Abstract
Over the last few years, there has been an increasing amount of evidence for the de novo emergence of protein-coding genes, i.e. out of non-coding DNA. Here, we review the current literature and summarize the state of the field. We focus specifically on open questions and challenges in the study of de novo protein-coding genes such as the identification and verification of de novo-emerged genes. The greatest obstacle to date is the lack of high-quality genomic data with very short divergence times which could help precisely pin down the location of origin of a de novo gene. We conclude that, while there is plenty of evidence from a genetics perspective, there is a lack of functional studies of bona fide de novo genes and almost no knowledge about protein structures and how they come about during the emergence of de novo protein-coding genes. We suggest that future studies should concentrate on the functional and structural characterization of de novo protein-coding genes as well as the detailed study of the emergence of functional de novo protein-coding genes.
Keywords: De novo protein evolution, novel genes, de novo genes, gene emergence, protein-coding genes
Introduction
The question of how new genes come about has been a major research theme in evolutionary biology since the discovery that different species’ genomes contain varying numbers of genes. This question is difficult to answer, since emerging genes cannot easily be “caught in the act”. Ohno 1 gave the first comprehensive answer: new genes can emerge via the duplication of old genes. Consequently, gene duplication was thought to be the only mechanism of gene birth for many years 2. However, the discovery of so-called orphan genes in newly sequenced genomes raised doubt about the general validity of Ohno’s model of gene duplication. Orphan genes are genes that lack detectable homologs outside of a species or lineage. To explain the presence of orphans under the assumption that new genes emerge only via duplication, one has to assume gene loss in all other lineages or a phase of highly accelerated evolution that leads to the loss of detectable sequence similarity 3. Yet convergent gene loss in many independent lineages is unlikely — especially given the high number of orphan genes — and it is difficult to explain why so many genes would experience prolonged phases of accelerated evolution 4. On the contrary, it would be expected that genes that do not experience any selective pressure — which is required here for accelerated evolution — would be pseudogenized eventually, i.e. not be transcribed anymore.
These inconsistencies and further observations suggested that there could be other mechanisms of gene emergence 5, 6, for example de novo gene emergence, a process in which a new gene evolves from a previously non-genic sequence. The product of this process can be an RNA gene or a protein-coding gene. The possibility of de novo gene emergence has long been disputed, with many claiming that it is impossible for an intergenic, random open reading frame (ORF) to encode a functional protein (reviewed in 4, 7). But, despite these open questions regarding the exact mechanism of de novo gene birth, many recent studies report de novo emergence of protein-coding genes 5, 6, 8– 19.
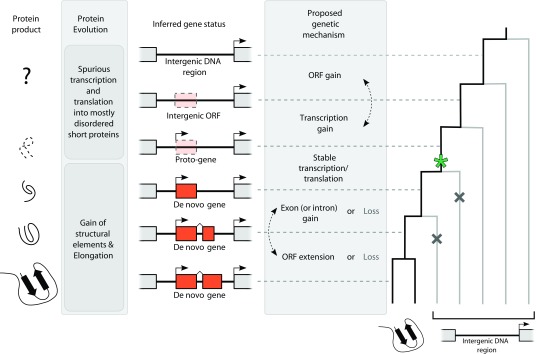
In general, genes without detectable homologs can be summarized under the term novel genes. These genes can also be called orphan genes, or — more precisely — species-/lineage-specific genes. The term de novo describes a specific subclass of novel genes, namely genes emerging from non-genic sequences 20. Additionally, one has to discriminate between functional genes and other classes of sequences. A de novo transcript can be any species-specific transcript that is homologous to an intergenic sequence in outgroups. De novo transcripts can be seen as putative de novo genes (see also Figure 1). The term protogene also describes intergenic transcripts or ORFs that are situated on a continuum between non-genic sequences and functional genes 21 (see also Figure 1). At the genic end of the spectrum, the term de novo gene describes a functional gene that has emerged de novo. De novo genes can either code for a protein or be functional as RNAs 22. Here, we will use the term de novo gene to describe de novo genes of unknown coding status and de novo protein-coding gene to describe de novo-emerged genes that likely produce a functional protein product.
Figure 1. Schematic depiction of de novo protein-coding gene emergence.
Shown is the hypothesis of a step-wise genic and structural maturation of an intergenic sequence towards a protein-coding gene. The steps are each shown as pictograms of protein and gene structure. An exemplary phylogenetic tree is shown to the right. The status of the protein/gene is projected onto the tree using grey, dotted lines. Gene emergence is depicted using a green star, gene loss using a grey X symbol. ORF, open reading frame.
Identification of de novo genes
The first step necessary to determine de novo status of a gene is to verify that no homologous sequences are present in outgroups. This homology search is often performed using BLAST or similar alignment search tools, for example against non-redundant protein databases containing all known protein sequences. Usually, an e-value cutoff between 10 −3 to 10 −5 is used for this step to ensure that no spurious, suboptimal alignments are taken into account 4. If this homology search does not find any homologs outside of the analyzed species, the query gene has successfully been confirmed to be a novel gene. This definition states only that there are no homologous sequences outside of a certain phylogenetic group. Calling a gene novel does not imply any knowledge about the emergence mechanism of the gene.
To additionally determine de novo gene origin, the homologous non-coding outgroup DNA sequence has to be retrieved 14, 23. The outgroup homologous sequence can be recovered using synteny information about the position of orthologous neighbor genes. Another possibility is searching the target gene sequence in outgroup genomes using alignment search tools such as BLAST 4, 23. A number of different types of de novo genes can be discriminated depending on the type of sequence that the genes likely emerged from 23.
Problems in de novo gene identification and annotation. In the past 24 and also more recently 25, 26, studies have raised questions regarding the reliability of homology-based searches of novel genes. Specifically, short and fast-evolving genes were proposed to lose detectable sequence similarity faster than other genes. As a result, shorter genes would be expected to be over-represented among young genes, thereby biasing the results of studies of genes of different ages 24– 26. Doubts have been raised as to which fraction of genes would actually be affected by this effect 27. Also, this should not be a problem for de novo genes defined by the methods summarized here. The possibility that the examined gene is actually a fast-evolving old gene is excluded, since for a confirmed de novo gene the homologous non-genic outgroup sequence has to be determined. Additionally, doubts have been raised regarding the accuracy of the initial claims of the unreliability of homology detection 28.
Another challenge is the previously mentioned identification of a non-coding sequence in an outgroup which is clearly homologous to the suspected de novo gene. In non-coding DNA, homology signals disappear very quickly, since non-coding sequences accumulate mutations faster than coding sequences. Because of this, it is often impossible to determine the homologous non-coding sequence in an outgroup. This problem increases with gene age. As a result, it is often not possible to determine the mechanism of origin, especially for older genes.
Additionally, there are methodological difficulties in the annotation of de novo and also all other types of novel genes 4. These problems could lead to a systematic underestimation of the number of de novo/novel genes. The problems are caused by genome annotation also being based on sequence homology 29. As de novo/novel proteins per definition do not possess any homologs, they cannot be annotated based on that criterion and their number is likely to be underestimated. Other common criteria such as minimum expression strength and the presence of multiple exons could also contribute to the problem, as these criteria do not represent intrinsic requirements for gene existence and are biased against de novo/novel genes 18. Nevertheless, the criteria might be necessary to prevent an over-annotation of spurious transcripts as genes, but they also make it impossible to identify all de novo genes. Recent studies on de novo protein-coding genes also employed such thresholds on exon number and expression strength to produce a more robust data set 15, 17, 18.
De novo gene emergence
Conceptually, de novo genes can evolve via two different mechanisms. The first mechanism is transcription-first, where an intergenic sequence gains transcription before evolving an ORF 20, 30. Recently, this has been shown to happen frequently when long non-coding RNAs (lncRNAs) become protein coding 17, 31, 32. Consequently, lncRNAs could represent an intermediate step in the evolution of a protein-coding gene 33. The second model is ORF-first, in which an intergenic ORF gains transcription 20, 30. Such a transcribed de novo ORF has been proposed to represent an intermediate step in gene emergence, a protogene ( Figure 1). High turnover of intergenic transcription 34 likely plays a role in de novo gene emergence by exposing novel transcripts to selection. Transposable elements can also play a role in de novo gene emergence 35. Additionally to whole proteins, terminal domains can also emerge de novo 33, 36. One model regarding the emergence of novel domains is the “grow slow and molt”, in which reading frames get extended gradually and eventually gain a structure and function 37, 38.
An additional process that could play a role during de novo protein-coding gene emergence is a (partial) revival of pseudogenized gene fragments. This possibility has already been proposed by Ohno 1. Regarding de novo protein-coding gene emergence, it seems possible that fragments of a pseudogenized gene that has been somewhat eroded by drift could become part of a de novo ORF later on. These fragments could provide a starting point for de novo protein emergence by providing remnants of structural elements. For all of these models, there are several consistent findings, but none of the models is, as yet, supported by a comprehensive set of data from diverse sources and corresponding experimental data.
De novo gene death. Orphan genes seem to generally have a high loss probability 14, 39 that seems to be negatively correlated with gene age 40, 41. The cause of this correlation is not yet well understood. It seems possible that young orphan genes have not yet gained a function or do not perform transient functions. It is also not clear yet how much of these findings can be transferred to de novo genes, as the studies on this topic examined all novel genes of different emergence mechanisms jointly.
De novo gene functions
A number of studies have examined the functions of orphan genes, some of which may represent de novo-emerged genes. Findings on orphan gene functions include involvement of orphan genes in the stress response 21, 42, rapid adaptation to changing environments as well as species-specific adaptations 43, 44, and limb regeneration 45. Additionally, novel genes were found to quickly gain interaction partners and become essential 39, 46.
Fewer studies, however, have examined the functions of systematically verified de novo-emerged genes. Generally, a high number of de novo genes was found to be expressed specifically in the testes, at least in Drosophila species 5, 6 and primates 18, as well as in plant pollen 16, 47. In the mouse, a de novo-emerged RNA gene was found to raise reproductive fitness 22. Another study found de novo genes to play a role in the Arabidopsis stress response 12. More specifically, one de novo ORF was found to play a role in male reproduction in Drosophila 48. Reinhardt et al. 48 also presented findings suggesting a role of de novo genes in developmental stages of Drosophila. However, these findings have to be interpreted carefully, as the RNAi method used has been shown to produce unreliable results 49, 50. A few other examples of functional de novo genes have been found 30, while others were not able to determine specific functions of identified de novo-emerged genes 15. The available data suggest that de novo-evolved genes can play a role in many different processes from reproduction to the stress response.
Recently, one study analyzed the function of two putative de novo protein-coding genes in Drosophila melanogaster 51. The two analyzed genes were found to be essential for male reproduction and to have testis-biased expression. Both genes are located inside introns of other, older genes with homologs in outgroups. However, the de novo origin of the analyzed genes could not be confirmed with certainty owing to the outgroup homologous sequences not being identifiable (see above for a general description of this problem).
Protein structure of de novo proteins
Little is known about the protein structures of de novo proteins. Some studies have found a high amount of intrinsic protein disorder 52 in very young genes 15, 51, 53, while others have not 21. A priori, it seems unlikely that de novo-emerging proteins have a well-defined protein structure. Intuitively, it seems more likely for random sequences to be intrinsically disordered instead (see Figure 1). Nevertheless, disordered regions can also be highly functional 52, 54 and could as such also represent an evolved state.
Also, contrary to intuition, at least semi-random (restricted alphabet) proteins appear to sometimes have a defined secondary structure 55, 56. Additionally, the existing protein structure families appear to have multiple origins 57. This finding suggests that the emergence of new protein structures is at least possible. Avoidance of misfolding and aggregation, on the other hand, have been proposed to be driving forces of protein evolution 58, 59. This observation and the existence of de novo protein-coding genes suggest that de novo proteins have the potential to exhibit a defined structure.
Open questions regarding de novo genes
Despite many advances in recent years, many open questions remain regarding de novo protein-coding genes. One understudied field is the functional characterization of protein-coding de novo-emerged genes. One non-coding RNA gene has been found to have a role in reproduction in the mouse 22, and additionally one likely protein-coding gene has been found to be essential for reproduction in Drosophila 48. However, beyond that, there is a substantial lack of data. Consequently, it remains unclear how de novo protein-coding genes gain their function and if there are some roles that they are more or less likely to carry out.
As described above, the structural characterization of de novo protein-coding genes is still an open question. Previously, ambiguous signals have been found regarding the role of intrinsic disorder in de novo-emerging protein-coding genes 15, 21. It would be important to experimentally verify the structure — or lack thereof — of de novo protein-coding genes. Here it is of major interest to determine the proportion of intergenic ORFs with folding potential and also what the implications are for the retention of such ORFs. This would allow further conclusions about de novo gene emergence: if most intergenic, random ORFs are foldable, function would seem to be the bottleneck of de novo protein-coding gene retention. On the other hand, if most confirmed de novo genes are folding, but most intergenic ORFs do not possess folding potential, folding potential would be a bottleneck of de novo protein-coding gene emergence and retention.
Another unsolved problem is how to find specific annotation thresholds for orphans/ de novo genes 4. As described above, a number of their properties make de novo genes difficult to annotate and to be distinguished from transcriptional noise. One solution would be to generate high-quality proteome data using e.g. mass spectrometry. However, this process is still highly expensive and might also not be able to generate a complete picture, since low-frequency peptides are hard to detect 60. Another method is ribosome profiling, which uses ribosome occupancy of sequences as a measure of translation. This method has been successfully used to show that some transcripts that were previously classified as non-coding could in fact be translated 61.
Additionally, patterns of selection, e.g. measured in the ratio of non-synonymous to synonymous mutations, can be used to infer the coding status of sequences. Genes with a higher fraction of synonymous mutations compared to non-synonymous mutations can be expected to be protein coding and under purifying selection 17, 20. However, these measures require a number of orthologs to be present, which makes them of limited use for novel genes. Another possibility is the use of population data for the same purpose, which circumvents the problem of the unavailability of orthologs for novel genes.
As it stands, studies mostly have to rely on arbitrary cutoffs 15, 17 and thus might miss a number of genes. It would be of major interest to be able to differentiate de novo genes and protogenes from transcriptional noise. Recent research has already shown that small ORFs (smORFs) can play a functional role 62, 63, and consequently it seems quite likely that also very short novel ORFs could be functional. This question also touches upon the problem of differentiating lncRNAs from protein-coding genes, which is often performed via an ORF length cutoff 17, 32.
Going forward, it is of major interest to fully characterize a large number of de novo genes in terms of evolutionary, functional, and structural history to be able to draw some general conclusion about their evolution. Specifically, it is of major interest to determine whether a functional role is an exception for protogenes or if most expressed ORFs have a functional impact which mostly does not affect the fitness of the organism at a significant level. If most expressed ORFs have only a negligible fitness effect, they would mostly evolve via drift. Two closely related questions are how and when de novo proteins gain their function: are de novo genes usually functional from the time point of their emergence, or do they gain a cellular task only after a period of drift?
Conclusions
In recent years, an increasing number of studies confirmed a major role of de novo gene emergence in the evolution of new protein-coding genes. The functional description of de novo-emerged genes is still lacking, but more general findings for orphan genes suggest that novel genes have a broad functional potential. However, the more detailed functional as well as structural characterization of de novo-emerged protein-coding genes remains one of the big open questions. An interesting recent finding was the confirmation of lncRNAs as an intermediate step in de novo protein-coding gene evolution. This finding offers a solution to two of the big questions in de novo gene evolution — how and why do intergenic sequences gain transcription? However, these findings also touch upon a difficult problem in studying de novo genes: how can protein-coding genes be distinguished from non-coding ones? This problem is exacerbated by recent findings that show that very short ORFs can also be functional 63. Tackling all of these problems and integrating them into detailed studies of the emergence, structure, and function of de novo protein-coding genes will provide new, interesting insights and allow for a deeper understanding of the inner workings of the evolution of de novo protein-coding genes.
Acknowledgements
The authors would like to thank Andreas Lange and the reviewers for valuable feedback on the manuscript.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
M. Mar Alba, UPF/IMIM, Barcelona, Spain
Tomislav Domazet-Lošo, Laboratory of Evolutionary Genetics, Ruđer Bošković Institute, Zagreb, Croatia
Chuan-Yun Li, The Institute of Molecular Medicine, Peking University, Beijing, China
Funding Statement
The author(s) declared that no grants were involved in supporting this work.
[version 1; referees: 3 approved]
References
- 1. Ohno S: Evolution by Gene Duplication.London: George Alien & Unwin Ltd. Berlin, Heidelberg and New York: Springer-Verlag.,1970. 10.1007/978-3-642-86659-3 [DOI] [Google Scholar]
- 2. Long M, Betrán E, Thornton K, et al. : The origin of new genes: glimpses from the young and old. Nat Rev Genet. 2003;4(11):865–75. 10.1038/nrg1204 [DOI] [PubMed] [Google Scholar]
- 3. Domazet-Loso T, Tautz D: An evolutionary analysis of orphan genes in Drosophila. Genome Res. 2003;13(10):2213–9. 10.1101/gr.1311003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tautz D, Domazet-Lošo T: The evolutionary origin of orphan genes. Nat Rev Genet. 2011;12(10):692–702. 10.1038/nrg3053 [DOI] [PubMed] [Google Scholar]
- 5. Levine MT, Jones CD, Kern AD, et al. : Novel genes derived from noncoding DNA in Drosophila melanogaster are frequently X-linked and exhibit testis-biased expression. Proc Natl Acad Sci USA. 2006;103(26):9935–9. 10.1073/pnas.0509809103 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 6. Begun DJ, Lindfors HA, Kern AD, et al. : Evidence for de novo evolution of testis-expressed genes in the Drosophila yakuba/Drosophila erecta clade. Genetics. 2007;176(2):1131–7. 10.1534/genetics.106.069245 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 7. Tautz D: The discovery of de novo gene evolution. Perspect Biol Med. 2014;57(1):149–61. 10.1353/pbm.2014.0006 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 8. Begun DJ, Lindfors HA, Thompson ME, et al. : Recently evolved genes identified from Drosophila yakuba and D. erecta accessory gland expressed sequence tags. Genetics. 2006;172(3):1675–81. 10.1534/genetics.105.050336 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 9. Cai J, Zhao R, Jiang H, et al. : De novo origination of a new protein-coding gene in Saccharomyces cerevisiae. Genetics. 2008;179(1):487–96. 10.1534/genetics.107.084491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Knowles DG, McLysaght A: Recent de novo origin of human protein-coding genes. Genome Res. 2009;19(10):1752–9. 10.1101/gr.095026.109 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 11. Toll-Riera M, Bosch N, Bellora N, et al. : Origin of primate orphan genes: a comparative genomics approach. Mol Biol Evol. 2009;26(3):603–12. 10.1093/molbev/msn281 [DOI] [PubMed] [Google Scholar]
- 12. Donoghue MT, Keshavaiah C, Swamidatta SH, et al. : Evolutionary origins of Brassicaceae specific genes in Arabidopsis thaliana. BMC Evol Biol. 2011;11:47. 10.1186/1471-2148-11-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu DD, Irwin DM, Zhang YP: De novo origin of human protein-coding genes. PLoS Genet. 2011;7(11):e1002379. 10.1371/journal.pgen.1002379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wissler L, Gadau J, Simola DF, et al. : Mechanisms and dynamics of orphan gene emergence in insect genomes. Genome Biol Evol. 2013;5(2):439–55. 10.1093/gbe/evt009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhao L, Saelao P, Jones CD, et al. : Origin and spread of de novo genes in Drosophila melanogaster populations. Science. 2014;343(6172):769–72. 10.1126/science.1248286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cui X, Lv Y, Chen M, et al. : Young Genes out of the Male: An Insight from Evolutionary Age Analysis of the Pollen Transcriptome. Mol Plant. 2015;8(6):935–45. 10.1016/j.molp.2014.12.008 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 17. Chen JY, Shen QS, Zhou WZ, et al. : Emergence, Retention and Selection: A Trilogy of Origination for Functional De Novo Proteins from Ancestral LncRNAsin Primates. PLoS Genet. 2015;11(7):e1005391. 10.1371/journal.pgen.1005391 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 18. Ruiz-Orera J, Hernandez-Rodriguez J, Chiva C, et al. : Origins of De Novo Genes in Human and Chimpanzee. PLoS Genet. 2015;11(12):e1005721. 10.1371/journal.pgen.1005721 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 19. Guerzoni D, McLysaght A: De Novo Genes Arise at a Slow but Steady Rate along the Primate Lineage and Have Been Subject to Incomplete Lineage Sorting. Genome Biol Evol. 2016;8(4):1222–32. 10.1093/gbe/evw074 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 20. Schlötterer C: Genes from scratch--the evolutionary fate of de novo genes. Trends Genet. 2015;31(4):215–9. 10.1016/j.tig.2015.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Carvunis AR, Rolland T, Wapinski I, et al. : Proto-genes and de novo gene birth. Nature. 2012;487(7407):370–4. 10.1038/nature11184 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 22. Heinen TJ, Staubach F, Häming D, et al. : Emergence of a new gene from an intergenic region. Curr Biol. 2009;19(18):1527–31. 10.1016/j.cub.2009.07.049 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 23. McLysaght A, Hurst LD: Open questions in the study of de novo genes: what, how and why. Nat Rev Genet. 2016;17(9):567–78. 10.1038/nrg.2016.78 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 24. Elhaik E, Sabath N, Graur D: The “inverse relationship between evolutionary rate and age of mammalian genes” is an artifact of increased genetic distance with rate of evolution and time of divergence. Mol Biol Evol. 2006;23(1):1–3. 10.1093/molbev/msj006 [DOI] [PubMed] [Google Scholar]
- 25. Moyers BA, Zhang J: Phylostratigraphic bias creates spurious patterns of genome evolution. Mol Biol Evol. 2015;32(1):258–67. 10.1093/molbev/msu286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Moyers BA, Zhang J: Evaluating Phylostratigraphic Evidence for Widespread De Novo Gene Birth in Genome Evolution. Mol Biol Evol. 2016;33(5):1245–56. 10.1093/molbev/msw008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Albà MM, Castresana J: On homology searches by protein Blast and the characterization of the age of genes. BMC Evol Biol. 2007;7:53. 10.1186/1471-2148-7-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Domazet-Loso T, Carvunis A, Alba MM, et al. : No evidence for phylostratigraphic bias impacting inferences on patterns of gene emergence and evolution. 2016. 10.1101/060756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yandell M, Ence D: A beginner’s guide to eukaryotic genome annotation. Nat Rev Genet. 2012;13(5):329–42. 10.1038/nrg3174 [DOI] [PubMed] [Google Scholar]
- 30. McLysaght A, Guerzoni D: New genes from non-coding sequence: the role of de novo protein-coding genes in eukaryotic evolutionary innovation. Philos Trans R Soc Lond B Biol Sci 2015;370(1678): 20140332. 10.1098/rstb.2014.0332 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 31. Ruiz-Orera J, Messeguer X, Subirana JA, et al. : Long non-coding RNAs as a source of new peptides. eLife. 2014;3:e03523. 10.7554/eLife.03523 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 32. Xie C, Zhang YE, Chen JY, et al. : Hominoid-specific de novo protein-coding genes originating from long non-coding RNAs. PLoS Genet. 2012;8(9):e1002942. 10.1371/journal.pgen.1002942 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 33. Bornberg-Bauer E, Albà MM: Dynamics and adaptive benefits of modular protein evolution. Curr Opin Struct Biol. 2013;23(3):459–66. 10.1016/j.sbi.2013.02.012 [DOI] [PubMed] [Google Scholar]
- 34. Neme R, Tautz D: Fast turnover of genome transcription across evolutionary time exposes entire non-coding DNA to de novo gene emergence. eLife. 2016;5:e09977. 10.7554/eLife.09977 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 35. Hoen DR, Bureau TE: Discovery of novel genes derived from transposable elements using integrative genomic analysis. Mol Biol Evol. 2015;32(6):1487–506. 10.1093/molbev/msv042 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 36. Andreatta ME, Levine JA, Foy SG, et al. : The Recent De Novo Origin of Protein C-Termini. Genome Biol Evol. 2015;7(6):1686–701. 10.1093/gbe/evv098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bornberg-Bauer E, Schmitz J, Heberlein M: Emergence of de novo proteins from ‘dark genomic matter’ by ‘grow slow and moult’. Biochem Soc Trans. 2015;43(5):867–73. 10.1042/BST20150089 [DOI] [PubMed] [Google Scholar]
- 38. Bitard-Feildel T, Heberlein M, Bornberg-Bauer E, et al. : Detection of orphan domains in Drosophila using “hydrophobic cluster analysis”. Biochimie. 2015;119:244–53. 10.1016/j.biochi.2015.02.019 [DOI] [PubMed] [Google Scholar]
- 39. Abrusan G: Integration of new genes into cellular networks, and their structural maturation. Genetics. 2013;195(4):1407–17. 10.1534/genetics.113.152256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Palmieri N, Kosiol C, Schlötterer C: The life cycle of Drosophila orphan genes. eLife. 2014;3:e01311. 10.7554/eLife.01311 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 41. Yang H, He BZ, Ma H, et al. : Expression profile and gene age jointly shaped the genome-wide distribution of premature termination codons in a Drosophila melanogaster population. Mol Biol Evol. 2015;32(1):216–28. 10.1093/molbev/msu299 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 42. Colbourne JK, Pfrender ME, Gilbert D, et al. : The ecoresponsive genome of Daphnia pulex. Science. 2011;331(6017):555–61. 10.1126/science.1197761 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 43. Khalturin K, Hemmrich G, Fraune S, et al. : More than just orphans: are taxonomically-restricted genes important in evolution? Trends Genet. 2009;25(9):404–13. 10.1016/j.tig.2009.07.006 [DOI] [PubMed] [Google Scholar]
- 44. Babonis LS, Martindale MQ, Ryan JF: Do novel genes drive morphological novelty? An investigation of the nematosomes in the sea anemone Nematostella vectensis. BMC Evol Biol. 2016;16(1):114. 10.1186/s12862-016-0683-3 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 45. Frobisch NB, Bickelmann C, Olori JC, et al. : Deep-time evolution of regeneration and preaxial polarity in tetrapod limb development. Nature. 2015;527(7577):231–4. 10.1038/nature15397 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 46. Zhang W, Landback P, Gschwend AR, et al. : New genes drive the evolution of gene interaction networks in the human and mouse genomes. Genome Biol. 2015;16:202. 10.1186/s13059-015-0772-4 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 47. Wu DD, Wang X, Li Y, et al. : “Out of pollen” hypothesis for origin of new genes in flowering plants: study from Arabidopsis thaliana. Genome Biol Evol. 2014;6(10):2822–9. 10.1093/gbe/evu206 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 48. Reinhardt JA, Wanjiru BM, Brant AT, et al. : De novo ORFs in Drosophila are important to organismal fitness and evolved rapidly from previously non-coding sequences. PLoS Genet. 2013;9(10):e1003860. 10.1371/journal.pgen.1003860 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 49. Green EW, Fedele G, Giorgini F, et al. : A Drosophila RNAi collection is subject to dominant phenotypic effects. Nat Methods. 2014;11(3):222–3. 10.1038/nmeth.2856 [DOI] [PubMed] [Google Scholar]
- 50. Vissers JH, Manning SA, Kulkarni A, et al. : A Drosophila RNAi library modulates Hippo pathway-dependent tissue growth. Nat Commun. 2016;7:10368. 10.1038/ncomms10368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gubala A, Schmitz JF, Kearns M, et al. : Two putative de novo evolved genes are essential for male fertility in Drosophila melanogaster . In press in Mol Bio Evol. 2016. [Google Scholar]
- 52. Tompa P: Unstructural biology coming of age. Curr Opin Struct Biol. 2011;21(3):419–25. 10.1016/j.sbi.2011.03.012 [DOI] [PubMed] [Google Scholar]
- 53. Kovacs E, Tompa P, Liliom K, et al. : Dual coding in alternative reading frames correlates with intrinsic protein disorder. Proc Natl Acad Sci U S A. 2010;107(12):5429–34. 10.1073/pnas.0907841107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Garner E, Romero P, Dunker AK, et al. : Predicting Binding Regions within Disordered Proteins. Genome Inform Ser Workshop Genome Inform. 1999;10:41–50. 10.11234/gi1990.10.41 [DOI] [PubMed] [Google Scholar]
- 55. Davidson AR, Lumb KJ, Sauer RT: Cooperatively folded proteins in random sequence libraries. Nat Struct Biol. 1995;2(10):856–64. 10.1038/nsb1095-856 [DOI] [PubMed] [Google Scholar]
- 56. Hecht MH, Das A, Go A, et al. : De novo proteins from designed combinatorial libraries. Protein Sci. 2004;13(7):1711–23. 10.1110/ps.04690804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Choi IG, Kim SH: Evolution of protein structural classes and protein sequence families. Proc Natl Acad Sci U S A. 2006;103(38):14056–61. 10.1073/pnas.0606239103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Monsellier E, Chiti F: Prevention of amyloid-like aggregation as a driving force of protein evolution. EMBO Rep. 2007;8(8):737–42. 10.1038/sj.embor.7401034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Geiler-Samerotte KA, Dion MF, Budnik BA, et al. : Misfolded proteins impose a dosage-dependent fitness cost and trigger a cytosolic unfolded protein response in yeast. Proc Natl Acad Sci U S A. 2011;108(2):680–5. 10.1073/pnas.1017570108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schulze WX, Usadel B: Quantitation in mass-spectrometry-based proteomics. Annu Rev Plant Biol. 2010;61:491–516. 10.1146/annurev-arplant-042809-112132 [DOI] [PubMed] [Google Scholar]
- 61. Wilson BA, Masel J: Putatively noncoding transcripts show extensive association with ribosomes. Genome Biol Evol. 2011;3:1245–52. 10.1093/gbe/evr099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Couso JP: Finding smORFs: getting closer. Genome Biol. 2015;16:189. 10.1186/s13059-015-0765-3 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 63. Mackowiak SD, Zauber H, Bielow C, et al. : Extensive identification and analysis of conserved small ORFs in animals. Genome Biol. 2015;16:179. 10.1186/s13059-015-0742-x [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation