities for lipids (Guerbette et al., 1999). In addition to this structural and functional diversity, LTPs also differ in their expression under conditions of abiotic (Yubero-Serrano et al., 2003) and biotic stress (Blilou et al., 2000; Gomès et al., 2003). The present article investigates the functional properties of a tobacco LTP1, which can be produced in high amounts in P. pastoris, and allows studies on a homologous plant/pathogen system. The sequence of this LTP1, which displays the cysteine signature, as well as its secondary and three-dimensional structures (Da Silva et al., unpublished results), indicate that this tobacco protein belongs to the type I family of LTPs.
Although tobacco LTP1 exhibits a broad binding spectrum toward FA, the chain length as well as the number and the geometry of double bonds strongly affect the affinity of the protein for FAs. Our results show that linear chain FAs are not easily accommodated in the hydrophobic tunnel of the tobacco LTP1, whereas cis-unsaturated FAs and JA, which exhibit more compacted structures, have a higher affinity for the protein. These results are in agreement with those reported for other lipid binding proteins like plant nsLTPs (Guerbette et al., 1999), elicitins (Osman et al., 2001a), or the brain FA-binding protein (Balendiran et al., 2000). Furthermore, Dixon's plot clearly shows that JA and TNS competed for the same site in the protein. These data provide the first evidence that a plant LTP is able to bind JA. This is particularly interesting with respect to the role of JA in plant signaling (Liechti and Farmer, 2002; Turner et al., 2002; Glazebrook et al., 2003) and with the recently described role of a putative LTP in SAR (Maldonado et al., 2002). This led us to investigate the role of LTP1-JA complex on plant responses.
JA, but Not LA, Modulates LTP1 Binding on the Elicitin Plasma Membrane Receptor
The tobacco LTP1 binds to specific protein sites located in the plasma membrane (Figure 3). Displacement experiments demonstrated that the specific binding sites for LTP1 and cryptogein are identical and that the LTP1 interaction with the binding sites is reversible, as for cryptogein (Wendehenne et al., 1995) or for wheat LTP1 (Buhot et al., 2001). LTP1-JA and LTP1-LA complexes bind to the sites previously characterized as the elicitin receptors. The binding curves of both complexes display the same sigmoidal shape and identical thermodynamic parameters (number of sites N and apparent dissociation constants KB, Table 1). However, the values of the allosteric constants, T, are ∼1000–1400 for the tobacco LTP1 loaded or not with LA (poorly active, this work), values that are similar to those previously described for a wheat LTP1 (inactive, Buhot et al., 2001).
On the contrary, those of cryptogein and of the LTP1-JA complex are 30 and 56, respectively, i.e., 20–40 times lower, suggesting that the active conformation of the receptor is favored by their binding. It also suggests that the formation of the LTP1-JA complex results in a conformational change of the LTP1, facilitating its recognition by the high-affinity sites. These observations are in agreement with the necessity for elicitins to bind a lipid molecule (a sterol) in order to interact with their receptors and with the shift of the binding curves of mutated elicitins previously reported: the lower the activity of the mutated protein is, the higher its binding curves shifted toward the higher free protein concentrations (Osman et al., 2001b). Altogether, this work demonstrates for the first time that a LTP-JA complex is bioactive and is recognized by the elicitin receptor, known to be involved in plant defense induction (Osman et al., 2001b).
The LTP1-JA Complex Induces Protection against P. parasitica
Localized exogenous application of the complex LTP1-JA on a decapitated tobacco plant provided long distance protection against P. parasitica spreading (Figure 4), whereas the LTP1-LA complex, LTP1, LA, or JA alone are very poorly efficient in this respect. Nevertheless, it is not possible to discriminate between the possibility that the LTP1-JA complex is the mobile signal leading to the induction of the protection in the distant leaves or that the binding of the complex on plasma membrane receptors is the initial step that induces the production of the mobile signal. Recently published lines of evidence make both hypotheses rather attractive, even if not proven yet. In this respect, a first indication is that the biologically active form of elicitins is a protein-lipid complex, which is recognized by specific biological receptors. Their activation triggers a SAR in tobacco plants (Osman et al., 2001b). Another part of this puzzle is the demonstration that a 9-kDa wheat LTP can bind to the elicitin receptors and interfere with elicitin (Buhot et al., 2001). Furthermore, a recent work proposes that DIR1, a putative LTP2, could be the translocator for release of the mobile signal involved in SAR in A. thaliana and strongly suggests that the protein works in cooperation with a ligand (Maldonado et al., 2002). However, the nature of the mobile signal remains unknown as well as its origin and destination. Finally, the fact that a tobacco LTP1-JA complex could induce protection in tobacco plants, together with its enhanced specific interaction with the elicitin receptors, highlights the necessity to identify the possible associations between LTPs (7 or 9 kDa) and putative hydrophobic ligands, native or induced in plants. Thus, to decipher the mechanism of the formation of these LTP-lipid complexes appears critical for understanding the acquisition of systemic resistance in planta.
Acknowledgments
We thank A. Jacquemard, G. Vago, and B.F. Maume for their help in the mathematical binding curve analyses; B. Industri and J. Fromentin for excellent technical assistance; and L. Suty and F. Simon-Plas for helpful discussions. We thank R. Thompson for revising the English and C. J. Lamb for critical reading and helpful comments on the manuscript. This work is supported by the Institut National dela Recherche Agronomique and the Centre National dela Recherche Scientifique.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04–07–0575. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–07–0575.
Abbreviations used: FA, fatty acid; JA, jasmonic acid; LA, linolenic acid; LTP, lipid transfer protein, SAR, systemic acquired resistance; TFA, trifluoroacetic acid, TNS, 2-p-toluidinonaphtalene-6-sulfonate.
References
- Arondel, V., Vergnolle, C., Cantrel, C., and Kader, J.-C. (2000). Lipid transfer proteins are encoded by a small multigene family in Arabidopsis thaliana. Plant Sci. 157, 1-12. [DOI] [PubMed] [Google Scholar]
- Balendiran, G.K., Schnütgen, F., Scapin, G., Börchers, T., Xhong, N., Lim, K., Godbout, R., Spener, F., and Sacchettini, J.C. (2000). Crystal structure and thermodynamic analysis of human brain fatty acid-binding protein. J. Biol. Chem. 275, 27045-27054. [DOI] [PubMed] [Google Scholar]
- Blée, E. (2002). Impact of phyto-oxylipins in plant defense. Trends Plant Sci. 7, 315-322. [DOI] [PubMed] [Google Scholar]
- Blein, J.-P., Milat, M.-L., and Ricci, P. (1991). Responses of cultured tobacco cells to cryptogein, a proteinaceous elicitor from Phytophthora cryptogea: possible plasmalemma involvement. Plant Physiol. 95, 486-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou, I., Ocampo, J.A., and Garcia-Garrido, J.M. (2000). Induction of Ltp (lipid transfer protein) and Pal (phenylalanine ammonia-lyase) gene expression in rice roots colonized by the arbuscular mycorrhizal fungus Glomus mosseae. J. Exp. Bot. 51, 1969-1977. [DOI] [PubMed] [Google Scholar]
- Bonnet, P., Bourdon, E., Ponchet, M., Blein, J.-P., and Ricci, P. (1996). Acquired resistance triggered by elicitins in tobacco and other plants. Eur. J. Plant Pathol. 102, 181-192. [Google Scholar]
- Bourgis, F., and Kader, J.-C. (1997). Lipid-transfer proteins: tools for manipulating membrane lipids. Physiol. Plant. 100, 78-84. [Google Scholar]
- Bourque, S., Binet, M.-N., Ponchet, M., Pugin, A., and Lebrun-Garcia, A. (1999). Characterization of the cryptogein binding sites on plant plasma membranes. J. Biol. Chem. 274, 34699-34705. [DOI] [PubMed] [Google Scholar]
- Bourque, S., Ponchet, M., Binet, M.-N., Ricci, P., Pugin, A., and Lebrun-Garcia, A. (1998). Comparison of binding properties and early biological effects of elicitins in tobacco cells. Plant Physiol. 118, 1317-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhot, N. et al. (2001). A lipid transfer protein binds to a receptor involved in the control of plant defence responses. FEBS Lett. 509, 27-30. [DOI] [PubMed] [Google Scholar]
- Cammue, B.P.A. et al. (1995). A potent antimicrobial protein from onion seeds showing sequence homology to plant lipid transfer proteins. Plant Physiol. 109, 445-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canevascini, S., Caderas, D., Mandel, T., Fleming, A.J., Dupuis, I., and Kuhlemeier, C. (1996). Tissue-specific expression and promoter analysis of the tobacco ltp1 gene. Plant Physiol. 112, 513-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho, A.O., Machado, O.L.T., Cunha, M.A., Santos, I.S., and Gomes, V.M. (2001). Antimicrobial peptides and immunolocalization of a LTP in Vigna unguiculata seeds. Plant Physiol. Biochem. 39, 137-146. [Google Scholar]
- Coutos-Thévenot, P. et al. (1993). Four 9-kDa Proteins excreted by somatic embryos of grapevine are isoforms of lipid-tranfer proteins. Eur. J. Biochem. 217, 885-889. [DOI] [PubMed] [Google Scholar]
- Dansen, T.B., Westerman, J., Wouters, F.S., Wanders, R.J.A., Van Hoek, A., Gadella, T.W.J., and Wirtz, K.W.A. (1999). High-affinity binding of very-long-chain fatty acyl-CoA esters to the peroxisomal non-specific lipid-transfer protein (sterol carrier protein-2). Biochem. J. 339, 193-199. [PMC free article] [PubMed] [Google Scholar]
- Douliez, J.-P., Michon, T., Elmorjani, K., and Marion, D. (2000). Structure, biological and technological functions of lipid transfer proteins and indolines, the major lipid binding proteins from cereal kernels. J. Cereal Sci. 32, 1-20. [Google Scholar]
- Elmorjani, K., Lurquin, V., Lelion, A., Rogniaux, H., and Marion, D. (2004). A bacterial expression system revisited for the recombinant production of cysteine-rich plant lipid transfer proteins. Biochem. Biophys. Res. Comm. 316, 1202-1209. [DOI] [PubMed] [Google Scholar]
- Fleming, A.J., Mandel, T., Hofmann, S., Sterk, P., De Vries, S.C., and Kuhlemeier, C. (1992). Expression pattern of a putative lipid transfer protein gene within the shoot apex. Plant J. 2, 855-862. [PubMed] [Google Scholar]
- Glazebrook, J., Chen, W.J., Estes, B., Chang, H.S., Nawrath, C., Métraux, J.-P., Zhu, T., and Katagiri, F. (2003). Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression phenotyping. Plant J. 34, 217-228. [DOI] [PubMed] [Google Scholar]
- Gomès, E., Sagot, E., Gaillard, C., Laquitaine, L., Poinsot, B., Sanejouand, H.-Y., Delrot, S., and Coutos-Thévenot, P. (2003). Non specific lipid-transfer protein genes expression in grape (Vitis sp.) cells in response to fungal elicitor treatments. Mol. Plant Microbe Interact. 16, 456-464. [DOI] [PubMed] [Google Scholar]
- Guerbette, F., Grosbois, M., Jolliot-Croquin, A., Kader, J.-C., and Zachowski, A. (1999). Comparison of lipid binding and transfer properties of two lipid transfer proteins from plant. Biochemistry 38, 14131-14137. [DOI] [PubMed] [Google Scholar]
- Kader, J.-C. (1975). Proteins and the intracellular exchange of lipids: stimulation of phospholipids exchange between mitochondria and microsomal fraction by protein isolated from potato tuber. Biochim. Biophys. Acta 380, 31-44. [PubMed] [Google Scholar]
- Kader, J.-C. (1996). Lipid-transfer protein in plants. Ann. Rev. Plant Physiol. Plant Mol. Biol. 47, 627-654. [DOI] [PubMed] [Google Scholar]
- Liechti, R., and Farmer, E.E. (2002). The jasmonate pathway. Science 296, 1649-1650. [DOI] [PubMed] [Google Scholar]
- Maldonado, A.M., Doerner, P., Dixon, R.A., Lamb, C.J., and Cameron, R.K. (2002). A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis. Nature 419, 399-403. [DOI] [PubMed] [Google Scholar]
- Marion, D., Douliez, J.-P., Gautier, M.-F., and Elmorjani, K. (2004). Plant lipid transfer proteins: relationships between allergenicity and structural, biological and technological properties. In: Plant Food Allergens, ed. E.N.C. Mills and S. P.W. Shewry. Oxford: Blackwell Publishing, 57-69.
- Meijer, E.A., De Vries, S.C., Sterk, P., Gadella, D.W., Wirtz, K.W., and Hendricks, T. (1993). Characterization of a non-specific lipid transfer protein EP2 from carrot (Daucus carota L.). Mol. Cell Biochem. 123, 159-166. [DOI] [PubMed] [Google Scholar]
- Mikès, V., Milat, M.-L., Ponchet, M., Panabières, F., Ricci, P., and Blein, J.-P. (1998). Elicitins, proteinaceous elicitors of plant defenses, are a new class of sterol carrier proteins. Biochem. Biophys. Res. Comm. 245, 133-139. [DOI] [PubMed] [Google Scholar]
- Molina, A., and Garcia-Olmedo, F. (1997). Enhanced tolerance to bacterial pathogens caused by the transgenic expression of barley lipid transfer protein LTP2. Plant J. 12, 669-675. [DOI] [PubMed] [Google Scholar]
- Molina, A., Segura, A., and Garcia-Olmedo, F. (1993). Lipid transfer proteins (nsLTP) from barley and maize leaves are potent inhibitors of bacterial and fungal plant pathogens. FEBS Lett. 316, 119-122. [DOI] [PubMed] [Google Scholar]
- Monod, J., Wyman, J., and Changeux, J.-P. (1965). On the nature of allosteric transitions: a plausible model. J. Mol. Biol 12, 88-118. [DOI] [PubMed] [Google Scholar]
- Osman, H., Mikès, V., Milat, M.-L., Ponchet, M., Marion, D., Prangé, T., Maume, B., Vauthrin, S., and Blein, J.-P. (2001a). Fatty acids bind to the fungal elicitor cryptogein and compete with sterols. FEBS Lett. 489, 55-58. [DOI] [PubMed] [Google Scholar]
- Osman, H. et al. (2001b). Mediation of elicitin activity is assumed by elicitin-sterol complexes. Mol. Biol. Cell 12, 2825-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponchet, M. et al. (1999). Are elicitins cryptograms in plant-Oomycetes communication? Cell. Mol. Life Sci. 56, 1020-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poznanski, J., Sodano, P., Suh, S.W., Lee, J.Y., Ptak, M., and Vovelle, F. (1999). Solution structure of a lipid transfer protein extracted from rice seeds: comparison with homologous proteins. Eur. J. Biochem. 259, 692-708. [DOI] [PubMed] [Google Scholar]
- Provencher, S.W., and Glockner, J. (1981). Estimation of globular protein secondary structure from circular dichroism. Biochemistry 20, 33-37. [DOI] [PubMed] [Google Scholar]
- Sterk, P., Booij, H., Schellekens, G.A., Van Kammen, A., and De Vries, S.C. (1991). Cell-specific expression of the carrot EP2 lipid transfer protein gene. Plant Cell 3, 907-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratmann, J.W. (2003). Long distance run in the wound response—jasmonic acid is pulling ahead. Trends Plant Sci. 8, 247-250. [DOI] [PubMed] [Google Scholar]
- Tassin-Moidrot, S., Caille, A., Douliez, J.-P., Marion, D., and Vovelle, F. (2000). The wide binding properties of a wheat nonspecific lipid transfer protein. Solution structure of a complex with prostaglandin B2. Eur. J. Biochem. 267, 1117-1124. [DOI] [PubMed] [Google Scholar]
- Thoma, S., Kanedo, Y., and Somerville, C. (1993). A non-specific lipid transfer protein from Arabidopsis is a cell wall protein. Plant J. 3, 427-436. [DOI] [PubMed] [Google Scholar]
- Turner, J.G., Ellis, C., and Devoto, A. (2002). The jasmonate signal pathway. Plant Cell 14, S153-S164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Loon, L.C., and Van Strien, E.A. (1999). The family of pathogenesis-related proteins, their activities, and comparative analysis of PR1-type proteins. Physiol. Mol. Plant Pathol. 55, 85-97. [Google Scholar]
- Vauthrin, S., Mikès, V., Milat, M.-L., Ponchet, M., Maume, B., Osman, H., and Blein, J.-P. (1999). Elicitins trap and transfer sterols from micelles, liposomes and plant plasma membranes. Biochim. Biophys. Acta 1419, 335-342. [DOI] [PubMed] [Google Scholar]
- Wasternack, C., and Parthier, B. (1997). Jasmonate-signalled plant gene expression. Trends Plant Sci. 2, 302-307. [Google Scholar]
- Wendehenne, D., Binet, M.-N., Blein, J.-P., Ricci, P., and Pugin, A. (1995). Evidence for specific, high-affinity binding sites for a proteinaceous elicitor in tobacco plasma membrane. FEBS Lett. 374, 203-207. [DOI] [PubMed] [Google Scholar]
- Yubero-Serrano, E.M., Moyano, E., Medina-Escobar, N., Munoz-Blanco, J., and Caballero, J.-L. (2003). Identification of a strawberry gene encoding a non-specific lipid transfer protein that responds to ABA, wounding and cold stress. J. Exp. Bot. 54, 1865-1877. [DOI] [PubMed] [Google Scholar]
- Zachowski, A., Guerbette, F., Grosbois, M., Jolliot-Croquin, A., and Kader, J.-C. (1998). Characterisation of acyl binding by a plant lipid-transfer protein. Eur. J. Biochem. 257, 443-448. [DOI] [PubMed] [Google Scholar]