Abstract
The mechanisms underlying completion of cytokinesis are still poorly understood. Here, we show that the Drosophila orthologue of mammalian Citron kinases is essential for the final events of the cytokinetic process. Flies bearing mutations in the Drosophila citron kinase (dck) gene were defective in both neuroblast and spermatocyte cytokinesis. In both cell types, early cytokinetic events such as central spindle assembly and contractile ring formation were completely normal. Moreover, cytokinetic rings constricted normally, leading to complete furrow ingression. However late telophases of both cell types displayed persistent midbodies associated with disorganized F actin and anillin structures. Similar defects were observed in dck RNA interference (RNAi) telophases, which, in addition to abnormal F actin and anillin rings, also displayed aberrant membrane protrusions at the cleavage site. Together, these results indicate that mutations in the dck gene result in morphologically abnormal intercellular bridges and in delayed resolution of these structures, suggesting that the wild-type function of dck is required for abscission at the end of cytokinesis. The phenotype of Dck-depleted cells is different from those observed in most Drosophila cytokinesis mutants but extraordinarily similar to that caused by anillin RNAi, suggesting that Dck and anillin are in the same pathway for completion of cytokinesis.
INTRODUCTION
Cytokinesis is the complex process by which two daughter cells physically separate at the end of mitosis or meiosis. In animal cells, cytokinesis is mediated by an actomyosin-based structure called contractile ring that assembles just beneath the equatorial cortex of the dividing cell. Constriction of this ring mediates furrow ingression, whereas membrane addition at the cleavage site ensures both proper furrow invagination and the final separation (abscission) of the daughter cells (Glotzer, 2003).
Genetic and biochemical analyses have identified many proteins required for contractile ring formation and regulation. These proteins include structural components of the ring such as actin, myosin II, the regulatory light chain of myosin II (RMLC), and anillin (reviewed by Straight and Field, 2000; Glotzer 2001). In addition, ring formation is regulated by Rho, its upstream activator Rho GEF, and its downstream effector Diaphanous (Dia), which interacts with profilin (reviewed by Wasserman, 1998; Prokopenko et al., 2000; Glotzer, 2001). Another protein that controls ring behavior is cofilin, a small actin-binding protein required for contractile ring disassembly (Gunsalus et al., 1995).
Completion of animal cell cytokinesis also relies on the central spindle (reviewed by Gatti et al., 2000; Glotzer, 2001), the dense network of overlapping antiparallel microtubules (MTs) that forms during late anaphase between the separating daughter nuclei (Mastronarde et al., 1993). Proper central spindle assembly depends on several proteins that accumulate at the central spindle midzone. These include plus-end–directed kinesin-like proteins, such as Drosophila Klp3A and the orthologues CHO1/MKLP1 in mammals, Pavarotti (Pav) in Drosophila, and ZEN-4 in Caenorhabditis elegans (Nislow et al., 1992; Williams et al., 1995; Adams et al., 1998; Powers et al., 1998; Raich et al., 1998). Pav, ZEN-4 and MKLP1 interact with orthologous Rho family GTPase-activating proteins (Drosophila RacGAP50C, C. elegans CYK-4, and human HsCYK-4), forming an evolutionarily conserved complex called centralspindlin that has MT bundling activity in vitro and is required for cytokinesis (Mishima et al., 2002; Somers and Saint, 2003). Other proteins that accumulate at the central spindle midzone are the orthologues PRC1 in mammals and Fascetto (Feo) in Drosophila, which also have MT bundling activity and are required for both central spindle formation and cytokinesis (Mollinari et al., 2002; Vernì et al., 2004).
Studies in Drosophila indicate that the central spindle MTs interact with the actomyosin ring throughout cytokinesis (Giansanti et al., 1998, 2004; Gatti et al., 2000; Somma et al., 2002). Mutants in the klp3A, pav, and RacGAP50C genes lack not only the central spindle but also the actomyosin ring (Williams et al., 1995; Adams et al., 1998; Giansanti et al., 1998; Somma et al., 2002). Mutations in rho1, pebble (pbl, a Rho GEF-coding gene), spaghetti squash (sqh, an RMLC-coding gene), diaphanous (dia), and chickadee (chic, a profilin-coding gene), all of which encode proteins involved in actomyosin ring formation and function, similarly disrupt the assembly of both the central spindle and the contractile ring (Giansanti et al., 1998, 2004; Prokopenko et al., 1999; Somma et al., 2002). The central spindle and the actomyosin ring thus seem to be mutually dependent structures, but the molecular basis for their interactions remains poorly understood.
In most animal cells, cleavage furrow ingression ensures an almost complete separation of daughter cells, which nevertheless remain interconnected for variable times by a cytoplasmic bridge, until abscission takes place (reviewed by Schweitzer and D'Souza-Schorey, 2004). It has been proposed that completion of cytokinesis involves three distinct steps: stabilization of the intercellular bridge, contractile ring disassembly, and membrane remodeling for abscission (Schweitzer and D'Souza-Schorey, 2004). However, only a few proteins have been described to date that are specifically required for these final stages of cytokinesis. This is not surprising, because most mutants that disrupt cytokinesis seem to affect earlier steps of the process. For example, the analysis of mutants in 22 different Drosophila genes required for spermatocyte cytokinesis yielded only two mutants defective in contractile ring disassembly; the remaining 20 mutants displayed defects in earlier steps of the process such as ring and central spindle formation or ring constriction (Giansanti et al., 2004). Thus, the molecular mechanisms underlying the last steps of cytokinesis remain largely obscure. To the best of our knowledge, no mutations have been identified to date that specifically affect intercellular bridge resolution.
Here, we show that mutations in the Drosophila citron kinase (dck) gene specifically disrupt the final stages of daughter cell separation. Citron kinase belongs to an evolutionarily conserved family of serine-threonine kinases. In vertebrate cells, this kinase is a target molecule for activated Rho (Rho GTP), accumulates at the cleavage furrow, and is required for cytokinesis (Madaule et al., 1998; Di Cunto et al., 2000, 2002; Madaule et al., 2000). However, studies on mammalian systems did not define the primary defect that disrupts cytokinesis in Citron kinase mutant cells. We have characterized the cytological defects elicited by mutations in the dck gene and performed dck RNAi in S2 tissue culture cells. Our results indicate that mutant neuroblasts and spermatocytes, and dck RNAi S2 cells can assemble normal cytokinetic structures, which behave normally for most of the cytokinetic process. These three cell types show defects only in late telophases, suggesting problems in abscission.
MATERIALS AND METHODS
Drosophila Stocks
The Drosophila citron kinase2 (dck2) mutant allele was isolated from a collection of 1600 ethylmethanesulfonate (EMS)-induced third chromosome late lethals, generated in Charles Zuker's laboratory (University of California, San Diego, CA). A cytological screen of larval brain preparations from these lethals, carried out in collaboration with Mike Goldberg's laboratory (Cornell University, Ithaca, NY), yielded ∼150 mitotic mutants, one of which is dck2. The dck1 allele, also called l(3)7m-62 or sticky1 (sti1), is described by Gatti and Baker (1989) and by FlyBase (http://flybase.bio.indiana.edu/). The dck3 allele is a P element insertion into the CG10522 gene designated as KG01697 (FlyBase); both the KG01697 stock and the third chromosome deficiency kit used for mapping dck were provided by the Bloomington Stock Center, Indiana University (Bloomington, IN). The dck mutations and the third chromosome deficiencies were maintained over the TM6B, Hu e Tb ca balancer. Homozygous and hemizygous mutant larvae were selected based on their non-Tubby phenotype. The Oregon R laboratory stock was used as wild-type control.
Antibodies, Western Blotting, and DNA Analysis
Tubulin was detected using a commercial monoclonal antibody (Sigma-Aldrich, St. Louis, MO). Rabbit anti-anillin401–828 (Field and Alberts, 1995), rabbit anti-Feo1–148 (Vernì et al., 2004), and rabbit anti-myosin (kindly provided by Chris Field, Harvard Medical School, Boston MA) have been described previously (Giansanti et al., 2001b; Vernì et al., 2004). To obtain a polyclonal antibody against Dck, the sequence corresponding to aa 471–600 of the CG10522-encoded protein was cloned into the BamHI site of pMAL-C2 vector. The MBP-Dck fusion protein was then injected into rabbits and the immune serum was first adsorbed on an MBP-CNBr-Sepharose column, and then affinity purified on an MBP-Dck CNBr-Sepharose column. Immunoblotting was performed according to Somma et al. (2002); the anti-Dck and the anti-tubulin antibodies were diluted 1:1000 and 1:2000, respectively. Genomic DNA was isolated by standard methods, amplified by polymerase chain reaction (PCR) and sequenced using an automatic DNA sequencer.
Cytology and Immunostaining
Aceto-orcein–stained chromosome preparations were obtained according to Gatti and Goldberg (1991). The mitotic parameters in control and mutant brains were estimated as described by Gatti and Baker (1989). For immunofluorescence experiments, brains were dissected from third instar larvae and fixed according to Bonaccorsi et al. (2000). For tubulin and either anillin, myosin, Feo, or Dck immunostaining, testes were dissected from third instar larvae or pupae and fixed according to Cenci et al. (1994); for tubulin or anillin immunostaining plus actin staining by phalloidin testes were fixed according to Gunsalus et al. (1995). S2 cells were fixed as described by Somma et al. (2002).
Brain, testis, and S2 cell preparations, after several rinses in phosphate-buffered saline (PBS), were incubated overnight at 4°C with any of the following rabbit primary antibodies diluted in PBS with 1% bovine serum albumin: anti-anillin (1:300), anti-myosin (1:250), anti-Feo (1:200), or anti-Dck (1:100). After two rinses in PBS (5 min each), primary antibodies were detected by a 1-h incubation at room temperature with Alexa 555-conjugated anti-rabbit IgG (Molecular Probes, Eugene, OR) diluted 1:400 in PBS. Slides were then incubated for 1 h at room temperature with a monoclonal anti-α-tubulin antibody (Sigma-Aldrich) diluted 1:200 in PBS, which was detected by Alexa 488-conjugated anti-mouse (Molecular Probes) diluted 1:400 in PBS. For anillin plus actin staining, testis preparations were immunostained for anillin as described above, rinsed in PBS, and then incubated for 2 h at 37°C with rodhamine-phalloidin (Molecular Probes) according to Gunsalus et al. (1995). Immunostained preparations were all mounted in the Vectashield medium H-1200 containing the DNA dye 4,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Burlingame, CA).
All preparations were examined using an Axioplan (Carl Zeiss, Jena, Germany) microscope equipped with an HBO 50-W mercury lamp for epifluorescence and with a cooled charge-coupled device (Photometrics, Tucson, AZ). DAPI, Alexa 555, and Alexa 488 fluorescence were detected using the 0.1, 15, and 0.9 filter sets (Carl Zeiss), respectively. Grayscale images were collected separately using the IPLab Spectrum software. Images were then converted to Photoshop (Adobe Systems, Mountain View, CA), pseudocolored, and merged.
RNA Interference (RNAi)
S2 cell cultures, double-stranded RNA (dsRNA) production and purification, and RNAi treatments were carried out as described previously (Somma et al., 2002). The primers used in the PCR reactions were 35 nt in length and contained a 5′ T7 RNA polymerase binding site (5′-TAATACGACTCACTATAGGGAGG-3′) flanked by CG10522-specific sequences (FlyBase ID FBgn0036295). The sense and antisense CG10522-specific sequences were TGGAGCTGAAGA and CGCTTGCAAATG, respectively.
RESULTS
Isolation of Mutations in the Drosophila dck Gene
We identified the Drosophila dck2 mutant allele by screening a collection of 1600 late lethal mutations induced by EMS in C. Zuker laboratory (see Materials and Methods). The basis of this screen is that homozygous mutants in essential mitotic genes can often survive until the end of the larval period by using the materials stored into the egg laid by their heterozygous mothers (Gatti and Baker 1989, Gatti and Goldberg 1991). Examination of orcein-stained larval brain squashes from dck2 homozygotes revealed the presence of a high frequency of polyploid cells (Table 1 and Supplemental Figure 1). Deficiency mapping showed that dck2 is uncovered by Df(3L)iro-2, Df(3L)F10, and Df(3L)E44, suggesting that the mutation maps to the polytene chromosome interval 69C; 69D1-2. Complementation tests between dck2 and known mutations located within this interval revealed that dck2 is allelic to both l(3)7m62 (henceforth called dck1) and to l(3)KG01697 (henceforth called dck3). l(3)7m62 is an EMS-induced lethal mutation that exhibits frequent polyploid cells in larval brains (Gatti and Baker, 1989); l(3)KG01697 is another lethal mutation induced by a P-element insertion within the 5′untranslated region (UTR) of the predicted gene CG10522 (FlyBase).
Table 1.
Frequency of polyploid cells and anaphase figures in dck mutant brains
| No. of metaphasesa
|
No. of anaphases
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Genotype | No. of cells scored | Diploid | Polyploid | Diploid | Polyploid | Polyploid figures (%) | Anaphases (%) | Mitotic indexb |
| dck1/dck1 | 254 | 81 | 127 | 19 | 27 | 60.6 | 18.1 | 0.68 |
| dck2/dck2 | 273 | 152 | 77 | 38 | 6 | 30.4 | 16.1 | 0.55 |
| dck2/Df(3L)E44 | 373 | 131 | 169 | 42 | 31 | 53.6 | 19.6 | 0.94 |
| dck3/Df(3L)E44 | 385 | 272 | 48 | 55 | 10 | 15.1 | 16.9 | 0.76 |
| Oregon R (control) | 282 | 227 | 0 | 55 | 0 | 0 | 19.5 | 0.70 |
This class includes both prometaphases and metaphases
The mitotic index corresponds to the average number of divisions per optic field. The optic field used in this analysis is the circular area seen under a phase contrast Neofluar 100× oil-immersion Zeiss objective, by using 10× oculars and the Optovar set at 1.25
Cytological analysis of squashed brain preparations from third instar larvae of dck1, dck2, and dck3 mutants showed that these mutations result in high proportions of polyploid cells (Table 1 and Supplemental Figure 1). Some of these polyploid cells were very big and contained >1000 chromosomes (our unpublished data; see Gatti and Baker, 1989). Chromosomes of mutant cells seemed to be morphologically normal, with no evidence of irregular chromosome condensation or chromosome breakage (our unpublished data; Gatti and Baker, 1989). The mitotic index, a parameter measuring the frequency of cells engaged in mitosis, was similar in mutant and in wild-type control brains (Table 1). The percentages of mitotic cells undergoing anaphase in dck mutants and in wild-type were also comparable (Table 1). Thus, the cytological phenotypes observed in dck mutants are very similar to those elicited by mutations that disrupt neuroblast cytokinesis (Karess et al., 1991; Castrillon and Wasserman, 1994; Gunsalus et al., 1995) and together strongly suggest that the wild-type function of dck is required for this process.
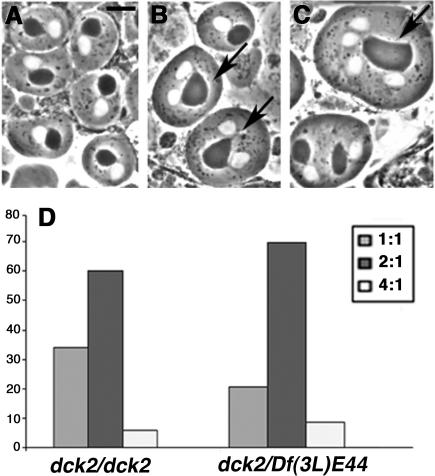
Further evidence that the wild-type function of dck is required for cytokinesis is provided by in vivo analysis of mutant spermatids. In wild-type, each newly formed (onion stage) spermatid consists of a round, phase-light nucleus associated with an equally sized, phase-dark mitochondrial derivative called nebenkern. Mutants that disrupt cytokinesis abrogate proper mitochondria partition between the daughter cells resulting in aberrant spermatids containing an abnormally large nebenkern associated with either two or four equally sized nuclei (Fuller, 1993). Cytological analysis of testes from dck3/dck3 and dck3/Df(3L)E44 males did not reveal defective spermatids, consistent with the relatively weak effect of the dck3 mutation on brain cell division (Table 1). All the mutant allelic combinations involving dck1 caused a strong proliferation defect of male germ cells, resulting in tiny testes virtually devoid of dividing spermatocytes. However, testes from dck2/dck2 and dck2/Df(3L)E44 males consistently exhibited both dividing spermatocytes and spermatids. A high proportion of these spermatids contained an abnormally large nebenkern associated with two or four normal-sized nuclei (Figure 1), a phenotype reflecting cytokinesis failures in either one or both meiotic divisions (Fuller, 1993; Giansanti et al., 2004).
Figure 1.
Abnormal spermatids observed in living dck2 mutant testes. (A) Wild-type spermatids showing nuclei (white circles) and nebenkern (dark circles) of similar sizes. (B and C) Abnormal spermatids in dck mutant males. (B) Two abnormal spermatids (arrows) with two nuclei of similar sizes associated with a single nebenkern which is twice the size of a normal nebenkern. (C) An abnormal spermatid containing four equally sized nuclei associated with only one nebenkern that is 4 times larger than a regular nebenkern (arrow). Bar, 10 μm. (D) Frequencies of abnormal spermatids in dck2/dck2 and dck2/Df(3L)E44 mutant males; 2:1 and 4:1 aberrant spermatids are shown in B and C, respectively. In wild-type males, the frequency of abnormal spermatids is virtually zero.
Molecular Analysis of dck Mutants
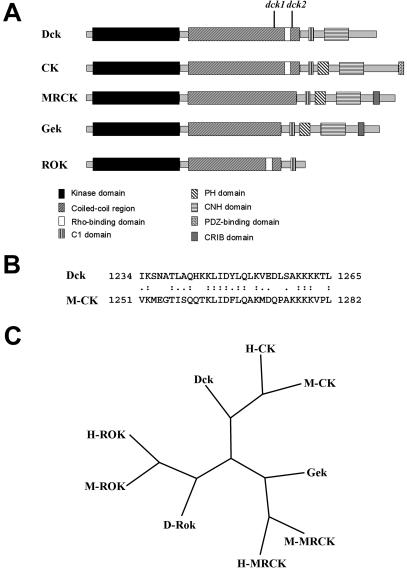
The dck3 mutant allele carries a P-element insertion within the 5′UTR of the predicted gene CG10522 (FlyBase). Additional evidence that dck corresponds to CG10522 is provided by the molecular analysis of the dck1 and dck2 mutant alleles. We compared the sequences of the CG10522 genes from these mutants with both the CG10522 sequence reported in FlyBase and the sequence of the CG10522 gene from an Oregon R wild-type stock. This analysis revealed that the dck1 allele contains a T-A transvection at nucleotide 4013, resulting in a stop codon within exon 5 of the gene; the dck2 mutant allele contains a C-T transition at nucleotide 4606 that introduces a stop codon within exon 6 of the CG10522 gene. Thus, the dck1 and dck2 mutations result in truncated proteins of 1092 and 1264 aa, respectively (Figure 2A).
Figure 2.
Organization and evolutionary conservation of Dck. (A) Schematic representation of domain organization of the Dck protein compared with mammalian CKs, mammalian MRCK, Drosophila Gek, and mammalian ROKs. dck1 and dck2 indicate the position of the stop codons found in the corresponding mutants. The domains of Dck and evolutionarily related proteins were determined by scanning the sequence with the SMART program (Schultz et al., 1998). (B) Sequence alignment of the putative Rho binding domain of Dck with its mouse homologous sequence. The putative Rho binding domain was identified by aligning the Dck and M-CK sequences with the Smith-Waterman algorithm by using the BLOSUM 80 substitution matrix. (C) Phylogenetic tree representing the position of Dck compared with the other Drosophila (D), human (H) and mouse (M) family members. The phylogenetic analysis was performed using the maximum parsimony algorithm in the PHYLIP suite. The tree represents the consensus of a bootstrap analysis performed with 1000 replicas.
The predicted CG10522 gene encodes a 1854-aa protein that shares homology with the conserved family of Rho-interacting serine/threonine kinases. This family includes the Rho-kinases (ROKs), the myotonin-related CDC42 binding kinase (MRCK; Leung et al., 1998), the Drosophila Gengis Khan protein (Gek; Luo et al., 1997), and mammalian Citron kinases (CKs; Madaule et al., 1995, 2000; Di Cunto et al., 1998). The domain organization of the CG10522-encoded polypeptide is very similar to Drosophila Gek and mammalian MRCK and CK proteins (Figure 2; Madaule et al., 1998). gek mutations exhibit abnormal accumulation of F-actin in Drosophila oocytes, suggesting that Gek is a Cdc42 effector for the regulation of actin polymerization (Luo et al., 1997). Moreover, analysis of gek mutants and RNAi experiments indicated that Gek is not required for cytokinesis (Wakefield, Somma, Bonaccorsi, and Gatti, unpublished data). Mammalian MRCK is another Cdc42 effector, which has been involved in the regulation of actin organization and filopodia extension but not cytokinesis (Leung et al., 1998). In contrast, CK is a Rho-effector protein required for mammalian cell cytokinesis (Madaule et al., 1998). It is encoded by a complex transcription unit that also produces Citron-N, a neuronal-specific isoform that binds the PDZ domain of synapsis-associated proteins such as PSD-95 (Furuyashiki et al., 1999; Zhang et al., 1999). However, the PDZ binding domain does not seem to be required for cytokinesis (Madaule et al., 1998).
The CG10522-encoded polypeptide, Gek, MRCK, and CK are all characterized by an N-terminal ser/thr kinase domain, a long coiled-coil region, a lipid binding (C1) domain, and a citron homology domain (Figure 2A). However, these proteins differ for the presence/absence of several domains. The CG10522-encoded polypeptide lacks a pleckstrin homology (PH) domain that is present in the Gek, CK, and MRCK proteins (Figure 2A). PH domains are found in membrane-associated proteins and have been implicated in protein–protein and protein–phospholipid interactions (reviewed by Rebecchi and Scarlata, 1998). MRCK and Gek are characterized by the presence of a carboxy terminal CDC42-binding CRIB domain that is absent in both CKs and the CG10522-encoded protein. In contrast, the region encompassing amino acids 1234–1265 of the CG10522-encoded protein shows high sequence similarity with the unique Rho-binding domain of CKs (Figure 2B) but has no sequence homology to either Gek or MRCKs. Further evidence for a close similarity between mammalian CKs and the protein product of the CG10522 gene is provided by a phylogenetic analysis of their kinase domains (Figure 2C). This analysis showed that the kinase domain of the CG10522-encoded protein is closely related to CKs, whereas the Gek kinase domain is similar to that of MRCK (Figure 2C). Thus, the functional and structural features of these proteins indicate that the CG10522-encoded protein is the fly orthologue of mammalian CKs, whereas Gek is the orthologue of MRCKs. We have therefore named the CG10522 gene dck.
Characterization of dck Mutant Phenotype
To obtain insight into the primary defect that disrupts cytokinesis in dck mutants, we stained brain preparations from dck2/dck2, dck1/dck2, and dck2/Df(3L)E44 mutant larvae for tubulin, DNA, and either myosin II, anillin, or Fascetto. Myosin II is a well-known component of the contractile ring; anillin is an actin-binding protein that accumulates in the cleavage furrow and is thought to mediate interactions between the contractile ring and the plasma membrane (Field and Alberts 1995; Giansanti et al., 1999; Somma et al., 2002). Feo, the Drosophila orthologue of human PRC1, is an MT-binding protein that concentrates at the central spindle midzone and is required for central spindle assembly (Vernì et al., 2004). A preliminary analysis of dck2/dck2, dck1/dck2, and dck2/Df(3L)E44 mutant brains revealed almost identical mutant phenotypes. We thus focused on dck2/Df(3L)E44 mutants for a detailed phenotypic analysis.
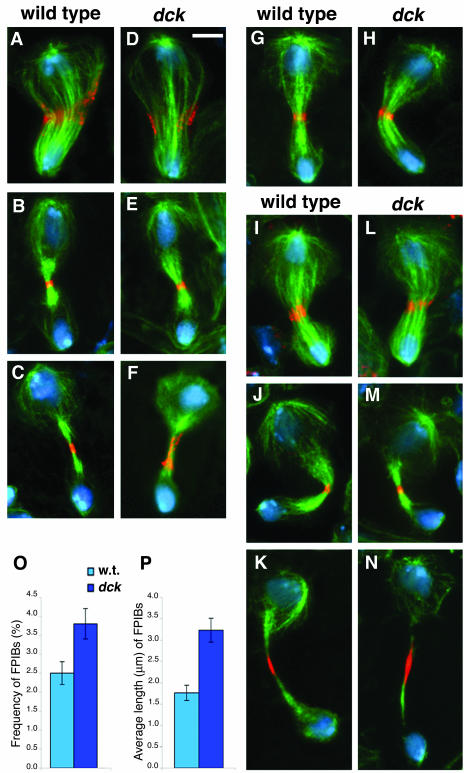
Drosophila brains from third instar larvae mostly contain neuroblasts (NBs) and ganglion mother cells (GMCs). NBs are stem cells that divide asymmetrically to give rise to another NB and to a smaller GMC. Wild-type NBs exhibit prominent asters at metaphase but the astral MTs associated with the presumptive GMC shorten dramatically during ana-telophase. Concomitantly, the GMC-associated centrosome becomes much smaller than the NB-associated one, and the central spindle is displaced toward the GMC nucleus (Giansanti et al., 2001b). Contractile ring components such as myosin II and anillin concentrate around the central spindle midzone, so that at telophase the cytokinetic ring is asymmetrically located between the two daughter cells (Figure 3). In late NB telophases, the myosin II and anillin signals are restricted to the cleavage area. After complete furrow ingression, the NB and the GMC remain physically connected by an intercellular bridge until abscission takes place. This intercellular bridge, henceforth designated as late midbody, often displays an anillin signal at its center. This signal is consistently more extended than that seen in telophases at earlier stages of cytokinesis, suggesting that anillin spreads along the midbody after completion of furrow ingression (Figure 3, compare B and C).
Figure 3.
Phenotypic analysis of dck mutant neuroblasts. (A–F) Wild-type (A–C) and dck mutant (D–F) telophases stained for tubulin (green), anillin (orange), and DNA (blue). A and D, early telophases; B and E, late telophases; C and F, very late telophases. Note the extended anillin signal associated with the midbody of the dck telophase figure shown in F. (G and H) Wild-type (G) and dck (H) mid/late telophases stained for tubulin (green), myosin II (orange), and DNA (blue). (I–N) Wild-type (I–K) and dck mutant (L-N) telophases stained for tubulin (green), Feo (orange), and DNA (blue). I and L, mid-telophases; J and M, late telophases; K and N, very late telophases. Note the extended Feo signals associated with late midbodies (K and N). Bar, 5 μm. (O) Frequency of Feopositive intercellular bridges (FPIBs, no. of bridges/no. interphase nuclei) ± SE in wild-type (w. t.) and dck mutant brains; p < 0.05. (P) Average length of FPIBs ± SEM in wild-type (w. t.) and dck mutant brains; p < 0.01. Note that the Feo-positive intercellular bridges are both longer and more frequent in dck mutant brains than in wild-type brains.
Although dck mutant brains displayed high frequencies of polyploid cells, mutant NBs (diploid) did not show any detectable defect in cell division until the very late steps of cytokinesis. In dck mutant NBs, metaphase and anaphase spindles were normal; telophase figures were also normal and showed regular central spindles displaced toward the GMC nucleus (Figure 3). In mutant telophases, anillin and myosin II were normally accumulated around the central spindle midzone and the cytokinetic ring was regularly constricted (Figure 3). The maximum degree of constriction achieved by the contractile ring of mutant NBs was comparable with that observed in wild-type cells, suggesting that mutant NBs are not defective in furrow ingression. However, in very late telophases of mutant NBs, the anillin signals associated with the intercellular bridges were often substantially more extended than in wild-type (Figure 3, compare C and F), suggesting a defect in completion of cytokinesis.
Immunostaining with an anti-Feo antibody confirmed that mutant NBs are defective in the very late steps of cytokinesis. Feo localization in mutant NBs was comparable to wild-type controls during most stages of NB division. Feo normally associated with central spindle midzone of anaphase and telophase figures of mutant NBs (Figure 3, I–N). However, the late midbodies of mutant NBs displayed Feo signals significantly longer than those associated with their wild-type counterparts (Figure 3, compare K and N, Figure 3P, and Supplemental Figure 2). In addition, the frequency of the Feo-positive intercellular bridges in mutant brains was significantly higher than in controls (Figure 3O), suggesting that completion of cytokinesis in dck mutants takes longer than in wild type.
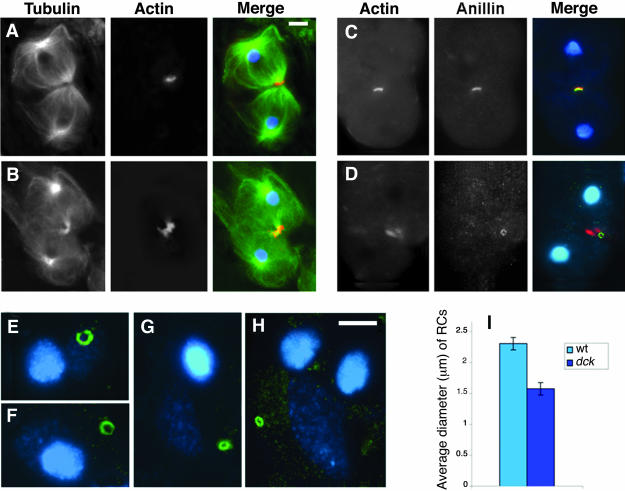
To further characterize the cytokinetic defect in dck mutants, we stained larval testis preparations for tubulin, DNA, and either actin, anillin, or Feo. Spermatocytes from dck2/dck2 and dck2/Df(3L)E44 males did not show any defect during most steps of meiotic division. They showed regular central spindles and regular actin and anillin rings, which constricted normally (our unpublished data). At the end of both meiotic divisions, however, very late telophase figures of dck mutant spermatocytes were often characterized by the presence of extended F-actin aggregates associated with the midbody (Figure 4B). Consistent with the observations on actin ring behavior, mutant spermatocytes stained for anillin were indistinguishable from their wild-type counterparts until the final stages of meiotic division (our unpublished data). However, late telophase figures often showed overconstricted anillin rings (Figure 4D), which resulted in comparatively small ring canals (Figure 4, E–H). The average diameter of the ring canals observed in mutant spermatid cysts was 1.6 ± 0.1 μm, whereas in Oregon R control this diameter was 2.3 ± 0.1 μm (Figure 4I).
Figure 4.
Cytological phenotype of dck mutant spermatocytes. (A and B) Late telophase I figures from wild-type (A) and dck mutant males (B) stained for tubulin (green), actin (orange), and DNA (blue). Note the extended actin ring associated with the midbody of the dck mutant cell. (C and D) Wild-type (C) and dck (D) late telophases stained for actin (red), anillin (green), and DNA (blue); overlapping of red and green signals results in a yellow color. (E–H) Examples of anillin-enriched ring canals observed in wild-type (E and F) and dck mutant spermatids (G and H); cells were stained for anillin (green) and DNA (blue). Bars, 5 μm. (I) Average diameters ± SEM of RCs from wild-type (w. t.) and dck mutant males; p < 0.01.
In contrast to the observations on NBs, dck mutant spermatocytes did not exhibit any detectable difference in Feo localization and behavior with respect to controls (our unpublished data).
Together, these results indicate that mutations in the dck gene lead to a defective distribution of some proteins associated with the late midbody and to a delay in the resolution of this structure, suggesting that the wild-type function of dck is specifically required for completion of cytokinesis.
Disruption of Dck by RNAi
To determine the consequences of complete ablation of the dck gene, we treated S2 tissue culture cells with dck dsRNA for either 72 or 96 h. These treatments caused a strong depletion of the Dck protein (Figure 6) and dramatic increases in the frequency of binucleated cells (Table 2), a phenotype diagnostic of failures in cytokinesis (Somma et al., 2002). These results are consistent with previous data showing that treatments with dck dsRNA disrupt cytokinesis in Drosophila cultured cells (Rogers et al., 2003; Kiger et al., 2003).
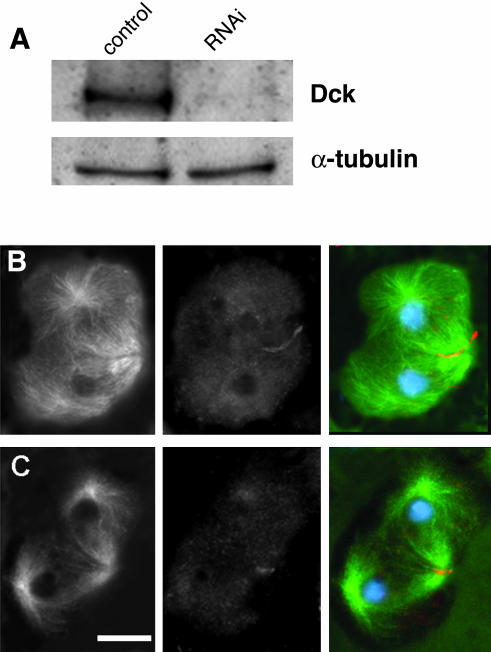
Figure 6.
Subcellular localization of the Dck protein. (A) Western blot from S2 cell extracts showing that the anti-Dck antibody recognizes a band of ∼210 kDa, which is absent in S2 cells treated for 96 h with dck dsRNA. The antibody reacted weakly with some additional bands of lower molecular weight, which did not disappear in RNAi cells (our unpublished data). Tubulin has been used as a loading control. (B and C) Wild-type primary spermatocytes stained for tubulin (green), Dck (orange), and DNA (blue); B, early telophase; C, late telophase. Note that Dck accumulates at the cleavage furrow. Bar, 10 μm.
Table 2.
Frequencies of mitotic figures and binucleated cells in control and dck RNAi cultures
| Frequencies relative to metaphases (%)
|
||||
|---|---|---|---|---|
| Treatment | No. of metaphasesa | Anaphases | Telophasesb | Binucleated cells (%)c |
| Control | 241 | 17.8 | 70.5 | 7.8 |
| dck RNAi 72 h | 120 | 20.0 | 100.0 | 30.8 |
| dck RNAi 96 h | 161 | 19.9 | 97.5 | 50.5 |
Includes both metaphases and prometaphases
Differences in the frequencies of telophases between control and RNAi cells are significative with the χ2 test (72 h, p < 0.05; 96 h, p < 0.05)
Cells (9105, 5687, and 4167) scored for control, RNAi 72 h, and RNAi 96 h, respectively
To characterize the cytokinesis defect caused by Dck depletion, RNAi cells were stained for tubulin, DNA, and either actin, anillin, or Feo. Cytological analysis showed that these RNAi cells display higher frequencies of telophases than untreated controls (Table 2), suggesting that Dck depletion increases the duration of telophase. In Dck-depleted cells, metaphase, anaphase, early and mid-telophase figures were all comparable with controls for both spindle and contractile ring morphology (our unpublished data). Late telophases showed morphologically normal central spindles but displayed a number of defects that were particularly evident in cells that seemed to be in very late stages of the cytokinetic process. These defects included disorganized actin and anillin rings (Figure 5, B, D, and E), abnormal membrane protrusions at the cleavage furrow (Figure 5, B, D, and E), and the absence of the dark band that marks the midzone of normal central spindles stained for tubulin (Figure 5, D and E). This dark band corresponds to the so-called Flemming body (Zeitlin and Sullivan, 2001), a proteinaceous matrix associated with the central spindle midzone, which prevents tubulin immunostaining (Sellitto and Kuriyama 1988; Matuliene and Kuriyama, 2002). Together, these results are consistent with observations on dck mutant NBs and spermatocytes and strongly suggest that the dck gene controls the last steps of cytokinesis.
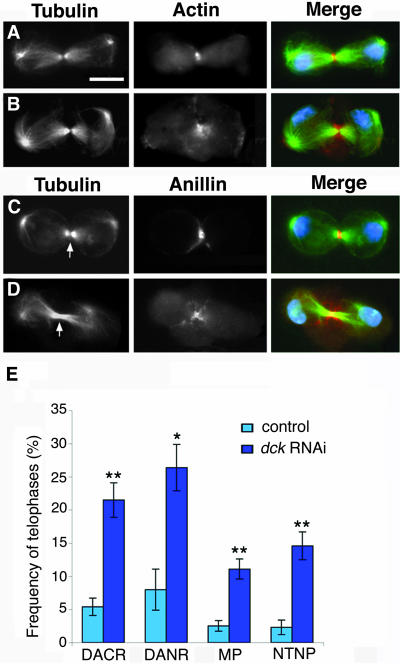
Figure 5.
Depletion of Dck by RNAi affects S2 cell cytokinesis. (A and B) Control (A) and dck RNAi (B) cells stained for tubulin (green), actin (orange), and DNA (blue). Note that the Dck-depleted cell displays a disorganized acting ring and membrane protrusions at the cleavage site. (C and D) Control (C) and dck RNAi (D) cells stained for tubulin (green), anillin (orange), and DNA (blue). The cell in D does not exhibit a tubulin negative band at the central spindle midzone (compare C and D, arrows) and shows an extended anillin signal and membrane protrusions at the cleavage site. Bar, 5 μm. (E) Frequencies (±SE) of telophases showing: disorganized actin rings (DACR), disorganized anillin rings (DANR), membrane protrusions at the cleavage site (MP), no tubulin negative band at the central spindle midzone (NTNB). These frequencies were calculated by examining 280, 75, 396, and 170 control telophases, and 256, 159, 459, and 277 dck RNAi telophases, respectively; *p < 0.005; **p < 0.001.
Intracellular Localization of Dck
To determine the intracellular localization of Dck, we raised an antibody against amino acid 471–600 of the protein. Western blot analysis of S2 cell extracts showed that this antibody recognizes a band of the expected size (mol. wt. = 211 kDa). This band was absent in extracts from cells treated for 96 h with dck dsRNA, confirming the specificity of the antibody (Figure 6A). Nonetheless, immunolocalization experiments in formaldehyde-fixed S2 cells resulted in a diffuse staining of both control and RNAi cells. Similar results were obtained with formaldehyde-fixed wild-type and mutant NBs. However, immunolocalization experiments on methanol-acetone–fixed testis preparations revealed that the Dck protein accumulates at the cleavage furrow during both meiotic divisions. Interestingly, Dck concentration at the furrow was detectable since late anaphase and persisted until late telophase (Figure 6, B and C). Spermatocytes from dck2/dck2 and dck2/Df(3L)E44 males did not exhibit any dectectable accumulation of Dck at the cleavage furrow. These results suggest that the formaldehyde fixation procedures used for both S2 and NB cell preparations do not preserve the Dck epitope(s) recognized by our antibody. However, this epitope(s) is probably preserved by fixation with methanol-acetone.
DISCUSSION
Sequence analysis has shown that the polypeptide encoded by the dck gene is closely related to mammalian Citron kinases. The Dck protein has an overall domain organization that is very similar to that of mammalian CKs and carries a domain with a high sequence similarity with the unique Rho-binding domain of these CKs (Figure 2). Consistent with this high degree of sequence homology, mammalian CKs and Drosophila Dck are both required for cytokinesis, suggesting that Dck is the fly orthologue of mammalian CKs.
We have analyzed the consequences of Dck depletion in larval NBs, S2 tissue culture cells, and spermatocytes. Spindle formation and chromosome segregation seemed to be completely normal in each cell type. In addition, the three cell types displayed regular central spindles and actomyosin rings, which constricted normally, leading to complete furrow ingression. However, each cell type showed a number of defects in late telophase figures and failed to complete cytokinesis.
Our results indicate that in larval NBs of dck mutants, there is an abnormal persistence and an altered morphology of the late midbody (intercellular bridge), suggesting that the wild-type function of dck is required for abscission. Late telophases of mutant NBs also exhibited equatorial anillin signals that were more extended than those seen in their wild-type counterparts. These extended anillin signals were never observed in early and mid-telophases of dck mutants, suggesting that anillin diffusion along the midbody is due to the disorganization of a previously well formed anillin ring.
Dck-depleted S2 cells showed cytokinesis phenotypes comparable with those observed in NBs. In dck RNAi cells, late telophases displayed disorganized and extended F actin and anillin rings and frequent membrane protrusions at the cleavage site. The defects in the actin and anillin rings were not observed in early and mid-telophases, suggesting that Dck depletion affects the stability but not the assembly of these cytokinetic structures. In addition, the frequency of telophases in dck RNAi cells was higher than in controls, suggesting that completion of cytokinesis is delayed in the absence of the dck function.
The phenotype of dck mutant spermatocytes was partially different from that observed in both NBs and S2 cells, but the cytokinesis defects were restricted to the late stages of the process as in mitotic cells. Late spermatocyte telophases often exhibited extended and disorganized actin rings similar to those seen in NB and S2 cell telophases. However, spermatocyte late telophases never showed extended equatorial anillin signals. Instead, these cells displayed overconstricted anillin rings, which resulted in ring canals smaller than their wild-type counterparts. The phenotypic difference in the anillin ring morphology between somatic and meiotic cells may reflect the peculiar cytokinetic process in spermatocytes. In Drosophila gonial cells and spermatocytes cytokinesis is incomplete, and arrested contractile rings develop into ring canals that are highly enriched in anillin (Hime et al., 1996; Giansanti et al., 1999). It is thus likely that in spermatocyte telophases of dck mutants anillin is recruited for the ring canal assembly pathway and does not accompany actin in its diffusion along the midbody.
It is clear that all cytokinesis defects caused by depletion of the Dck protein are restricted to late telophases. In addition, most of these defects seem to be due to a progressive disorganization of structures that formed and behaved normally in earlier stages of cytokinesis. These observations raise the question of whether these defects are the cause of cytokinesis disruption or merely the consequence of delayed abscission. For example, one can imagine that a late deformation of the F actin ring can interfere with the abscission process, preventing the final separation of the daughter cells. Alternatively, it is conceivable that the failure of mutant cells to complete cytokinesis can lead to an abnormal persistence of the intercellular bridge and to a progressive disorganization of the contractile apparatus. Our current results do not allow discrimination between these alternatives. It is also possible that some of the observed phenotypes are indeed responsible for failure in cytokinesis, whereas others are the consequence of the abnormal persistence of the intercellular bridge. dck mutant spermatocytes displayed a cytokinesis defect that was never observed in other mutants that disrupt meiotic cytokinesis of Drosophila males. The phenotypic analysis of mutants in 22 cytokinesis genes revealed that they can be subdivided into four different classes: genes required for central spindle assembly, anillin localization, and F actin ring formation; genes required for both central spindle and F actin ring formation but not for anillin localization; genes required for F actin ring constriction; and genes required for actin ring disassembly. The phenotype of dck mutants is similar to that elicited by mutations in the twinstar (tsr) and bird nest soup (bns) genes, which are required for actin ring disassembly. However, in tsr and bns mutant spermatocytes the actin ring not only fails to disassemble but also overgrows, forming a large and persistent actin aggregate (Gunsalus et al., 1995; Giansanti et al., 2004). In dck spermatocytes, F actin rings do not seem to overgrow, but only exhibit a late disorganization, consisting in a diffusion of actin along the midbody. Thus, dck mutants identify a fifth class of genes involved in spermatocyte cytokinesis: those required for the very late events of the process.
The phenotype of Dck-depleted cells is also different from most phenotypes observed after RNAi for cytokinesis genes. For example, cells depleted of either the Pavarotti kinesin-like protein, the Rho1 GTPase, the Rho GTPase activating protein RacGap50, the Rho GEF encoded by pbl, the RMLC encoded by sqh, Syntaxin1, or the Drosophila homologue of PRC1 encoded by feo all exhibit defective central spindles and contractile rings (Somma et al., 2002; Vernì et al., 2004). Cells depleted of the cofilin encoded by the tsr gene exhibit normal central spindles and contractile rings in early and late telophases, but these rings overgrow, resulting in large masses of F actin that remain associated with the cleavage site in late telophases (Somma et al., 2002). However, the phenotype of Dck-depleted cells is extraordinarily similar to that caused by anillin RNAi. In anillin-depleted cells, late telophases exhibit disorganized actin rings and frequent membrane protrusions at the cleavage furrow, as well as central spindles lacking the dark band at their midzones (Somma et al., 2002; Somma, unpublished observations). Because anillin contains an actin-binding domain and a PH domain, these observations have led to the hypothesis that anillin interacts with both the plasma membrane and the actin-based contractile ring, regulating membrane–ring interactions during late stages of cytokinesis (Somma et al., 2002). The finding that anillin- and Dck-depleted cells display comparable phenotypes suggests these two proteins are in the same pathway for completion of S2 cell cytokinesis.
An elucidation of the role of Drosophila Citron kinase requires identification of its substrates. Studies on mammalian cells have shown that citron kinase phosphorylates the regulatory myosin light chain at both threonine-18 and serine-19 residues in vitro (Yamashiro et al., 2003). The Drosophila RMLC is phosphorylated at the threonine-20 and serine-21 residues, which are equivalent to threonine-18 and serine-19 of the mammalian protein, respectively. Mutant sqh genes in which both threonine-20 and serine-21 have been replaced by alanines, behave like severe sqh mutants and disrupt cytokinesis in female germ cells (Jordan and Karess, 1997). However, the analysis of spermatocytes and S2 cells has shown that the phenotypes elicited by sqh ablation are very different from those seen in Dck-depleted cells: the defects observed in sqh mutants are in early steps of cytokinesis such as central spindle and contractile ring formation (Giansanti et al., 2001a; Somma et al., 2002), whereas those associated with dck mutations pertain to the last steps of the process. The simplest interpretation of these results is that Drosophila citron kinase does not phosphorylate the Sqh protein. However, it is also possible that Sqh has a dual function during Drosophila cytokinesis: a Dck-independent function in the early stages of the process and a late function that requires phosphorylation by Dck.
Regardless of whether Sqh is a Dck substrate, the Dck targets that need to be phosphorylated in order to ensure completion of cytokinesis are likely to be very few. The finding that a cytokinesis defect comparable with that observed in dck mutant spermatocytes was not observed in any of the 22 cytokinesis mutants that have been characterized to date (Giansanti et al., 2004), strongly suggests that very few proteins are required for the final stages of cytokinesis. At the moment, the best candidate substrate for Dck is anillin, which possesses many serine and threonine residues that can be phosphorylated. Anillin and Dck colocalize at the spermatocyte cleavage furrow throughout ana-telophase, and the anillin depletion phenotype in S2 cells parallels the one caused by Dck depletion. Unfortunately, the NB and spermatocyte phenotypes observed in dck mutants could not be compared with those elicited by mutations in anillin-coding gene. The early lethality associated with scraps (scra), the only extant mutant in the anillin coding gene (FlyBase), has thus far precluded phenotypic analysis of mutant NBs and spermatocytes.
Supplementary Material
Acknowledgments
We are extremely grateful to Charles Zuker for the lethal stocks used to isolate the dck2 mutant allele, and to Barbara Fasulo and Patrizia Somma for help in the RNAi experiments. We also thank Chris Field for the anti-anillin and anti-myosin II antibodies. This work was supported by grants from Fondo per gli Investimenti della Ricerca di Base (RBNE01KXC9-004), Progetto Strategico Genomica Funzionale (L449/97), and Centro di Eccellenza di Biologia e Medicina Molecolare to M.G.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04–06–0536. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–06–0536.
The online version of this article contains supplementary material accessible through http://www.molbiolcell.org.
References
- Adams, R.R., Tavares, A.A., Salzberg, A., Bellen, H.J., and Glover, D.M. (1998). Pavarotti encodes a kinesin-like protein required to organize the central spindle and contractile ring for cytokinesis. Genes Dev. 12, 1483-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaccorsi, S., Giansanti, M.G., and Gatti, M. (2000). Spindle assembly in Drosophila neuroblasts and ganglion mother cells. Nat. Cell Biol. 2, 54-56. [DOI] [PubMed] [Google Scholar]
- Castrillon, D.H., and Wasserman, S.A. (1994). Diaphanous is required for cytokinesis in Drosophila and shares domains of similarity with the products of the limb deformity gene. Development 120, 3367-3377. [DOI] [PubMed] [Google Scholar]
- Cenci, G., Bonaccorsi, S., Pisano, C., Vernì, F., and Gatti, M. (1994). Chromatin and microtubule organization during premeiotic, meiotic and early postmeiotic stages of Drosophila melanogaster spermatogenesis. J. Cell Sci. 10, 3521-3534. [DOI] [PubMed] [Google Scholar]
- Di Cunto, F., Calautti, E., Hsiao, J.J., Ong, L., Topley, G., Turco, E., and Dotto, G.P. (1998). Citron rho-interacting kinase, a novel tissue-specific ser/thr kinase encompassing the Rho-Rac-binding protein Citron. J. Biol. Chem. 273, 29706-29711. [DOI] [PubMed] [Google Scholar]
- Di Cunto, F., Imarisio, S., Camera, P., Boitani, C., Altruda, F., and Silengo, L. (2002). Essential role of citron kinase in cytokinesis of spermatogenic precursors. J. Cell Sci. 115, 4819-4826. [DOI] [PubMed] [Google Scholar]
- Di Cunto, F., et al. (2000). Defective neurogenesis in citron kinase knockout mice by altered cytokinesis and massive apoptosis. Neuron 28, 115-127. [DOI] [PubMed] [Google Scholar]
- Field, C., and Alberts, B.M. (1995). Anillin, a contractile ring protein that cycles from the nucleus to the cell cortex. J. Cell Biol. 131, 165-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller, M. (1993). Spermatogenesis. In: Development of Drosophila, ed. A. Martinez-Arias and M. Bate, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 71-147.
- Furuyashiki, M., Fujisawa, K., Fujita, A., Madaule, P., Uchino, S., Mishina, M., Bito, H., and Narumiya, S. (1999). Citron, a Rho-target, interacts with PSD-95/SAP-90 at glutamatergic synapses in the thalamus. J. Neurosci. 19, 109-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti, M., and Baker, B.S. (1989). Genes controlling essential cell-cycle functions in Drosophila melanogaster. Genes Dev. 3, 438-453. [DOI] [PubMed] [Google Scholar]
- Gatti, M., Giansanti, M.G., and Bonaccorsi, S. (2000). Relationships between the central spindle and the contractile ring during cytokinesis in animal cells. Microsc. Res. Tech. 49, 202-208. [DOI] [PubMed] [Google Scholar]
- Gatti, M., and Goldberg, M.L. (1991). Mutations affecting cell division in Drosophila. Methods Cell Biol. 35, 543-585. [DOI] [PubMed] [Google Scholar]
- Giansanti, M.G., Bonaccorsi, S., Bucciarelli, E., and Gatti, M. (2001a). Drosophila male meiosis as a model system for the study of cytokinesis in animal cells. Cell Struct. Funct. 26, 631-639 [DOI] [PubMed] [Google Scholar]
- Giansanti, M.G., Bonaccorsi, S., and Gatti, M. (1999). The role of anillin in meiotic cytokinesis of Drosophila males. J. Cell Sci. 112, 2323-2334. [DOI] [PubMed] [Google Scholar]
- Giansanti, M.G., Bonaccorsi, S., Williams, B., Williams, E.V., Santolamazza, C., Goldberg, M.L., and Gatti, M. (1998). Cooperative interactions between the central spindle and the contractile ring during Drosophila cytokinesis. Genes Dev. 12, 396-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giansanti, M.G., Farkas, R.M., Bonaccorsi, S., Lindsley, D.L., Wakimoto, B.T., Fuller, M.T., and Gatti, M. (2004). Genetic dissection of meiotic cytokinesis in Drosophila males. Mol. Biol. Cell 15, 2509-2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giansanti, M.G., Gatti, M., and Bonaccorsi, S. (2001b). The role of centrosomes and astral microtubules during asymmetric division of Drosophila neuroblasts. Development 128, 1137-1145. [DOI] [PubMed] [Google Scholar]
- Glotzer, M. (2001). Animal cell cytokinesis. Annu. Rev. Cell Dev. Biol. 17, 351-386. [DOI] [PubMed] [Google Scholar]
- Glotzer, M. (2003). Cytokinesis: progress on all fronts. Curr. Opin. Cell Biol. 15, 684-690. [DOI] [PubMed] [Google Scholar]
- Gunsalus, K., Bonaccorsi, S., Williams, E., Vernì, F., Gatti, M., and Goldberg, M.L. (1995). Mutations in twinstar, a Drosophila gene encoding a Cofilin/ADF homologue, result in defects in centrosome migration and cytokinesis J. Cell Biol. 131, 1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hime, G.R., Brill, J.A., and Fuller, M.T. (1996). Assembly of ring canals in the male germ line from structural components of the contractile ring. J. Cell Sci. 109, 2779-2788. [DOI] [PubMed] [Google Scholar]
- Jordan, P., and Karess, R. (1997). Myosin Light Chain-activating phosphorylation sites are required for oogenesis in Drosophila. J. Cell Biol. 139, 1805-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karess, R.E., Edwards, K.A., Kulkarni, S., Aguilera, I., and Kiehart, D.P. (1991). The regulatory light chain of nonmuscle myosin is encoded by spaghetti-squash, a gene required for cytokinesis in Drosophila. Cell 65, 1177-1189. [DOI] [PubMed] [Google Scholar]
- Kiger, A., Baum, B., Jones, S., Jones, M., Coulson, A.C.E., and Perrimon, N. (2003). A functional genomic analysis of cell morphology using RNA interference. J. Biol. 2, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung, T., Chen, X.Q., Tan, I., Manser, E., and Lim, L. (1998). Myotonic dystrophy kinase-related Cdc42-binding kinase acts as a Cdc42 effector in promoting cytoskeletal reorganization. Mol. Cell. Biol. 18, 130-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, L., Lee, T., Tsai, L., Tang, G., Jan, L.Y., and Jan, Y.N. (1997). Gengis Khan (Gek) as a putative effector for Drosophila Cdc-42 and regulator of actin polymerization. Proc. Natl. Acad. Sci. USA 94, 12963-12968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madaule, P., Eda, M., Watanabe, N., Fujisawa, K., Matsuoka, T., Bito, H., Ishizaki, T., and Narumiya, S. (1998). Role of citron kinase as a target of small GTPase Rho in cytokinesis. Nature 394, 491-494. [DOI] [PubMed] [Google Scholar]
- Madaule, P., Furuyashiki, T., Eda, M., Bito, H., Ishizaki, T., and Narumiya, S. (2000). Citron, a Rho target that affects contractility during cytokinesis. Microsc. Res. Tech. 49, 123-126. [DOI] [PubMed] [Google Scholar]
- Madaule, P., Furuyashiki, T., Reid, T., Ishizaki, T., Watanabe, G., Morii, N., and Narumiya, S. (1995). A novel partner for the GTP-bound forms of rho and rac. FEBS Lett. 377, 243-248. [DOI] [PubMed] [Google Scholar]
- Mastronarde, D.N., McDonald, K.L., Dijng, R., and McIntosh, J.R. (1993). Interpolar spindle microtubules in PtK cells. J. Cell Biol. 123, 1474-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matuliene, J., and Kuriyama, R. (2002). Kinesin-like protein CHO1 is required for the formation of the midbody matrix and the completion of cytokinesis in mammalian cells. Mol. Biol. Cell 13, 1832-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima, M., Kaitna, S., and Glotzer, M. (2002). Central spindle assembly and cytokinesis require a kinesin-like protein/RhoGAP complex with microtubule bundling activity. Dev. Cell 2, 41-54. [DOI] [PubMed] [Google Scholar]
- Mollinari, C., Kleman, J.P., Jiang, W., Schoehn, G., Hunter, T., and Margolis, R.L. (2002). PRC1 is a microtubule binding and bundling protein essential to maintain the mitotic spindle midzone. J. Cell Biol. 157, 1175-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nislow, C., Lombillo, V.A., Kuriyama, R., and McIntosh, J.R. (1992). A plus-end-directed motor enzyme that moves antiparallel microtubules in vitro localizes to the interzone of mitotic spindles. Nature 359, 543-547. [DOI] [PubMed] [Google Scholar]
- Powers, J., Bossinger, O., Rose, D., Strome, S., and Saxton, W. (1998). A nematode kinesin required for cleavage furrow advancement. Curr. Biol. 8, 1133-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokopenko, S.N., Brumby, A., O'Keefe, L., Prior, L., He, Y., Saint, R., and Bellen, H.J. (1999). A putative exchange factor for Rho1 GTPase is required for initiation of cytokinesis in Drosophila. Genes Dev. 13, 2301-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokopenko, S.N., Saint, R., and Bellen, H.J. (2000). Untying the Gordian knot of cytokinesis. Role of small G proteins and their regulators. J. Cell Biol. 148, 843-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raich, W.B., Moran, A.N., Rothman, J.H., and Hardin, J. (1998). Cytokinesis and midzone microtubule organization in Caenorhabditis elegans require the kinesin-like protein ZEN-4. Mol. Biol. Cell 9, 2037-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebecchi, M.J., and Scarlata, S. (1998). Pleckstrin homology domains: a common fold with diverse functions. Annu. Rev. Biophys. Biomol. Struct. 27, 503-528. [DOI] [PubMed] [Google Scholar]
- Rogers, S.L., Wiedemann, U., Stuurman, N., and Vale, R.D. (2003). Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J. Cell Biol. 162, 1079-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, J., Milpetz, F., Bork, P., and Ponting, C.P. (1998). SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 95, 5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer, J.K., and D'Souza-Schorey, C. (2004). Finishing the job: cytoskeletal and membrane events bring cytokinesis to an end. Exp. Cell Res. 295, 1-8. [DOI] [PubMed] [Google Scholar]
- Sellitto, C., and Kuriyama, R. (1988). Distribution of a matrix component of the midbody during the cell cycle in Chinese hamster ovary cells. J. Cell Biol. 106, 431-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers, W.G., and Saint, R. (2003). A RhoGEF and Rho Family GTPase-Activating protein complex links the contractile ring to cortical microtubules at the onset of cytokinesis. Dev. Cell 4, 29-39. [DOI] [PubMed] [Google Scholar]
- Somma, M.P., Fasulo, B., Cenci, G., Cundari, E., and Gatti, M. (2002). Molecular dissection of cytokinesis by RNA interference in Drosophila cultured cells. Mol. Biol. Cell 13, 2448-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight, A.F., and Field, C.M. (2000). Microtubules, membranes and cytokinesis. Curr. Biol. 10, 760-770. [DOI] [PubMed] [Google Scholar]
- Vernì, F., Somma, M.P., Gunsalus, K.C., Bonaccorsi, S., Belloni, G., Goldberg, M.L., and Gatti, M. (2004). Feo, the Drosophila homologue of PRC1, is required for central spindle formation and cytokinesis. Curr. Biol. 14, 1569-1575. [DOI] [PubMed] [Google Scholar]
- Wasserman, S. (1998). FH proteins as cytoskeletal organizers. Trends Cell Biol. 8, 111-115. [DOI] [PubMed] [Google Scholar]
- Williams, B.W., Riedy, M.F., Williams, E.V., Gatti, M., and Goldberg, M.L. (1995). The Drosophila Kinesin-like protein KLP3A is a midbody component required for central spindle assembly and initiation of cytokinesis. J. Cell Biol. 129, 709-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro, S., Totsukawa, G., Yamakita, Y., Sasaki, Y., Madaule, P., Ishizaki, T., Narumiya, S., and Matsumura, F. (2003). Citron kinase, a Rho-dependent kinase, induces di-phosphorylation of regulatory light chain of myosin II. Mol. Biol. Cell 14, 1745-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlin, S.G., and Sullivan, K.F. (2001). Animal cytokinesis: breaking up is hard to do. Curr. Biol. 11, 514-516. [DOI] [PubMed] [Google Scholar]
- Zhang, W., Vazquez, L., Apperson, M., and Kennedy, M.B. (1999). Citron binds to PSD-95 at glutamatergic synapses on inhibitory neurons in the hippocampus. J. Neurosci. 19, 96-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.