Abstract
To investigate a putative link between genetically determined variations in Toll-like receptor 2 (TLR2) and the occurrence of severe Staphylococcus aureus infection, the functional Arg753Gln single-nucleotide polymorphism and the GT repeat microsatellite in the TLR2 gene were examined in a large case-control study. No associations with disease or mortality attributable to these features were found.
Staphylococcus aureus is responsible for a growing burden of disease in both community and hospital settings, causing a wide variety of infections, ranging from superficial skin infections to severe, often fatal, systemic infections. Toll-like receptors (TLRs) are a central part of the innate immune response to bacterial infection (12). They recognize pathogen-associated molecular patterns found on or in the cell walls of gram-negative and gram-positive bacteria but not on the surfaces of mammalian cells. TLR2 in particular has been implicated in the innate immune response to S. aureus infection, recognizing both staphylococcal peptidoglycan and lipoteichoic acid (1-3, 7). TLR2- and MyD88-deficient mice demonstrated an impaired cytokine response following exposure to a preparation of S. aureus peptidoglycan and had a higher mortality than that of wild-type mice after inoculation with S. aureus (9, 10).
Whether genetically determined variation in TLR2 determines susceptibility to staphylococcal infection in human populations is unknown. To address this issue, we examined the roles of the published TLR2 gene polymorphisms in a large case-control study of severe disease caused by S. aureus.
Three putatively functional polymorphisms have been described to occur in the human TLR2 gene: the Arg753Gln and Arg677Trp single-nucleotide polymorphisms (SNPs) (4, 5) and a microsatellite GT repeat polymorphism in intron 2 (13). The Arg677Trp SNP has been associated with lepromatous leprosy (4) but, as it is not present in Caucasian patients, was not examined in the present study. The Arg753Gln polymorphism was identified in a group of healthy Caucasian blood donors and, in tissue culture, was associated with a reduced response to different bacterial lipoproteins (5). This polymorphism has recently been shown to be significantly associated with tuberculosis (8), and in a population of septic shock patients, there was a weak association with infections caused by gram-positive organisms, particularly with staphylococcal septic shock (P = 0.056) (5).
A highly polymorphic GT repeat has been found 100 bp upstream of the translation start site in the TLR2 gene (described by Yim et al. [13] and discovered independently by S. Segal [personal communication]). This microsatellite is functional; alleles with high and low numbers of GT repeats have greater promoter activity than those with medium numbers of repeats (reference 13 and D. Wyllie [personal communication]).
We recruited 420 consecutive patients with severe S. aureus infection presenting to the microbiology department of the John Radcliffe Hospital in Oxfordshire, United Kingdom, between July 1997 and July 1999. Patients exhibited an illness consistent with invasive S. aureus disease, and S. aureus was isolated from a patient specimen from a normally sterile site, such as blood, cerebrospinal fluid, or a deep abscess (Table 1). Overall, 244 patients (57%) were male and the mean age was 56 years (confidence interval, 53 to 58 years; range, 0 to 94 years). Patients were classified as having community-acquired disease if S. aureus was isolated within 24 h of admission to the hospital and the patient had not been admitted to the hospital in the preceding 6 months (n = 112). Attributable mortality was defined as death occurring apparently as a direct consequence of the staphylococcal infection. Blood for control DNA was collected anonymously from the umbilical cords of 696 healthy neonates born in the John Radcliffe Hospital over the same 2 years (case/control ratio, 1:1.66). The possibility of maternal-DNA contamination was excluded by examination of microsatellite markers. All individuals of non-Caucasian ancestry were excluded from both cases and controls. Ethical approval was obtained prospectively from the Central Oxford Research Ethics Committee.
TABLE 1.
Severe S. aureus disease cases
| Diagnosis | No. (%) of patients in indicated disease group(s)
|
||
|---|---|---|---|
| Community- acquired infection (n = 112) | Hospital- acquired infection (n = 308) | Both | |
| Device-related septicemia | 5 (4) | 154 (50) | 159 (38) |
| Soft tissue infection with septicemia | 29 (26) | 39 (13) | 68 (16) |
| Osteomyelitis | 17 (15) | 34 (11) | 51 (12) |
| Unexplained septicemia | 16 (14) | 20 (6) | 36 (9) |
| Prosthetic joint infection | 5 (4) | 25 (8) | 30 (7) |
| Septic arthritis | 13 (12) | 7 (2) | 20 (5) |
| Endovascular infection | 12 (11) | 4 (1) | 16 (4) |
| Secondary meningitis | 1 (1) | 11 (4) | 12 (3) |
| Deep abscess (including empyema) | 5 (4) | 7 (2) | 12 (3) |
| Pneumonia | 5 (4) | 4 (1) | 9 (2) |
| Urinary tract infection with septicemia | 3 (3) | 3 (1) | 6 (1) |
| Toxic shock syndrome | 1 (1) | 0 | 1 |
This study has the power to detect with an odds ratio of 1.8 an effect of the Arg753Gln heterozygote on patients' susceptibility to severe staphylococcal disease, assuming a background allele frequency of 0.025, with a 5% significance level (2α of 0.05) and power of 80% (β of 0.80). Analysis of genotypes within the patient cohort has a similar power to detect an effect of the presence of the heterozygote on the death of a patient, with an odds ratio of 3.2 (higher because of the smaller size of the overall study and the relatively small number of attributable deaths). For the microsatellite polymorphism, the study is able to detect a 30% increase in the frequency of long and short (as opposed to medium-sized) alleles in patients compared to that in controls (2α of 0.05 and β of 0.80). The present study size allows us to detect a 75% increase in low-activity allele frequency in the attributable-death group compared with that in survivors.
DNA extraction from blood was carried out using Nucleon II kits (Scotlab Bioscience, Buckingham, United Kingdom), and patient and control DNA interspersed randomly in deep-well plates prior to genotyping. The scientists performing the genotyping were blind to whether samples were from patients or controls.
A restriction fragment length polymorphism analysis was designed to detect the SNP at base 2251 coding for the Arg753Gln polymorphism. The published primers were used (5) in a PCR mixture containing 50 ng of genomic DNA, 1× PCR buffer, 2 mM MgCl2, 0.32 mM deoxynucleoside triphosphates, a 0.12 μM concentration of each primer, and 0.2 U of AmpliTaq Gold DNA polymerase/μl. The PCR cycles consisted of a hot start at 94°C for 14 min, and then 40 cycles of 94°C for 15 s, 58°C for 30 s, and 72°C for 30 s, followed by 72°C for 2 min on a Tetrad Thermal cycler (MJ Research, Boston, Mass.). Overnight digestion was carried out at 37°C with 2 U of PstI from New England BioLabs (Hitchin, United Kingdom). The presence of wild-type homozygotes resulted in one band (430 bp), heterozygotes resulted in three bands (430, 281, and 149 bp), and mutant homozygotes resulted in two bands (281 and 149 bp). Each digestion was controlled with known homozygotes and heterozygotes.
For typing the microsatellite, the allele primers used were 5′-TATCCCCATTCATTCGTTCCATT-3′ (forward) and 5′FAM-GACCCCCAAGACCCACACC-3′ (reverse; contains the fluorescein marker 6-carboxyfluoroscein [FAM]). Sizes were determined with a PCR identical to that for SNP detection. The resulting fragments were run on an ABI 3700 sequencer.
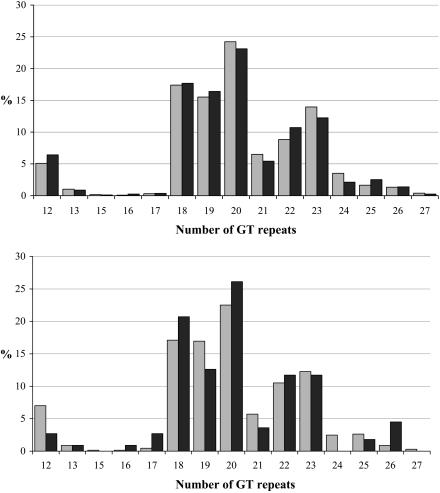
The genotype frequencies of the Arg753Gln SNP were similar to those described recently, with no observed homozygote mutants (5) (Table 2). There was no association between the heterozygote genotype and disease. The genotype frequencies for the disease subgroups were identical with 106 G/G genotypes (95%) and 6 G/A genotypes (5%) in the community-acquired-disease group and with 294 G/G genotypes (95%) and 14 G/A genotypes (5%) in the hospital-acquired-disease group. In the 56 cases in which death attributable to S. aureus infection occurred, there were no genotypic associations (Table 2). Similarly, there were no associations between the distribution of GT repeat alleles and disease or attributable mortality (Fig. 1). We divided the microsatellite alleles into two groups: those with putatively high promoter activity (medium-sized alleles having between 18 and 22 GT repeats) and those with putatively low activity (<18 or >22 GT repeats). The overall frequencies (95% confidence interval) of putatively low-activity alleles was 0.27 (0.24 to 0.30) for patients, compared to 0.28 (0.25 to 0.30) for controls (P = 0.68), 0.23 (0.16 to 0.32) for the patients in the attributable-death group, and 0.27 (0.24 to 0.31) for survivors (P = 0.41). There were no significant differences in the composite genotypic frequencies of putatively high- and low-activity alleles between patients and controls or between survivors and patients with attributable fatal cases (Table 3). Location of disease acquisition (community [112 patients] or hospital [308 patients]) had no significant effect on the genotype distribution of either polymorphism. We applied a statistical procedure which includes the expectation maximization algorithm (the “hapipf” command in the Stata 8.0 [Stata Corp., College Station, Tex.] statistical program) to determine phase, and we used log-linear modeling to examine for linkage disequilibrium and disease association between the Arg753Gln SNP and the GT repeat alleles. There was a significant linkage disequilibrium between the two loci (P = <0.001) but no association with either disease or attributable mortality with the computed haplotypes (6).
TABLE 2.
Arg753Gln genotyping results
| Genotype | No. (%) of patientsa
|
|||
|---|---|---|---|---|
| Cases | Controls | With attributable deaths | Without attributable deaths | |
| G/G | 400 (95) | 664 (95) | 53 (95) | 347 (95) |
| G/A | 20 (5) | 32 (5) | 3 (5) | 17 (5) |
| A/A | 0 | 0 | 0 | 0 |
| Total | 420 | 696 | 56 | 364 |
P = 0.89 (Fisher's exact test) for values for cases and controls. P = 0.74 (Fisher's exact test) for values for patients with and without attributable deaths.
FIG. 1.
(Upper) TLR2 microsatellite allele frequencies of cases (n = 396) versus those of controls (n = 638). The controls are represented by the lighter bars, and the cases are represented by the darker bars. (Lower) TLR2 microsatellite allele frequencies in the attributable-death group (n = 54) versus those in the group without death attributable to S. aureus (n = 342). The deaths which were not attributable to S. aureus are represented by the lighter bars, and the attributable deaths are represented by the darker bars.
TABLE 3.
Microsatellite genotype results based on functional composite genotypes
| Genotypea | No. (%) of patientsb
|
|||
|---|---|---|---|---|
| Cases | Controls | With attributable deaths | Without attributable deaths | |
| L/L | 14 (3) | 19 (3) | 0 | 14 (4) |
| H/L | 174 (44) | 280 (44) | 24 (44) | 150 (44) |
| H/H | 208 (53) | 339 (53) | 30 (56) | 178 (52) |
| Total | 396 | 638 | 54 | 342 |
L, short or long alleles with putatively low promotor activity; H, medium-length alleles with putatively high promotor activity.
P = 0.89 (Fisher's exact test) for values for cases and controls. P = 0.39 (Fisher's exact test) for values for patients with and without attributable deaths.
No associations were found between polymorphisms in the TLR2 genes and serious morbidity or mortality caused by S. aureus. For the assessment of susceptibility to severe disease, this case-control study is reasonably powerful, and although it is possible that an association has been missed, it is unlikely that any effect on susceptibility is large. However, within the cases, the study is able to detect only moderate to large effects on disease progression. The lack of association with the Arg753Gln polymorphism is consistent with recent in vitro evidence that the presence of only one wild-type allele is required for a full cytokine response to S. aureus lipoteichoic acid (11).
Acknowledgments
We are grateful to the staff of the John Radcliffe Hospital Microbiology Department and to Alex Smarason and the midwives at the Oxford Women's Centre for helping with patient recruitment and sample collection. We thank David Wyllie for providing functional data about the TLR2 microsatellite.
This study was funded by the Wellcome Trust.
REFERENCES
- 1.Esen, N., F. Y. Tanga, J. A. DeLeo, and T. Kielian. 2004. Toll-like receptor 2 (TLR2) mediates astrocyte activation in response to the Gram-positive bacterium Staphylococcus aureus. J. Neurochem. 88:746-758. [DOI] [PubMed] [Google Scholar]
- 2.Fujita, M., T. Into, M. Yasuda, T. Okusawa, S. Hamahira, Y. Kuroki, A. Eto, T. Nisizawa, M. Morita, and K. Shibata. 2003. Involvement of leucine residues at positions 107, 112, and 115 in a leucine-rich repeat motif of human Toll-like receptor 2 in the recognition of diacylated lipoproteins and lipopeptides and Staphylococcus aureus peptidoglycans. J. Immunol. 171:3675-3683. [DOI] [PubMed] [Google Scholar]
- 3.Iwaki, D., H. Mitsuzawa, S. Murakami, H. Sano, M. Konishi, T. Akino, and Y. Kuroki. 2002. The extracellular toll-like receptor 2 domain directly binds peptidoglycan derived from Staphylococcus aureus. J. Biol. Chem. 277:24315-24320. [DOI] [PubMed] [Google Scholar]
- 4.Kang, T. J., and G. T. Chae. 2001. Detection of Toll-like receptor 2 (TLR2) mutation in the lepromatous leprosy patients. FEMS Immunol. Med. Microbiol. 31:53-58. [DOI] [PubMed] [Google Scholar]
- 5.Lorenz, E., J. P. Mira, K. L. Cornish, N. C. Arbour, and D. A. Schwartz. 2000. A novel polymorphism in the toll-like receptor 2 gene and its potential association with staphylococcal infection. Infect. Immun. 68:6398-6401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mander, A. 2001. Haplotype analysis in population-based association studies. Stata J. 1:58-75. [Google Scholar]
- 7.Mempel, M., V. Voelcker, G. Kollisch, C. Plank, R. Rad, M. Gerhard, C. Schnopp, P. Fraunberger, A. K. Walli, J. Ring, D. Abeck, and M. Ollert. 2003. Toll-like receptor expression in human keratinocytes: nuclear factor kappaB controlled gene activation by Staphylococcus aureus is toll-like receptor 2 but not toll-like receptor 4 or platelet activating factor receptor dependent. J. Investig. Dermatol. 121:1389-1396. [DOI] [PubMed] [Google Scholar]
- 8.Ogus, A. C., B. Yoldas, T. Ozdemir, A. Uguz, S. Olcen, I. Keser, M. Coskun, A. Cilli, and O. Yegin. 2004. The Arg753GLn polymorphism of the human toll-like receptor 2 gene in tuberculosis disease. Eur. Respir. J. 23:219-223. [DOI] [PubMed] [Google Scholar]
- 9.Takeuchi, O., K. Hoshino, and S. Akira. 2000. Cutting edge: TLR2-deficient and MyD88-deficient mice are highly susceptible to Staphylococcus aureus infection. J. Immunol. 165:5392-5396. [DOI] [PubMed] [Google Scholar]
- 10.Takeuchi, O., K. Hoshino, T. Kawai, H. Sanjo, H. Takada, T. Ogawa, K. Takeda, and S. Akira. 1999. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity 11:443-451. [DOI] [PubMed] [Google Scholar]
- 11.von Aulock, S., N. W. Schröder, S. Traub, K. Gueinzius, E. Lorenz, T. Hartung, R. R. Schumann, and C. Hermann. 2004. Heterozygous Toll-like receptor 2 polymorphism does not affect lipoteichoic acid-induced chemokine and inflammatory responses. Infect. Immun. 72:1828-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weber, J. R., P. Moreillon, and E. I. Tuomanen. 2003. Innate sensors for Gram-positive bacteria. Curr. Opin. Immunol. 15:408-415. [DOI] [PubMed] [Google Scholar]
- 13.Yim, J. J., L. Ding, A. A. Schaffer, G. Y. Park, Y. S. Shim, and S. M. Holland. 2004. A microsatellite polymorphism in intron 2 of human Toll-like receptor 2 gene: functional implications and racial differences. FEMS Immunol. Med. Microbiol. 40:163-169. [DOI] [PubMed] [Google Scholar]