Abstract
One of the most daunting challenges of nanomedicine is the finding of appropriate targeting agents to deliver suitable payloads precisely to cells affected by malignancies. Even more complex is the ability to ensure that the nanosystems enter those cells. Here, we use 2 nm (metal core) gold nanoparticles to target human hepatocellular carcinoma (HepG2) cells stably transfected with the SERPINB3 (SB3) protein. The nanoparticles were coated with a 85:15 mixture of thiols featuring, respectively, a phosphoryl choline (to ensure water solubility and biocompatibility) and a 28-mer peptide corresponding to the amino acid sequence 21–47 of the hepatitis B virus-PreS1 protein (PreS1(21–47)). Conjugation of the peptide was performed via the maleimide–thiol reaction in methanol, allowing the use of a limited amount of the targeting molecule. This is an efficient procedure also in the perspective of selecting libraries of new targeting agents. The rationale behind the selection of the peptide is that SB3, which is undetectable in normal hepatocytes, is overexpressed in hepatocellular carcinoma and in hepatoblastoma and has been proposed as a target of the hepatitis B virus (HBV). For the latter, the key recognition element is the PreS1(21–47) peptide, which is a fragment of one of the proteins composing the viral envelope. The ability of the conjugated nanoparticles to bind the target protein SB3, expressed in liver cancer cells, was investigated by surface plasmon resonance analysis and in vitro via cellular uptake analysis followed by atomic absorption analysis of digested samples. The results showed that the PreS1(21–47) peptide is a suitable targeting agent for cells overexpressing the SB3 protein. Even more important is the evidence that the gold nanoparticles are internalized by the cells. The comparison between the surface plasmon resonance analysis and the cellular uptake studies suggests that the presentation of the protein on the cell surface is critical for efficient recognition.
Introduction
Nanomedicine has developed as a platform to allow, in principle, sophisticated and smart drug delivery within the size window of a submicroscopic system that enables delicate and complex interactions with cancer cells and their biological milieu. The size scale of the nanosystems (1–100 nm), closer to proteins and viruses than to molecules, changes the nature of the interactions and, consequently, distribution in the biological environment with respect to traditional drugs.1 The self-organized nature of the nanoparticles prepared by bottom-up approaches allows the exploration of new therapeutic and diagnostic modalities,2,3 also via the exploitation of the multivalent and multifunctional properties of the systems.4
Notwithstanding such advantages, very few nanosystems are currently used in the clinical practice.5,6 This is because several issues still need to be addressed for the development of effective nanotheranostic agents, among which targeting is one of the most relevant. First-generation nanomedicine agents (including the few liposomal preparations currently approved, which represent the large majority of the nanomedicine agents entered in the clinical use) were based on the enhanced permeation and retention (EPR) effect. Here, the leakiness of the immature tumor vasculature, combined with poor lymphatic drainage, causes relatively large (10–100 nm) entities to preferentially accumulate in the cancer tissues.7,8 However, EPR effect is not general. Furthermore, nanoparticles larger than the threshold of the renal filters (5 nm) cannot be easily cleared from the organism, leading to long-term accumulation.9 These problems led to the development of second-generation nanosystems based on active targeting strategies, which include the conjugation of nanosystems with antibodies or ligands for receptors overexpressed by the target cells.10 For this reason, the selection of new targeting agents is very important and requires a strict collaboration between chemists, biologists, and medical doctors. Furthermore, even when targeting has been achieved, internalization of the nanosystem cannot be taken for granted.11 Several factors affect nanoparticle uptake by cells, including charge12 and the presence of specific peptides (e.g., TAT).13 Hence, the synthesis of nanoparticles not only able to target specific cells but also characterized by an enhanced uptake by these cells would represent a significant achievement.
In this paper, we show how 2 nm diameter monolayer-protected gold nanoparticles conjugated with a 28-mer peptide designed for the targeting of SERPINB3 expressing cells (the PreS1(21–47) fragment) not only bind to the selected target but also are internalized into the cells. We do not address their ultimate localization within the cells as this is not a major issue when the aim is cancer cells targeting and, eventually, their killing.
SERPINB3 (SB3, also known as Squamous Cell Carcinoma Antigen 1, SCCA1) is a soluble serine protease inhibitor of the ovalbumin–serin protease family (ov-serpins). This protein is frequently up-regulated in several malignancies of epithelial origin and of the liver. Indeed, it is undetectable in normal hepatocytes, but it is overexpressed in hepatocellular carcinoma (HCC) and in hepatoblastoma.14−18 A few years ago, some researchers19−23 have demonstrated that SB3 is also a target of the hepatitis B virus (HBV), and a key recognition element is the PreS1(21–47) peptide, which is a fragment of one of the proteins composing the viral envelope. Other target proteins have been suggested for the HBV capsid,24 particularly for the PreS1 region. Our results confirm that SB3 is one of them, and the PreS1(21–47) peptide represents a new potential targeting agent for not only hepatic cancer but also for cargo internalization into cells. The implications for possible applications in cancer therapy are obvious.
Results and Discussion
In this study, 2 nm metal core gold nanoparticles (AuNPs) coated with a monolayer of thiols were used as candidate nanosystem.25,26 The monolayer of organic molecules that protects the nanocluster is made by thiols strongly bound to the gold surface. These thiols typically bear at the opposite end functional groups that tailor the biological properties of the nanoparticles. Preparation of the nanoparticles is much simpler than that of functional polymers or dendrimers because the single functional units can be synthesized separately and then self-assembled on the gold core. However, their nondynamic nature ensures higher stability with respect to liposomes and lipid nanoparticles. For these reasons, these nanoparticles represent probably the most suitable platform for the preparation of multivalent functional nanosystems for the ease of preparation, the stability, and the very low toxicity.27
Synthesis and Characterization of the AuNPs
We used a two-phase, two-step synthetic procedure to prepare the monolayer-coated nanoparticles.28 Briefly, the gold cores are prepared in toluene by reduction of HAuCl4 with NaBH4 in the presence of tetrabutylammonium bromide as a phase-transfer agent and dioctylamine as capping agent. The nanoparticle size is controlled by the amount of dioctylamine, which acts as a weak surface stabilizer. Subsequent addition of a thiol, or of a mixture of thiols, results into the displacement of the amine and the formation of the final protecting monolayer. There are several advantages in the use of this method. First, because the metal core synthesis and thiol monolayer formation are separated, several samples of nanoparticles with the same core size and dispersion and different coatings can be prepared by batch splitting. Second, a good control of the monolayer composition can be achieved. Third, the use of the weaker stabilizing agent (dioctylamine) ensures a smaller size dispersion.
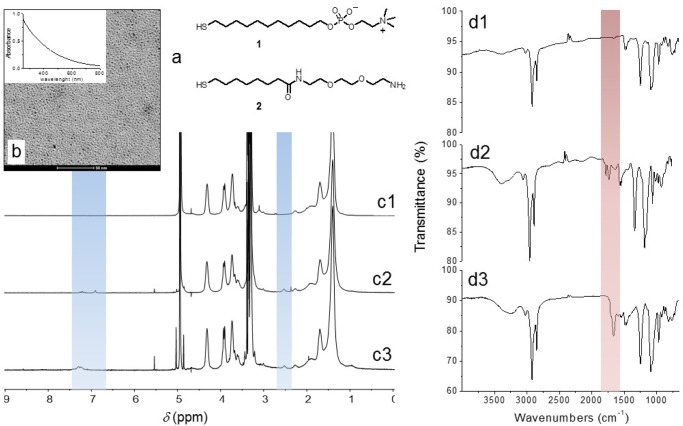
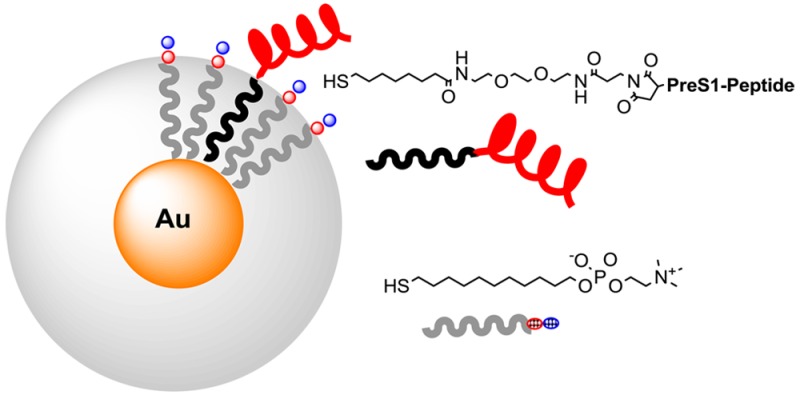
Thiols 1 and 2 were selected as coating elements (Figure 1a). Thiol 1, bearing the zwitterionic and biocompatible phosphorylcholine headgroup, was supposed to give the nanoparticles “stealth” properties by decreasing proteins absorption.29,30 Thiol 2 features a terminal primary amino group easily exploitable for conjugation of the targeting agents. The 1-to-2 ratio in the nanoparticles was adjusted around 10:1 to give them targeting properties without affecting multivalency and the ability to escape RES capture. Nanoparticles purification was performed by gel-permeation chromatography. Both 1- and 1/2-coated nanoparticles were found to be fully soluble in water and saline solutions for several weeks. The size of the metal cores from TEM analysis resulted to be 1.8 ± 0.5 and 1.9 ± 0.5 nm for 1-, and 1/2-coated nanoparticles, respectively (see Figure 1b for 1/2-AuNP). Such a value is confirmed by the UV–vis spectrum of the nanoparticles, which reveals the absence of the plasmonic absorption band at 520 nm (Figure 1b), typical of gold nanoparticles with a metal core diameter larger than 3 nm. Monolayer purity and composition was assessed by 1H NMR (see the Supporting Information). NMR analysis after iodine decomposition for the 1/2-coated nanoparticles yielded a 1 to 2 ratio of 92:8. The average formulas for the two nanoparticles are, accordingly, Au180170 and Au18016327, respectively.31,32
Figure 1.
AuNP characterization: (a) thiols used for nanoparticles passivation, (b) TEM picture of 1/2-AuNP (inset: UV–vis spectrum in water); (c) 1H NMR spectra of 1/2-AuNP (1), 1/2-AuNP after conjugation with MPBS (2), and PreS1-AuNPs (3); (d) Fourier transfer infrared (FT-IR) spectra of the same samples. The blue and pink strips in the spectra highlight the regions showing the relevant modifications described in the text as a consequence of the conjugation.
The CysPreS1(21–47) peptide (CPNFDWDPNNSNAGFAPDLQHDPFFGLP) presents a cysteine at the C terminus of the PreS1(21–47) sequence. The presence of the thiol group in the cysteine side chain allows the easy conjugation to the nanoparticles via maleimide–thiol addition. In principle, one could consider direct conjugation of the peptide to the gold surface. However, this would have resulted in its partial burying in the monolayer constituted by zwitterionic thiol 1 and a loss of peptide surface suitable for binding to the target. The 1/2-AuNPs were reacted with the cross-linker MPBS in DMF–methanol, to introduce the maleimide functional group, and subsequently conjugated with the CysPreS1(21–47) peptide in methanol to obtain the PreS1-AuNPs. Purification was performed by size-exclusion chromatography, and effective conjugation was confirmed by nuclear magnetic resonance (NMR) and Fourier transform infrared (FT-IR) spectroscopy (Figure 1c,d). Assuming a quantitative conversion of 2 in the peptide conjugate and on the basis of the thiols ratio on the nanoparticles reported above, there will be, on average, seven peptides per AuNP.
In the NMR spectra of 1/2-coated nanoparticles, small signals at 6.5 and 2.3 ppm appeared after MPBS conjugation, arising, respectively, from the protons of the vinyl group and of the methylene adjacent to the carbonyl group of the cross-linker. Upon reaction with the CysPreS1 derivative, the maleimide signal at 6.5 ppm disappeared to be substituted by a broad signal at 7.1 ppm, arising from the aromatic side chains of the Phe and Trp residues of PreS1, thus confirming the full conversion of the maleimide group into the conjugate. The FT-IR analysis provided a clear-cut evidence of the presence of the peptide in the purified nanoparticle sample (Figure 1d). Unconjugated particles showed a weak absorption at 1680 nm due to the amide group of thiol 2. After coupling with MPBS, a new signal at 1720 nm appeared due to the carbonyl groups of the maleimide. Finally, after conjugation with the peptide, absorption at 1680 strongly increased due to the large number of amide groups present in the peptide.
Interaction of the AuNPs with the Target Protein SERPINB3
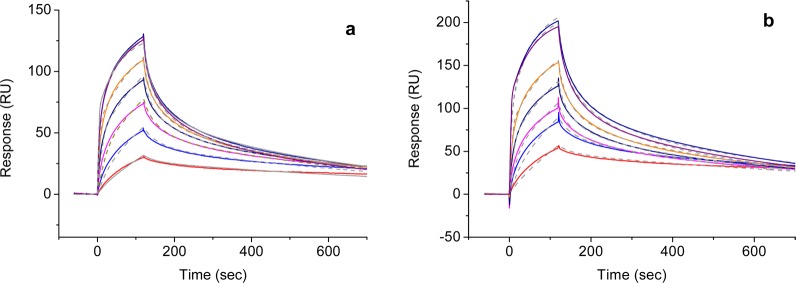
The ability of the conjugated peptide to interact with the target SB3 protein was first assessed by surface plasmon resonance (SPR) analysis, which allows for the direct measurement of the binding events. The SB3 protein was immobilized on a dextrane-coated gold chip via amide coupling (about 2 ng/mm2) and exposed to increasing concentrations of CysPreS1, 1-AuNPs, and PreS1-AuNPs. Experiments performed with the CysPreS1 peptide did not reveal any interaction (see the Supporting Information). Such an outcome was likely the result of the low molecular weight of the peptide, which reduced the amplitude of the instrument response, and of the possible low affinity of the peptide for SB3 at the concentration interval explored.20,23 However, 1-AuNPs revealed a quite-strong binding (Figure 2). Association and dissociation profiles were rather complex and could be fitted only with a kinetic model, which includes two different binding modes of the nanoparticles to SB3 (see the Supporting Information). Fitting results indicated that both binding events were characterized by very low dissociation constants (Kd,1 = 1.1 × 10–11 M and Kd,2 = 1.8 × 10–11 M) but featured different binding and release rates and interfered with each other. The first binding mode was characterized by fast association and dissociation rates (kin = 8.8 × 109 M–1sec–1 and kout = 9.5 × 10–2 s–1) and a low amount of binding sites (8% at low nanoparticle concentrations). The second type of binding sites featured slower binding and dissociation (kin = 1 × 108 M–1sec–1 and kout = 2 × 10–3 s–1). Such sites were predominant (92%) at low nanoparticles concentrations and decreased progressively when the amount of AuNPs injected was increased. Apparently saturation of the first binding sites by increasing the nanoparticle concentration disfavored the binding to the second. The presence of the PreS1 peptide in PreS1-AuNp led to greater binding (the maximum RU value reached is 200 versus 130 for 1-AuNP) but did not appear to increase the affinity for the protein (Kd,1 = 1.1 × 10–11 M and Kd,2 = 1.6 × 10–11 M). This is probably due to the fact that the highly nonspecific affinity of the monolayer made by 1 masked the specific binding of PreS1 to SB3 (Figure 2). Control experiments performed using nonfunctionalized chips or chips functionalized with different proteins confirmed the specific affinity of SB3 for 1-AuNP (see the Supporting Information). This was very disappointing, considering the reported29 small affinity of zwitterionic nanoparticles to proteins. These data do not allow drawing any conclusion on the advantage of functionalizing the nanoparticles with the PreS1 peptide as a targeting agent. We can only state that it does not affect it negatively.
Figure 2.
SPR analysis of the interaction between SB3 and nanoparticles: (a) kinetic assays (multicycle) of 1-AuNP and fitting as heterogeneous ligand; (b) kinetic assays (multicycle) of PreS1-AuNP and fitting as heterogeneous ligand (colored lines are experimental sensorgrams corresponding to different nanoparticle concentrations, namely red, 0.005 nM; blue, 0.010 nM; magenta, 0.020 nM; black, 0.038 mM, orange, 0.078 nm; and dark red and dark blue, 0 0.157 mM; gray dashed lines are the best-fitting in both graphs).
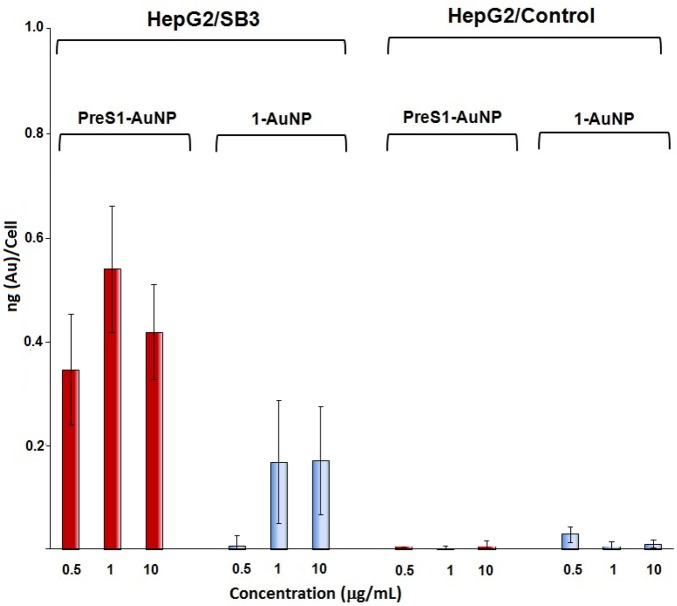
A more-realistic evaluation of ability of the PreS1-AuNp to selectively bind cancer cells was performed by using human HepG2 cells stably transfected with SB3 (HepG2/SB3 cells). Western blot experiments (see the Supporting Information) indicate that, indeed, only HepG2/SB3 cells show a remarkable amount of SB3. These cells were incubated with increasing concentrations of PreS1-AuNPs using HepG2 and 1-AuNPs as control cells and nanoparticles, respectively. The results (Figure 3) revealed a remarkably higher binding of PreS1-AuNP nanoparticles in the HepG2/SB3 cells compared to the controls. In particular, HepG2 cells bound neither PreS1-AuNPs nor 1-AuNPs nanoparticles, confirming the stealth features of the phosphorylcholine coating in the absence of the SB3 protein on the cells. However, SB3-transfected cells bound both nanoparticles, although at a different extent. In fact, the amount of PreS1-AuNPs bound to HepG2/SB3 cells was substantially higher than that of 1-AuNPs at any dose and, in particular, at the lower (0.5 μg/mL) concentration used. On one hand, these data indicated an increased uptake of the nanoparticles conjugated with the targeting peptide mediated by its interaction with the SB3 protein. On the other hand, they also confirmed the affinity of the phosphorylcholine coating for SB3 revealed by the SPR experiments. The better affinity of the PreS1-AuNPs for HepG2/SB3 cells compared to 1-AuNPs in these experiments indicated a lower unspecific interaction of the zwitterionic component of the monolayer or, more likely, a more-appropriate presentation of SB3 on the cells surface than on the chips used for the SPR experiments. It is not infrequent that the immobilization of a protein on the surface for an SPR experiment alters its interaction with the target.
Figure 3.
Binding specificity analysis of nanoparticles to HepG2/SB3 cells and controls (HepG2/control). Conditions: 1 h incubation at 37 °C and 5% CO2. Bars represent mean values ± SD of two different sets of experiments (two runs each). The gold cellular content was determined by atomic absorption analysis of cellular pellets. The X axis shows the different concentration of AuNPs utilized.
The dose–response trend was quite flat, suggesting that a threshold uptake of the nanoparticles is already reached at low concentrations. Consequently, we selected 1 μg/mL as the best experimental concentration at 1 h incubation to assess the fate of the nanoparticles and, in particular, their internalization into the cells. As reported in Figure 4, binding of PreS1-AuNP was more than 6 times higher than that of the nanoparticles without the peptide (1-AuNp). Trypsin treatment of HepG2/SB3 cells, which, as we have as previously reported allows to differentiate, at least in part, between HBV binding and internalization21 did not cause any decrease of the amount of gold bound to the cells. This suggests that the nanoparticles were internalized and not just bound to the cell surface. This adds another possibility to explain the difference observed in the SPR and in vitro experiments. In fact, it could be that binding is not much enhanced by SB3 presence on the surface of the cells, but whenever it occurs, it induces efficient internalization of the nanoparticles in them. We cannot exclude that this is due, at least in part, to an overall difference in endocytosis activity induced by SerpinB3. In any case, the presence of the PreS1 peptide is a requisite for the process to occur.
Figure 4.
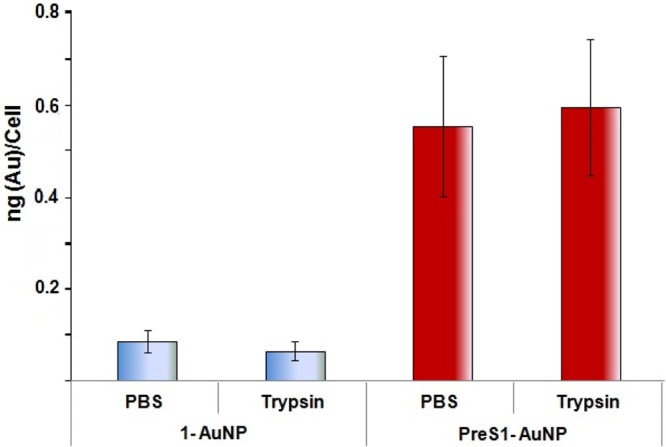
Nanoparticles uptake by HepG2/SB3 cells. Conditions: 1 h incubation at 1 μg/mL concentration with or without 5 min treatment with trypsin at 37 °C. Negative controls were treated with PBS. The gold cellular content was determined by atomic absorption analysis of cellular pellets. Bars represent mean values ± SD of three different experiments.
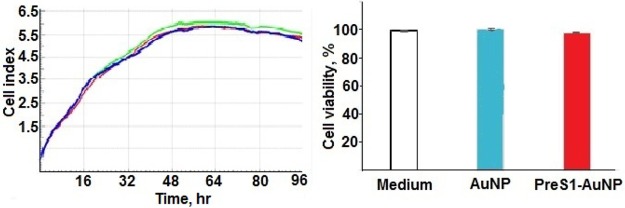
Cellular toxicity of the AuNPs was investigated by monitoring the cell growth curves upon incubation with and without the nanoparticles. All of the curves were identical over the time interval investigated (96 h), indicating no cytotoxicity induced by the nanoparticles (Figure 5).
Figure 5.
Left panel: time course of the cell growth in the medium (blue trace) in the presence of 1 μg/mL of 1-AuNP (red) or PreS1-AuNP (green); the cell index is a relative measure of cell number present in a well. Results are expressed as mean of automatically calculated cell indexes of three different experiments. Right panel: cell viability of the same solutions after 24 h.
Conclusions
In conclusion, we reported here the facile preparation of stable, water-soluble 2 nm gold nanoparticles that can be easily conjugated with a peptide bearing a thiol group.33−35 With respect to other conjugation methods, the use of the maleimide–thiol reaction in methanol allows the targeting molecule to be fully exposed on the surface of the nanoparticle, thus ensuring a better presentation to the target. The data we have obtained put in evidence that the PreS1(21–47) peptide is a suitable targeting agent for cells overexpressing the SB3 protein. Even more important is the evidence that the gold nanoparticles are internalized by the cells. The mechanism of internalization of the virus is far from being understood.36,37 It appears that the PreS1-SB3 interaction is not the only factor. For instance, Hao et al.38 have suggested that a ferritin light chain (FTL) is involved in the formation of a putative preS-FTL-SB3 triple complex. The SPR results, if we accept the hypothesis of a wrong orientation of SB3 on the chip surface, indicate a precise presentation of the protein to the virus surface is required for recognition. Harrison et al.22 have shown that the binding event does not involve the reactive site loop of SB3. This could imply that the binding occurs at the surface of the protein, supporting the suggestion that a wrong orientation is the cause of the unselective binding in the SPR experiments. Regardless the precise mechanism of binding and internalization, our results are important in the view of a possible use of these nanoparticles as carriers for delivering and internalizing a suitable payload in hepatocellular carcinoma treatment. Nanoparticle delivery to malignant tissues and cells is an hot topic, and efficiency in the process, particularly as tumors are concerned, has been shown recently to still be very challenging.39
Experimental Procedures
Syntheses
Thiols 1 and 2 were prepared following previously published procedures.30,40 Peptide PreS1(21–47) was purchased from JTP, Germany. Its purity was checked by HPLC and found to be better than 99%. Conjugation to the nanoparticles and functionalization of the peptide were performed following standard protocols. Details are reported in the Supporting Information.
Nanoparticles Characterization
Nanoparticles 1H NMR spectra were recorded using a Bruker AV300 spectrometer operating at 300 MHz. About 10 mg of nanoparticle were dissolved in 0.6 mL of CD3OD, and the spectrum was recorded. TEM images were recorded on a Jeol 300 PX electron microscope. A single drop of sample solution was placed on the sample grid, and the solvent was allowed to evaporate. UV–visible spectra and kinetic traces were recorded on a Cary 50 spectrophotometer equipped with thermostated multiple cell holders. IR spectra were recorded on a Nicolet 5700 FT-IR spectrophotometer and reported in cm–1. AuNPs were dissolved in methanol at the concentration of 10 mg/mL, and 20 μL of the solution was cast onto a NaCl plate by an Eppendorf micropipette and dried using a hot plate. NMR spectra and TEM images are in Figure 1 and in the Supporting Information.
Surface Plasmon Resonance Analysis
SPR analyses were performed using a double channel Biacore X100 instrument (GE Healthcare, Uppsala, Sweden). Recombinant SB3 was produced after cloning human SB3 cDNA in the directional expression vector pET101 (Invitrogen Carlsbad, CA), as previously described.41 A dextrane-coated gold chip (CM5) was activated by flowing a 1:1 mixture of 0.2 M N-ethyl-N-(3-(dimethylamino)propyl) carbodiimide and 0.05 M N-hydroxysuccinimide in water. A continuous flow of HEPES pH 7.4 was maintained. SB3 (50 μg/mL) in 10 mM sodium acetate (pH 5) was immobilized on the activated chip surface at a flow rate of 10 μL/min to obtain an immobilization level of “Response Bound” of 1661 RU and “Response Final” of 1840 RU. Excess of activated carboxylic groups on the chip was blocked with ethanolamine. The control channel was treated following the same protocol without protein immobilization. Kinetic assays were performed with both PreS1AuNPs and 1-AuNP as the control, following the instrument built-in standard protocol using HBS-EP+ buffer at pH 7.4 (0.1 M HEPES, 1.5 M NaCl, 30 mM EDTA, and 0.5% v/v P20 surfactant) by subsequent 180 s injections of nanoparticle solutions at increasing concentration (0.005–0.157 nM) using NaCl 3 M as regeneration solution. Details are reported in the Supporting Information.
Cellular Uptake and Viability Analysis
To analyze the ability of nanoparticles to selectively bind SB3 in cells manipulated to overexpress this serpin, in vitro experiments were carried out. HepG2 cells, derived from a human hepatoma (LGC Standards S.r.l., Sesto San Giovanni, MI, Italy) were stably transfected with a plasmid vector carrying the wild-type SB3 human gene (HepG2/SB3) or with the plasmid vector alone (HepG2/empty vector) (pcDNA3.1D/V5-His-TOPO, Invitrogen Life Technologies, NY), as previously reported.42 In preliminary experiments, both cell lines were tested for SerpinB3 protein content by Western blot, and only HepG2/SB3 showed a remarkable amount of this serpin (Figure S19). Both cell lines were initially incubated for 1 h with increasing amounts (0.5, 1, and 10 μg/mL) of PreS1-AuNPs or 1-AuNPs, as a negative control, to define the best experimental conditions and to test the specificity of functionalized nanoparticles binding to SB3 target protein. To evaluate if PreS1-AuNPs were internalized into SB3-expressing cells, trypsin treatment was carried out for 5′ at 37 °C after 1 h incubation. Negative controls were treated with PBS for the same time frame. Real-time monitoring of the potential cytotoxicity of nanoparticles was carried out using the xCELLigence DP instrument (Roche Diagnostics, Mannheim, Germany), as described in the supplier’s instruction manual. HepG2/SERPINB3 cells were seeded (50 000 cells/well) in the E-Plate 96, maintained at 37 °C in a 5% CO2 atmosphere and treated with 1 μg/mL of PreS1AuNPs or of 1-AuNPs or with medium. Cell cultures not treated with nanoparticles were used as the control. The cell index values (a quantitative measure of the cell number present in a well) were achieved using the RTC software (version 1.2, Roche Diagnostics) and monitored every 10 min for 96 h.
Atomic Absorption Analysis
To analyze the amount of nanoparticles entered into the cells, atomic absorption analysis was carried out. The digestion of cellular pellets was accomplished by addition of 0.5 mL of aqua regia (HCl 36%: HNO3 3% = 3:1) to each cell sample followed by heating at 70 °C for 1 h in a sealed vial. Subsequently, an aliquot of 250 μL of digestion solution was transferred into a 15 mL polypropylene tube and filled to a total volume of 10 mL with a 3% solution of HNO3. Gold content was determined by standard atomic absorption measurements.
Western Blot Analysis
A total of 50 μg of total protein content from each cellular extract, obtained by cellular protein extraction using RIPA lysis buffer, was loaded on 10% polyacrylamide gel. Antigenic detection was carried out by enhanced chemiluminescence (Amersham, Arlington Height, IL), and densitometric analysis was assessed using a VersaDoc Imaging system (Bio-Rad Laboratories, Hercules, CA). The following antibodies were used: monoclonal anti-SERPINB3 antibody (0.5 μg/mL, R&D SYSTEMS Minneapolis, MN), monoclonal anti-β-actin antibody (1:1000, Sigma-Aldrich, St. Louis, MO) and anti-mouse horseradish peroxidase conjugated secondary antibody (1:1000, Amersham Bioscience, Arlington Height, IL).
Acknowledgments
Financial support for this research has been provided by the European Union (ERC Starting Grants Project MOSAIC 259014), the Italian Ministry of Research (FIRB 2011, RBAP114AMK RINAME), and the University of Padova (Strategic Project NAMECA).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.bioconjchem.6b00441.
Additional details on experimental procedures, the synthesis of thiols, the synthesis and characterization of monolayer protected gold nanoparticles, surface plasmon resonance analysis, and Western blot experiments. Figures showing TEM, 1H-NMR, UV, and IR spectra and sensogram and kinetic assay results. (PDF)
Author Present Address
§ Sikkim Manipal Institute of Technology, Department of Chemistry, Rangpo, India.
The authors declare no competing financial interest.
Supplementary Material
References
- Crommelin D. J. A.; Scherphof G.; Storm G. (1995) Active targeting with particulate carrier systems in the blood. Adv. Drug Delivery Rev. 17, 49–60. 10.1016/0169-409X(95)00040-E. [DOI] [Google Scholar]
- Ferrari M. (2005) Cancer nanotechnology: opportunities and challenges. Nat. Rev. Cancer 5, 161–171. 10.1038/nrc1566. [DOI] [PubMed] [Google Scholar]
- Petros R.; DeSimone J. (2010) Strategies in the design of nanoparticles for therapeutic applications. Nat. Rev. Drug Discovery 9, 615–627. 10.1038/nrd2591. [DOI] [PubMed] [Google Scholar]
- Godin B.; Tasciotti E.; Liu X.; Serda R. E.; Ferrari M. (2011) Multistage nanovectors: from concept to novel imaging contrast agents and therapeutics. Acc. Chem. Res. 44, 979–989. 10.1021/ar200077p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J.; Xiao Z.; Kamaly N.; Farokhzad O. C. (2011) Self-assembled targeted nanoparticles: evolution of technologies and bench to bedside translation. Acc. Chem. Res. 44, 1123–1134. 10.1021/ar200054n. [DOI] [PubMed] [Google Scholar]
- Landesman-Milo D.; Peer D. (2016) Transforming nanomedicines from lab scale production to novel clinical modality. Bioconjugate Chem. 27, 855–862. 10.1021/acs.bioconjchem.5b00607. [DOI] [PubMed] [Google Scholar]
- Maeda H.; Bharate G. Y.; Daruwalla J. (2009) Polymeric drugs for efficient tumor-targeted drug delivery based on EPR-effect. Eur. J. Pharm. Biopharm. 71, 409–419. 10.1016/j.ejpb.2008.11.010. [DOI] [PubMed] [Google Scholar]
- Iyer A. K.; Khaled G.; Fang J.; Maeda H. (2006) Exploiting the enhanced permeability and retention effect for tumor targeting. Drug Discovery Today 11, 812–818. 10.1016/j.drudis.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Choi H. S.; Liu W.; Misra P.; Tanaka E.; Zimmer J. P.; Ipe B. I.; Bawendi M. G.; Frangioni J. V. (2007) Renal clearance of quantum dots. Nat. Biotechnol. 25, 1165–1170. 10.1038/nbt1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z.; Al Zaki A.; Hui J. Z.; Muzykantov V. R.; Tsourkas A. (2012) Multifunctional nanoparticles: cost versus benefit of adding targeting and imaging capabilities. Science 338, 903–910. 10.1126/science.1226338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévy R.; Shaheen U.; Cesbron Y.; Sée V. (2010) Gold nanoparticles delivery in mammalian live cells: a critical review. Nano Rev. 1, 4889–4906. 10.3402/nano.v1i0.4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha K.; Kim S. T.; Yan B.; Miranda O. R.; Alfonso F. S.; Shlosman D.; Rotello V. M. (2013) Surface functionality of nanoparticles determines cellular uptake mechanisms in mammalian cells. Small 9, 300–305. 10.1002/smll.201201129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesbron Y.; Shaheen U.; Free P.; Lévy R. (2015) TAT and HA2 facilitate cellular uptake of gold nanoparticles but do not lead to cytosolic localisation. PLoS One 10, e0121683. 10.1371/journal.pone.0121683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman G. A.; Bird P. I.; Carrell R. W.; Church F. C.; Coughlin P. B.; Gettins P. G. W.; Irving J. A.; Lomas D. A.; Luke C. J.; Moyer R. W.; et al. (2001) The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions and a revised nomenclature. J. Biol. Chem. 276, 33293–33296. 10.1074/jbc.R100016200. [DOI] [PubMed] [Google Scholar]
- Pontisso P.; Calabrese F.; Benvegnù L.; Lise M.; Belluco C.; Ruvoletto M. G.; De Falco S.; Marino M.; Valente M.; Nitti D.; et al. (2004) Overexpression of squamous cell carcinoma antigen variants in hepatocellular carcinoma. Br. J. Cancer 90, 833–837. 10.1038/sj.bjc.6601543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guido M.; Roskams T.; Pontisso P.; Fassan M.; Thung S. N.; Giacomelli L.; Sergio A.; Farinati F.; Cillo U.; Rugge M. (2008) Squamous cell carcinoma antigen in human liver carcinogenesis. J. Clin. Pathol. 61, 445–447. 10.1136/jcp.2007.051383. [DOI] [PubMed] [Google Scholar]
- Turato C.; Buendia M. A.; Fabre M.; Redon M. J.; Branchereau S.; Quarta S.; Ruvoletto M.; Perilongo G.; Grotzer M. A.; Gatta A.; et al. (2012) Over-expression of SERPINB3 in hepatoblastoma: A possible insight into the genesis of this tumour?. Eur. J. Cancer 48, 1219–1226. 10.1016/j.ejca.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Turato C.; Vitale A.; Fasolato S.; Ruvoletto M.; Terrin L.; Quarta S.; Morales R. R.; Biasiolo A.; Zanus G.; Zali N.; et al. (2014) SERPINB3 is associated with TGF-beta1 and cytoplasmic beta-catenin expression in hepatocellular carcinomas with poor prognosis. Br. J. Cancer 110, 2708–2715. 10.1038/bjc.2014.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Falco S.; Ruvoletto M. G.; Verdoliva A.; Ruvo M.; Raucci A.; Marino M.; Senatore S.; Cassani G.; Alberti A.; Pontisso P.; et al. (2001) Cloning and expression of a novel hepatitis B virus-binding protein from HepG2 cells. J. Biol. Chem. 276, 36613–36623. 10.1074/jbc.M102377200. [DOI] [PubMed] [Google Scholar]
- Pontisso P.; Petit M. A.; Bankowski M. J.; Peeples M. E. (1989) Human liver plasma membranes contain receptors for the hepatitis B virus Pre-S1 region and, via polymerized human serum albumin, for the Pre-S2 region. J. Virol. 8, 1981–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvoletto M. G.; Tono N.; Carollo D.; Vilei T.; Trentin L.; Muraca M.; Marino M.; Gatta A.; Fassina G.; Pontisso P. (2004) Surface expression of squamous cell carcinoma antigen (SCCA) can be increased by the preS1(21–47) sequence of hepatitis B virus. J. Gen. Virol. 85, 621–624. 10.1099/vir.0.19130-0. [DOI] [PubMed] [Google Scholar]
- Moore P. L.; Ong S.; Harrison T. J. (2003) Squamous cell carcinoma antigen 1-mediated binding of hepatitis B virus to hepatocytes does not involve the hepatic serpin clearance system. J. Biol. Chem. 278, 46709–46717. 10.1074/jbc.M302842200. [DOI] [PubMed] [Google Scholar]
- Kasuya T.; Nomura S.; Matsuzaki T.; Jung J.; Yamada T.; Tatematsu K.; Okajima T.; Tanizawa K.; Kuroda S. (2008) Expression of squamous cell carcinoma antigen-1 in liver enhances the uptake of hepatitis B virus envelope-derived bio-nanocapsules in transgenic rats. FEBS J. 275, 5714–5724. 10.1111/j.1742-4658.2008.06698.x. [DOI] [PubMed] [Google Scholar]
- Li W.; Urban S. (2016) Entry of hepatitis B and hepatitis D virus into hepatocytes: Basic insights and clinical implications. J. Hepatol. 64 (1 Suppl), S32–40. 10.1016/j.jhep.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancin F.; Prins L. J.; Scrimin P. (2013) Catalysis on gold-nanoparticle-passivating monolayers. Curr. Opin. Colloid Interface Sci. 18, 61–69. 10.1016/j.cocis.2012.12.003. [DOI] [Google Scholar]
- Pasquato L.; Pengo P.; Scrimin P. (2005) Nanozymes: functional nanoparticle-based catalysts. Supramol. Chem. 17, 163–171. 10.1080/10610270412331328817. [DOI] [Google Scholar]
- Kim S.; Saha K.; Kim C.; Rotello V. (2013) The role of surface functionality in determining nanoparticle cytotoxicity. Acc. Chem. Res. 46, 681–91. 10.1021/ar3000647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manea F.; Bindoli C.; Polizzi S.; Lay L.; Scrimin P. (2008) Expeditious synthesis of water-soluble, monolayer-protected gold nanoparticles of controlled size and monolayer composition. Langmuir 24, 4120–4124. 10.1021/la703558y. [DOI] [PubMed] [Google Scholar]
- Moyano D.; Saha K.; Prakash G.; Yan B.; Kong H.; Yazdani M.; Rotello V. (2014) Fabrication of corona-free nanoparticles with tunable hydrophobicity. ACS Nano 8, 6748–6755. 10.1021/nn5006478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmlin R. E.; Chen X.; Chapman R. G.; Takayama S.; Whitesides G. M. (2001) Zwitterionic SAMs that resist nonspecific adsorption of protein from aqueous buffer. Langmuir 17, 2841–2850. 10.1021/la0015258. [DOI] [PubMed] [Google Scholar]
- Guarino G.; Rastrelli F.; Scrimin P.; Mancin F. (2012) Lanthanide-based NMR: a tool to investigate component distribution in mixed-monolayer-protected nanoparticles. J. Am. Chem. Soc. 134, 7200–7203. 10.1021/ja211030y. [DOI] [PubMed] [Google Scholar]
- Guarino G.; Rastrelli F.; Mancin F. (2012) Mapping the nanoparticles coating monolayer with NMR pseudo-contact shifts. Chem. Commun. 48, 1523–1525. 10.1039/C1CC14726A. [DOI] [PubMed] [Google Scholar]
- Gobbo P.; Workentin M. S. (2012) Improved methodology for the preparation of water-soluble maleimide-functionalized small gold nanoparticles. Langmuir 28, 12357–12363. 10.1021/la302168g. [DOI] [PubMed] [Google Scholar]
- Zhu J.; Waengler C.; Lennox R. B.; Schirrmacher R. (2012) Preparation of water-soluble maleimide-functionalized 3 nm gold nanoparticles: a new bioconjugation template. Langmuir 28, 5508–5512. 10.1021/la300316j. [DOI] [PubMed] [Google Scholar]
- Oh E.; Susumu K.; Blanco-Canosa J. B.; Medintz I. L.; Dawson P. E.; Mattoussi H. (2010) Preparation of stable maleimide-functionalized Au nanoparticles and their use in counting surface ligands. Small 6, 1273–1278. 10.1002/smll.201000279. [DOI] [PubMed] [Google Scholar]
- Paran N.; Cooper A.; Shaul Y. (2003) Interaction of hepatitis B virus with cells. Rev. Med. Virol. 13, 137–143. 10.1002/rmv.376. [DOI] [PubMed] [Google Scholar]
- Ezzikouri S.; Ozawa M.; Kohara M.; Elmdaghri N.; Benjelloun S.; Tsukiyama-Kohara K. (2014) Recent insights into hepatitis B virus–host interactions. J. Med. Virol. 86, 925–932. 10.1002/jmv.23916. [DOI] [PubMed] [Google Scholar]
- Hao Z.; Zheng L.; Kluwe L.; Huang W. (2012) Ferritin light chain and squamous cell carcinoma antigen 1 are coreceptors for cellular attachment and entry of hepatitis B virus. Int. J. Nanomed. 7, 827–834. 10.2147/IJN.S27803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm S.; Tavares A. J.; Dai Q.; Ohta S.; Audet J.; Dvorak H. F.; Chan W. C. W. (2016) Analysis of nanoparticle delivery to tumours. Nat. Rev. Mater. 1, 16014. 10.1038/natrevmats.2016.14. [DOI] [Google Scholar]
- Diez-Castellnou M.; Mancin F.; Scrimin P. (2014) Efficient phosphodiester cleaving nanozymes resulting from multivalency and local medium polarity control. J. Am. Chem. Soc. 136, 1158–1161. 10.1021/ja411969e. [DOI] [PubMed] [Google Scholar]
- Turato C.; Calabrese F.; Biasiolo A.; Quarta S.; Ruvoletto M.; Tono N.; Paccagnella D.; Fassina G.; Merkel C.; Harrison T. J.; et al. (2010) SERPINB3 modulates TGF-beta expression in chronic liver disease. Lab. Invest. 90, 1016–23. 10.1038/labinvest.2010.55. [DOI] [PubMed] [Google Scholar]
- Quarta S.; Vidalino L.; Turato C.; Ruvoletto M.; Calabrese F.; Valente M.; Cannito S.; Fassina G.; Parola M.; Gatta A.; et al. (2010) SERPINB3 induces epithelial-mesenchymal transition. J. Pathol. 221, 343–356. 10.1002/path.2708. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.