Abstract
We recently reported a highly protective attenuated live virus vaccine for equine infectious anemia virus (EIAV) based on a proviral construct (EIAVUKΔS2) with a genetically engineered mutation in the viral S2 gene that eliminates expression of this accessory protein. While the EIAVUKΔS2 vaccine provides protection from detectable infection by experimental challenge with highly virulent virus, the potential for commercial application of this vaccine is complicated by the fact that horses inoculated with the EIAVUKΔS2 vaccine strain become seropositive in various reference diagnostic assays based on detection of antibodies to virion core or envelope proteins. To address this issue, we describe here the development and optimization of a new serologic EIAV diagnostic enzyme-linked immunosorbent assay (ELISA) to detect serum antibodies to the EIAV S2 protein that are produced in infected horses but not in horses inoculated with the EIAVUKΔS2 vaccine virus. The test S2 protein antigen was developed using the S2 gene sequence from the EIAVUK strain of virus and a series of modifications to facilitate production and purification of the diagnostic antigen, designated HS2G. Using this HS2G as antigen, we describe the development of an affinity ELISA that provides a sensitive and specific detection of S2-specific serum antibodies in experimentally and field-infected horses (22 of 24), without detectable reactivity with immune serum from uninfected (12 of 12) or vaccinated (29 of 29) horses. These data indicate that the S2-based diagnostic ELISA has the potential to accurately differentiate horses infected with EIAV from horses inoculated with an attenuated EIAV vaccine strain with a mutant S2 gene.
The macrophage-tropic animal lentivirus equine infectious anemia virus (EIAV) causes a persistent infection in horses and has been recognized as an important veterinary infectious disease for over 150 years (14, 24, 29). EIAV, transmitted by blood-feeding insects and fomites such as contaminated needles, is characterized by recurring cycles of viremia and clinical episodes that include fever, anemia, thrombocytopenia, edema, and wasting. Following 6 to 12 months of this chronic disease stage, most animals progress to an inapparent stage absent overt clinical disease (9, 14, 24, 29). Because of the economic importance of controlling EIAV infections, combined with the successful use of EIAV as an animal lentivirus model for human immunodeficiency virus type 1, the development of an effective vaccine for EIAV has been aggressively pursued (8, 16, 19, 25, 26, 33).
This laboratory has previously described the development of a live attenuated EIAV vaccine based on an engineered proviral construct with a mutated S2 gene, EIAVUKΔS2 vaccine (19). The EIAV S2 gene is located in the pol-env intergenic region, overlaps the amino terminus of the EIAV envelope proteins, and is synthesized in the late phase of the viral replication cycle (4, 6). While the biological function of this 65-amino-acid accessory protein is unknown, previous studies have indicated that S2 is not essential for EIAV replication in vitro (21, 28) but that the absence of S2 severely reduces EIAV replication and virulence in experimentally infected horses (20). In vivo studies of EIAVUKΔS2 as a potential attenuated viral vaccine demonstrated protection from infection in vaccinated horses subjected to low-dose multiexposure intravenous virulent virus challenge (to mimic field exposure) or to a single high-dose intravenous virus challenge (19).
In light of these promising initial observations on the efficacy of a live attenuated EIAV vaccine, the EIAVUKΔS2 vaccine may represent an effective vaccine for the control of EIAV infections. Thus, the development of a diagnostic assay that can effectively identify and differentiate EIAV-infected animals from EIAVUKΔS2-vaccinated horses has become a necessary advancement. In countries where veterinary regulatory policies are established, EIAV infections are currently controlled by detection and destruction or isolation of virus-infected horses. Approved diagnostic assays for EIAV infection are based on detection of serum antibodies to the capsid protein p26, transmembrane glycoprotein gp45, and surface glycoprotein gp90 in various assay formats (1, 3, 10, 15, 18, 22, 30-32). At present, the United States Department of Agriculture primary enzyme immunoassay (EIA) testing systems include the agar gel immunodiffusion assay (AGID or Coggins test) and enzyme-linked immunosorbent assay (ELISA), both of which are based on the detection of serum antibodies to EIAV p26 or gp45 (5, 13). Attenuated proviral vaccines containing a mutated S2 accessory gene still express the full complement of EIAV core and envelope proteins. Therefore, horses inoculated with EIAVUKΔS2 become seropositive in currently used diagnostic assays for EIAV infection. This is not a current problem but would become an issue if the EIAVUKΔS2 vaccine were approved for use in horses, as it would complicate current regulatory procedures used to control infection and disease in horses.
To address this important regulatory issue, we sought to develop and evaluate the reliability of an EIAV S2-based serological assay in detecting EIAV-infected horses and differentiating them from vaccinated horses inoculated with EIAVUKΔS2. We describe in the present report the engineering and production of a novel recombinant S2 protein antigen and the optimization of an S2 antigen-based ELISA for the detection of reactive antibodies in serum from experimentally and field-infected horses, without reactivity to antibodies in immune serum from horses inoculated with the attenuated EIAV vaccine. These data demonstrate for the first time the potential of an S2-based serological diagnostic assay to accurately identify and differentiate horses infected with wild-type EIAV from horses inoculated with S2 attenuated vaccine virus.
MATERIALS AND METHODS
Subcloning of S2 from EIAVUK.
To achieve adequate levels of recombinant S2 protein production, a series of cloned S2 gene constructs were developed and evaluated for protein expression properties. These constructs are summarized in Table 1.
TABLE 1.
Plasmids, promoters, and primers used for S2 antigen expression and for RT-PCR assays of expression levels
| Antigen | Plasmid | Promoter | Primera | Length (bp) |
|---|---|---|---|---|
| EIAVUK S2 | pcDNAS2 | CMV | FS2 | 210 |
| RS2 | ||||
| EIAVUK S2 | pcDNAS2 | VVb-T7 | FS2 | 210 |
| RS2 | ||||
| OptS2 | pOptS2 | CMV | FOptS2 | 210 |
| ROptS2 | ||||
| OptS2 | pOptS2 | VV-T7 | FOptS2 | 210 |
| ROptS2 | ||||
| OptS2 | pOptS2GFP | CMV | FOptS2 | 210 |
| ROptS2 | ||||
| OptS2GFP | pOptS2GFP | CMV | FOptS2 | 930 |
| RNcGFP | ||||
| EGFP | pEGFP-N3 | CMV | FKpGFP | 732 |
| RNcGFP |
F, forward primer; R, reverse primer.
VV, vaccinia virus.
The pcDNAS2 construct (Table 1) was developed to express histidine peptide-tagged S2 protein from the native EIAV S2 gene sequences. For construction of pcDNAS2 expression vector (Table 1), the EIAV S2 gene sequence was amplified from EIAVUK (6) by PCR with Taq DNA polymerase (Invitrogen, Carlsbad, Calif.) with primers FS2 (GGATTATTTGGTAAAGGG) and RS2 (TCATTTCTTGGTCTCTTG). The PCR products were ligated into pcDNA4/HisMAX (Invitrogen) by TA cloning, and a six-histidine peptide was fused to the 5′ end of S2. This vector allows for expression of a His6-tagged S2 protein from the cytomegalovirus (CMV) and phage T7 promoters.
The pOptS2 construct was designed to express recombinant histidine peptide-tagged S2 from an S2 gene modified for codon optimization. For construction of pOptS2 expression vector, the Codon Usage Database (www.kazusa.or.jp) was used to optimize the codons of the EIAVUK S2 gene sequence for maximum expression in mammalian and Escherichia coli cells (Midland Reagent Company, Midland, Tex.). This synthesized codon-optimized S2 sequence (Fig. 1) was subcloned into the pcDNA3.1/mycHis vector (Invitrogen) by using the NheI and HindIII restriction enzyme sites, and a His6 peptide was fused to the 3′ end of S2. This vector allows for expression of the histidine peptide-tagged S2 from the CMV and phage T7 promoters.
FIG. 1.
Sequence alignment of the parental and codon-optimized EIAVUK S2 gene sequences. The parental S2 sequence is as reported in the work of Cook et al. (6). The codon-optimized sequence was derived using the Codon Optimization Database (www.kazusa.or.jp).
The pOptS2GFP construct (Table 1) was designed to express recombinant histidine peptide-tagged S2 as a fusion protein with enhanced green fluorescent protein (EGFP). To construct the pOptS2GFP expression vector, the codon-optimized S2 gene sequence was amplified from the pOptS2 plasmid by PCR with the Pfu Turbo polymerase (Stratagene, La Jolla, Calif.) and primers FOptS2 (GGCCGTGCTAGCATGGGCCTGTTCGGCAAG) and ROptS2 (CTAGTGGGTACCCTTCTTGGTCTCCTGCTTGCGGCG). The digested fragments were ligated into the pEGFP-N3 plasmid (Clontech, Palo Alto, Calif.), which contained the EGFP, through the NheI and KpnI restriction enzyme sites. Expression from the pOptS2GFP vector was driven by the CMV promoter, and the EIAV S2-EGFP fusion protein was designated HS2G.
In addition to the expression plasmids described above for expression of mammalian cells, expression plasmids were also developed for production of recombinant proteins in E. coli. In the construction of the p80LOS2GFP expression vector for expressing the HS2G fusion protein in E. coli, the codon-optimized S2 gene sequence was amplified from the pOptS2 plasmid by PCR with primers FS2SacI (GTACCAGAGCTCATGGGCCTGTTCGGCAAG) and ROptS2 (shown above). The resulting fragments were double digested with the SacI and KpnI restriction enzymes and ligated into the pQE-Trisystem plasmid (Qiagen, Valencia, Calif.) to generate the intermediate vector pOS2QE. Next, the EGFP gene sequence was amplified from the pEGFP-N3 plasmid by using the primers FHindGFP (ATGGTGAAGCTTATCGAGGGAAGGGTGAGCAAGGGCGAGGAGCTGTTC) and RXhoIG (GAATTCCTCGAGCTTGTACAGCTCGTCCATGCCGAG) and subcloned into the pOS2QE plasmid by using the HindIII and XhoI restriction enzyme sites. This plasmid was designated pOS2GFP. The S2-EGFP gene sequence was then excised from the pOS2GFP plasmid and ligated into the pQE-80L plasmid (Qiagen) by using the SacI and SalI restriction enzyme sites. Expression of the HS2G fusion protein from the final p80LOS2GFP expression vector was driven by the CMV promoter. All expression vectors were verified by DNA sequencing analyses.
Transfection and protein expression in COS7 cells.
Monkey kidney (COS7) cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, penicillin (200 U/ml), and streptomycin (100 μg/ml) in a humidified atmosphere with 5% CO2 at 37°C. COS7 cells were transfected with the S2 expression vectors by using the PolyFect transfection reagent according to the manufacturer's protocol (Qiagen). When expression was to be driven by the T7 promoter, 80% confluent COS7 cells that had been seeded for 24 h were infected by recombinant vaccinia virus vTF7-3, which synthesizes bacteriophage T7 RNA polymerase at a multiplicity of infection of 30 for 30 min prior to transfection, as described elsewhere (7).
Protein expression in E. coli.
Previous studies in our lab indicated a high cytotoxic effect of expressed S2 protein in E. coli (unpublished data). To minimize this cytotoxic effect, the E. coli strain XL1-Blue (Stratagene) was utilized for S2 antigen expression. The presence of the lacIq mutation efficiently blocks transcription by causing production of high amounts of the lac repressor. Following transformation with the p80LOS2GFP expression vector, the transcription and expression of HS2G were rapidly induced by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside), which bound the lac repressor proteins, thus preventing their binding with lacO, to permit the initiation of transcription.
Following transformation with the p80LOS2GFP expression vector, the recombinant E. coli XL:p80LOS2GFP cells were precultured in 5 ml of Luria broth (LB) medium supplemented with 2 g of glucose/liter and 100 μg of ampicillin/ml overnight at 37°C with vigorous shaking. The overnight culture was used to inoculate 100 ml of LB medium. The expression of HS2G protein was induced by addition of 1 mM IPTG to the log-phase culture. The bacterial pellets were harvested at 4 h postinduction by centrifugation at 6,000 × g for 10 min and stored at −80°C until needed.
Purification of HS2G under denaturing conditions.
In preliminary experiments, it was indicated that HS2G protein expressed in E. coli was localized predominantly in relatively insoluble inclusion bodies. To recover HS2G from these inclusion bodies, we employed 8 M urea to denature the proteins. In brief, bacterial pellets were lysed using buffer A (100 mM NaH2PO4, 10 mM Tris-HCl, and 8 M urea, pH 8.0) at 5 ml/g (wet weight) of cells. After 45 min of shaking, nucleic acids and cell debris were removed by centrifugation at 10,000 × g for 15 min. The lysates were loaded onto a 5-ml IMAC column (Amersham, Piscataway, N.J.) that was preequilibrated with buffer A. The column was then washed with wash buffer (100 mM NaH2PO4, 10 mM Tris-HCl, 8 M urea, pH 6.3), and the denatured HS2G protein was eluted using the elution buffer (100 mM NaH2PO4, 10 mM Tris-HCl, and 8 M urea, pH 4.9). Fractions were monitored at 280 nm, and those fractions containing protein were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot assay.
Protein assay.
Protein concentrations were determined using the Bio-Rad protein assay reagent, and bovine gamma globulin (immunoglobulin G [IgG]) was used as a standard (Bio-Rad Laboratories, Hercules, Calif.).
Protein refolding.
HS2G proteins purified under denaturing conditions were refolded by stepwise dialysis against buffer 1 (6 M urea, 0.5 M NaCl, 0.1 mM dithiothreitol, and 0.5 mM phenylmethylsulfonyl fluoride) at 4°C. This dialysis buffer was then diluted stepwise against buffer 2 (25 mM Tris-HCl [pH 7.5] and 150 mM NaCl) until the urea concentration decreased to 2 M over 1 day, at which time the proteins were dialyzed against buffer 3 (25 mM Tris-HCl [pH 7.5], 0.1 M NaCl, and 5% glycerol).
SDS-PAGE and Western blot analysis of recombinant S2 and HS2G proteins in cell lysates.
For SDS-PAGE analysis of purified S2 proteins, samples were solubilized in protein sample buffer (2.5% SDS, 1% β-mercaptoethanol [pH 6.8], 0.05 M Tris-HCl, 0.01% bromophoenol blue, 3 mM EDTA, and 10% glycerol) prior to being heated at 95°C for 5 min. The proteins were then resolved on a 4 to 15% polyacrylamide gel (Bio-Rad) and visualized by Coomassie blue R-250 staining.
For analysis of intracellular S2 protein expression by Western blotting, cell lysates were first resolved on a precast 4 to 20% polyacrylamide gel (Bio-Rad) and then transferred electrophoretically to a polyvinylidene difluoride membrane (Millipore, Billerica, Mass.) at 100 V for 1 h. The membrane was first blocked for 1 h with Tween-phosphate-buffered saline (PBS; 1.37 M NaCl, 27 mM KCl, 81 mM Na2HPO4 · 12H2O, 15 mM KH2PO4, and 0.1% Tween 20) containing 5% powdered nonfat milk and then incubated with primary antibodies (rabbit anti-S2 serum [1:200] or GFP monoclonal antibody [1:2,000]; Promega, Madison, Wis.) at room temperature for 1 h. Following three 5-min washes with Tween-PBS, membranes were incubated with secondary antibodies (horseradish peroxidase [HRP]-conjugated anti-rabbit IgG [1:10,000] or HRP-conjugated anti-mouse IgG [1:4,000]; Sigma, St. Louis, Mo.) for 1 h and then treated with three final washes. Specific proteins were visualized with Western lightning chemiluminescence reagent plus substrate mixture (Perkin-Elmer, Boston, Mass.).
Quantification of S2-specific mRNA by RT-PCR assay.
To evaluate the transcription levels of the various S2 gene constructs in transfected cells, COS7 cells were transfected with the S2 expression plasmids outlined in Table 1 and harvested 48 h posttransfection. The mRNA from transfected cells was then prepared using a one-step reverse transcription-PCR (RT-PCR) system (Pierce, Rockford, Ill.), and the assay was performed according to the manufacturer's instructions. Levels of β-actin mRNA were assayed separately as an internal reference. The β-actin primer sets were as follows: forward primer, 5′-ATGGATGATGATATCGCCGC-3′; reverse primer, 5′-GAGTCCATCACGATGCCAGT-3′. Levels of mRNA in the cell lysates were measured and quantified using Kodak 1-D gel electrophoresis analysis software.
Sources of immune sera.
A panel of 65 immune serum samples was obtained from EIAV test-positive field horses and from horses experimentally infected with the attenuated EIAVUKΔS2 vaccine strain (19). All field and experimental horses were shown to be seropositive in United States Department of Agriculture reference AGID (Coggins) assays. Control horse sera were obtained from uninfected horses and confirmed as seronegative in the Coggins AGID assay. Rabbit antiserum to S2 protein was produced by immunization of rabbits with a synthetic S2 peptide preparation.
Detection of S2-specific serum antibodies by ELISA.
Serum reactivity to the recombinant purified HS2G protein was assayed in a standard ELISA format and compared to reactivity in an affinity ELISA format. For the standard ELISA procedure, 96-well microplates (Immunolon 2; Dynatech Laboratories, Inc., Chantilly, Va.) were coated overnight with HS2G protein diluted in 0.05 M sodium bicarbonate buffer (pH 9.6) to a final concentration of 10 μg/ml. The following day the plates were washed using PBS-Tween (0.05%) and blocked with blocking buffer (PBS containing 5% calf serum, 5% nonfat dry milk, and 0.25% Tween 20) for 1 h at room temperature. After washing with PBS-Tween, 100 μl of 1:25-diluted horse serum in blocking buffer was applied to the plates and incubated for 30 min at room temperature. The plates were washed again and incubated with 100 μl of 1:35,000-diluted anti-horse IgG HRP-conjugated antibody (Sigma) for 45 min at room temperature. Following a final PBS-Tween wash, 100 μl of the substrate 3,3′,5,5′-tetramethylbenzidine (TMB) (Sigma) was added to each well, and color was allowed to develop for 10 min before the reaction was stopped by the addition of 100 μl of 2 M H2SO4. The extent of the reactions was determined spectrophotometrically with a microplate reader (MR5000; Dynatech Laboratories Inc.) at 450 nm with the software BIOLINX. In the analyses with rabbit anti-S2 serum as primary antibody in the ELISA, the rabbit immune serum was 1:200 diluted in blocking buffer, and anti-rabbit IgG HRP-conjugated antibody diluted 1:10,000 (Sigma) was used as the secondary antibody.
For the affinity ELISA procedure (NN-ELISA), HS2G protein was diluted in PBS-bovine serum albumin (BSA; 0.2%) buffer to a final concentration of 10 μg/ml. Ni-NTA HisSorb plates (Qiagen) were coated with 100 μl of diluted HS2G per well by gentle shaking for 1.5 h at room temperature. Blocking was not required as the plates were preblocked with BSA by the manufacturer. PBS was used as wash buffer, and PBS-BSA was employed to dilute primary and secondary antibodies. All subsequent procedures were as described above for the standard format.
Horse sera known to be negative or positive for EIAV antibodies were tested in parallel in each assay as controls. Values for test serum samples were derived by subtracting the adsorption values obtained at an optical density at 450 nm (OD450) of the negative control serum sample. The cutoffs of HS2G ELISA and p26 ELISA were 0.3 and 0.1, respectively. We defined these cutoff levels based on our preliminary ELISA study with a panel of serum samples known to be seropositive and seronegative. All data were representative of three independent experiments.
Production of recombinant EIAV p26.
For the production of EIAV p26 capsid antigen, the p26 gene of EIAV was subcloned from EIAVUK, modified with a His6 tag for purification, and inserted into the pQE-80L expression plasmid as described above. E. coli cells were transformed with the p26 expression plasmid and precultured in 5 ml of LB medium supplemented with 2 g of glucose/liter and 100 μg of ampicillin/ml overnight at 37°C with vigorous shaking. The overnight culture was used to inoculate 100 ml of LB medium. The expression of p26 protein was induced by addition of 1 mM IPTG to the log-phase culture. The bacterial pellets were harvested at 4 h postinduction by centrifugation at 6,000 × g for 10 min. The cells were lysed with a bacterial protein extraction reagent (Pierce), filtered through 0.45-μm-pore-size filters, and applied to a column equilibrated with the binding buffer (10 mM Na2HPO4 · 2H2O, 10 mM NaH2PO4 · H2O, and 0.5 M NaCl [pH 7.4]) according to the manufacturer's directions (Amersham). The column was then washed with the wash buffer containing 10 mM Na2HPO4 · 2H2O, 10 mM NaH2PO4 · H2O, 0.5 M NaCl, and 10 mM imidazole (pH 7.4) at a flow rate of 5 ml/min. The p26 protein was eluted using the elution buffer (10 mM Na2HPO4 · 2H2O, 10 mM NaH2PO4 · H2O, 0.5 M NaCl, and 250 mM imidazole [pH 7.4]). To examine the quality of the purified p26 protein, fractions collected from the OD280 determinations were measured for their protein content and analyzed by SDS-PAGE and Western blot assays.
Detection of serum antibodies to p26.
For p26-specific NN-ELISA procedures, purified recombinant p26 antigen was dissolved in PBS-BSA (0.2%) to a final concentration of 5 μg/ml and horse serum was diluted to 1:200 in PBS-BSA. The other procedures for the p26 assay were as described above for the NN-ELISA to HS2G.
RESULTS
Expression of recombinant S2 antigen in mammalian cells.
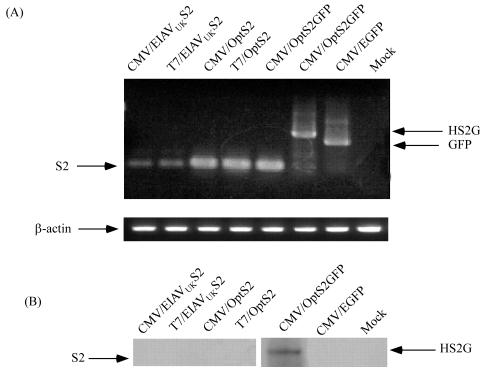
To achieve reproducible and adequate expression of a recombinant S2 antigen, a series of S2 gene expression constructs were developed and evaluated. Comparative transcription and protein expression levels from this series of antigen expression constructs are summarized in Fig. 2. Our first attempts to express the EIAV S2 protein began with the pcDNAS2 expression plasmid containing the EIAVUK S2 sequence. This plasmid was transfected into the mammalian cell line COS7, and mRNA expression (Fig. 2A) and recombinant S2 protein production (Fig. 2B) were evaluated by RT-PCR and Western blot analyses, respectively. These assays indicated a lack of detectable S2 protein expression in the transfected cells, despite the presence of detectable S2 mRNA from both the T7 and CMV promoters.
FIG. 2.
Analysis of the expression level of S2 mRNA and protein in transfected COS7 cells. (A) COS7 cells were transfected with the individual expression plasmids outlined in Table 1. Cell lysate was prepared from 1.5 × 105 cells harvested 48 h posttransfection, and mRNA was extracted and analyzed by RT-PCR with the primers indicated in Table 1. (B) Western blot analysis of S2 expression in COS7 cells. Approximately 3.2 × 106 cells were harvested, lysed, and immunoblotted using rabbit anti-S2 antibodies.
The transcription and translation efficiencies of the pcDNAS2 expression plasmid were next evaluated as a possible reason for lack of expression. It was determined that the equine S2 arginine codons AGG and AGA are less frequently utilized in COS and E. coli cells, probably due to a lower abundance of corresponding tRNAs. Accordingly, we constructed the pOptS2 expression plasmid by using the optimized EIAVUK S2 (OptS2) codon sequence (Fig. 1), and following transfection of COS7 cells, S2 mRNA and protein expression were evaluated using both the T7 and CMV promoters (Fig. 2). The results of these assays indicated that the codon optimization enhanced S2 mRNA expression by the T7 or CMV promoters by 3.2-fold compared to the parental S2 gene but that this increased mRNA expression still failed to yield detectable S2 protein in the transfected cells.
These observations suggested a potential instability of the expressed S2 protein in transfected cells. Small peptides are often degraded in vivo, and as S2 has a molecular mass of only 7.2 kDa (28), degradation was considered a possible reason for the lack of S2 expression in vitro. To stabilize and protect our expressed protein from degradation, a fusion protein was constructed by attaching the EGFP to the carboxyl terminus of the S2 protein (see Materials and Methods). This fusion protein was designated HS2G, and the plasmid expressing this fusion protein was termed pOptS2GFP. To assess the efficacy of this expression construct, COS7 cells were transfected as described above and assayed for mRNA and recombinant protein expression. The data in Fig. 2 demonstrate substantial mRNA production in the transfected cells and, for the first time, detectable levels of the recombinant S2 protein as part of the HS2G fusion protein by immunoblotting with the rabbit anti-S2 serum. The expression of the fusion protein was also verified using mouse anti-EGFP as a primary antibody (data not shown). These results indicate that the HS2G fusion protein is successfully recognized by anti-S2 antibodies and anti-EGFP. All combined, these data suggest that construction of the HS2G fusion protein successfully prevented degradation of the S2 protein in COS7 cells.
Production and purification of S2 antigen from E. coli.
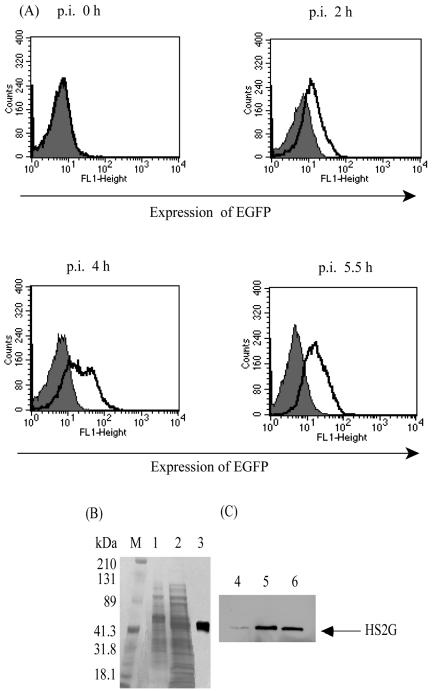
In parallel with our development of an S2 antigen expression system in mammalian COS7 cells as a basis for investigating S2 function in viral replication, we also examined the expression levels of the various recombinant antigen constructs in E. coli to provide a high-yield production system for a potential diagnostic antigen. For these purposes, the various engineered S2 genes were cloned into the prokaryotic expression plasmid pQE-80L, and their expression levels were evaluated in transformed E. coli. As observed with expression in COS7 cells, we observed in preliminary experiments that there was no detectable S2 antigen produced in E. coli cells transformed with either the parental S2 gene or the codon-optimized S2 gene, while substantial levels of the recombinant HS2G fusion protein were detected in E. coli transformed with the p80LOS2GFP expression plasmid (data not shown). Figure 3 summarizes representative data on HS2G expression in transformed E. coli cells. As described in Materials and Methods, p80LOS2GFP plasmid was transformed into E. coli XL cells. The expression of HS2G was verified by flow cytometric analysis as shown in Fig. 3A. Approximately 60% of recombinant cells expressed HS2G protein at 4 h postinduction with IPTG. This experimental study also demonstrated the optimal time frame for cell harvest to be 4 h post-IPTG induction because it presented the highest expression level of the fusion protein over the postinduction period 0 to 5.5 h.
FIG. 3.
Expression of HS2G in E. coli XL:p80LOS2GFP. (A) Flow cytometric analysis of the expression of HS2G in E. coli transformed with XL:p80LOS2GFP (bold lines). Wild-type E. coli served as a control (shaded). (B) Coomassie blue visualization of HS2G protein resolved by SDS-4 to 20% PAGE. E. coli collected 4 h postinduction was lysed under different conditions. Lane 1, cell lysate prepared under native conditions; lane 2, cell lysate prepared under denaturing conditions; lane 3, HS2G purified under denaturing conditions; lane M, standard protein markers. p.i., postinduction. (C) Immunoblot of HS2G with rabbit anti-EIAV S2 antibody as primary antibody (1:200 dilution) and anti-rabbit IgG HRP as the secondary antibody (1:10,000 dilution). Lane 4, HS2G in the cell lysate prepared under native conditions; lane 5, HS2G in the cell lysate prepared under denaturing conditions; lane 6, HS2G purified under denaturing conditions.
To evaluate the possible methods for extracting the HS2G protein from the transformed bacteria, soluble proteins were extracted from a transformed E. coli cell pellet by lysis with a commercially available bacterial protein extraction reagent (Pierce), filtered, and evaluated by SDS-PAGE (lane 1, Fig. 3B) and Western blotting (lane 4, Fig. 3C), respectively. To extract insoluble proteins from the transformed cells, an identical E. coli cell pellet was lysed using 8 M urea (Materials and Methods) and the extracted proteins were evaluated by both SDS-PAGE (lane 2, Fig. 3B) and Western blotting (lane 5, Fig. 3C). The data in Fig. 3B and C indicate that the HS2G protein expressed in transformed E. coli was predominantly in the insoluble fraction of the cell pellet, thus requiring 8 M urea for extraction. The extracted HS2G protein was then purified from the total cellular extract by affinity chromatography on a nickel-charged IMAC column according to the manufacturer's procedures. This purification procedure yielded a total of about 4.5 mg of HS2G protein from 500 ml of culture broth without detectable contaminating proteins (lane 3, Fig. 3B, and lane 6, Fig. 3C). To address the concern that denatured HS2G protein may be less reactive with serum antibodies and thus a less useful antigen in an ELISA, the affinity column-purified denatured HS2G protein was refolded by stepwise dialysis, as described in Materials and Methods. Typically, the final refolded HS2G protein yield was about 50% of the starting protein antigen recovered from the affinity column.
Development and optimization of an HS2G-based ELISA.
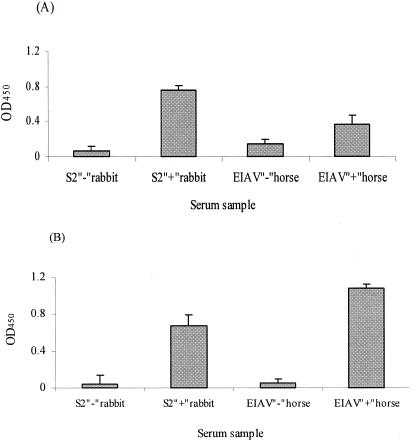
Having achieved the necessary goal of S2 antigen production, we next sought to evaluate the utility of an S2-based serological assay to detect EIAV-infected horses and differentiate them from horses inoculated with attenuated vaccine virus lacking S2 expression. To address this issue, the reactivity of the purified HS2G protein was initially examined in a standard ELISA for reactivity with a reference rabbit immune serum and with a reference EIAV-positive horse serum. The data in Fig. 4A demonstrate that HS2G protein displayed significant reactivity to the rabbit anti-S2 polyclonal antibodies, but relatively low-level reactivity was observed with the reference horse serum. The relatively low level of S2-specific antibody reactivity was consistent with previous reports of low titers of antibody to S2-specific synthetic peptides in ELISA formats (2, 24).
FIG. 4.
Evaluation of the reactivity of antigens to rabbit anti-S2 polyclonal antibodies and EIAV-infected horse serum in standard ELISA and NN-ELISA. Shown are HS2G reactivities to rabbit anti-S2 polyclonal antibodies (1:200 dilution) and to EIAV-infected horse immune serum (1:25 dilution) in the standard ELISA (A) or the affinity NN-ELISA (B) format.
To increase the sensitivity of the HS2G in vitro ELISA system, we modified the standard ELISA by employing Ni-NTA HisSorb 96-well ELISA plates to affinity adsorb the HS2G protein antigen through the His6 tag of the fusion protein. Thus, HS2G protein was applied as a coating to the Ni-NTA plates, and the reactivity of the adsorbed antigen with the reference rabbit and horse immune sera was evaluated. As shown in Fig. 4B, a level of reactivity of HS2G to rabbit anti-S2 antibodies was achieved in NN-ELISA similar to that in the standard ELISA above. In contrast, however, the reactivity of the horse immune serum in the NN-ELISA was increased by about threefold compared to the standard ELISA format, while the background reactivity with the negative control horse serum was reduced by about twofold in the NN-ELISA compared to the standard ELISA procedure. These data indicated the potential of the NN-ELISA as a format for further development and assessment.
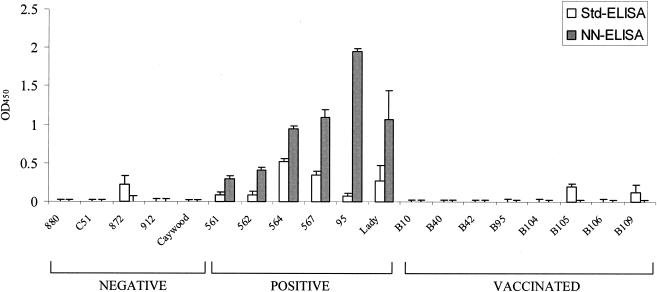
To evaluate in more detail the sensitivity and specificity of HS2G protein and to validate the application of the NN-ELISA system for identifying EIAV-infected horses, we appraised the reactivity of a coded panel of reference sera from EIAV-negative horses, EIAV-infected horses, and EIAVUKΔS2-vaccinated horses to HS2G in the NN-ELISA system and compared these results to those obtained from the standard ELISA system (Fig. 5). The results indicated that the NN-ELISA was superior to the standard ELISA procedure in both the sensitivity and the specificity of serum reactivity. For example, all five of the control serum samples from uninfected horses were seronegative in the NN-ELISA, while one of the uninfected horse serum samples displayed reactivity in the standard ELISA format. Similarly, all eight of the immune serum samples from EIAVUKΔS2-vaccinated horses were seronegative in the NN-ELISA format, while two of these vaccinated serum samples displayed low levels of seroreactivity in the standard ELISA. Taken together, these observations demonstrate the absence of nonspecific seroreactivity to the HS2G antigen in the context of the NN-ELISA format, providing the specificity required to avoid false indications of EIAV-infected horses. In addition to the superior specificity, the NN-ELISA also yielded substantially higher levels of reactivity of all six reference immune serum samples from EIAV-infected horses compared to the standard ELISA. The seroreactivity levels achieved in the NN-ELISA format for the reference immune serum samples ranged from 3- to 20-fold higher than the respective serum reactivity realized in the standard ELISA. Importantly, for three of the EIAV-positive immune serum samples (horses 561, 562, and 95), the NN-ELISA provided a strongly positive signal, whereas the standard ELISA reactivity bordered on a negative level. Thus, these data demonstrate the preferred sensitivity and specificity of the NN-ELISA for accurately detecting EIAV-infected horses with relatively low levels of S2-specific serum antibodies, while reliably distinguishing uninfected or vaccinated horses lacking S2-specific serum antibodies.
FIG. 5.
Comparison of HS2G reactivities to horse sera with the standard ELISA (Std-ELISA) and NN-ELISA (HS2G-NN) formats. Equal amounts of purified HS2G protein were analyzed in parallel in either the standard or affinity ELISA format against a panel of reference horse serum samples as described in Materials and Methods. Negative, uninfected horses shown to be seronegative in standard EIAV p26 diagnostic assays; positive, virus-infected horses shown to be seropositive in standard EIAV p26 diagnostic assays; vaccinated, horses experimentally inoculated with attenuated S2 vaccine strains of EIAV and seropositive in standard p26 diagnostic assays. All horse serum samples were used at a dilution of 1:25.
Application of NN-ELISA as a diagnostic assay for EIAV infection.
Having identified the NN-ELISA as the superior format as the basis for the S2 serological diagnostic assay, we next sought to compare the sensitivity and specificity of this assay with those of an ELISA detecting serum antibodies to the EIAV capsid protein p26. For these comparisons, we assembled a coded panel of reference sera from seven uninfected horses, 18 field-infected horses, and 21 EIAVUKΔS2-vaccinated horses (Fig. 6). All of the field-infected horses and vaccinates were confirmed as seropositive in the Vira-CHEK p26 ELISA (Synbiotics Corp.), while all of the uninfected horses were seronegative in this diagnostic assay (data not shown). The p26 serum antibody reactivity observed in our ELISA (Fig. 6) was completely in agreement with the Vira-CHEK assay in identifying virus-infected and uninfected horses, thus validating our p26 ELISA for comparison to the NN-ELISA diagnostic assay.
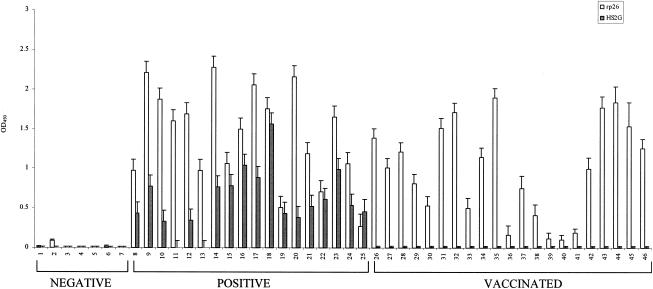
FIG. 6.
Evaluation of HS2G NN-ELISA to identify serum samples from uninfected, infected, and vaccinated horses. The panel of reference serum samples included uninfected horses (samples 1 to 7), horses persistently infected with EIAV (samples 8 to 25), and horses vaccinated with attenuated S2 vaccine strains (samples 26 to 46). All horse sera were used at a dilution of 1:25 in HS2G NN-ELISA. The affinity ELISA procedures for detecting serum antibodies to the HS2G antigen or the EIAV p26 protein were as described in Materials and Methods.
The NN-ELISA seroreactivity data in Fig. 6 demonstrated a high level of sensitivity and specificity for identifying EIAV-infected horses and distinguishing them from uninfected horses and EIAVUKΔS2-vaccinated horses. All seven of the uninfected horse serum samples were found to be nonreactive in the NN-ELISA format. Similarly, all 21 of the immune serum samples from the vaccinated horses failed to display any detectable reactivity in the NN-ELISA, confirming the lack of S2-specific antibodies in these horses. In contrast to these seronegative sera, the NN-ELISA identified 16 of 18 EIAV-infected horses as seropositive, with two infected horse serum samples (samples 11 and 13) failing to display serum antibody reactivity to the HS2G antigen.
Combining the NN-ELISA serological data presented in Fig. 5 and 6, it can be calculated that the NN-ELISA was 100% accurate in identifying S2 antibody-negative serum from uninfected horses (12 of 12); there were no detectable false positives. The NN-ELISA achieved 92% sensitivity in identifying EIAV-infected horses (22 of 24) by S2 serum antibody reactivity. While the present studies utilize a relatively limited number of test samples, the data clearly indicate the sensitivity and specificity of the NN-ELISA as a potential diagnostic assay for detecting EIAV-infected horses and distinguishing them from horses inoculated with attenuated EIAV vaccine strains lacking S2 gene expression.
DISCUSSION
EIAV infection of horses is a global veterinary concern and a disease for which there is currently no vaccine. Therefore, control of this viral infection is achieved in certain countries by the destruction of EIAV-infected horses identified by serological diagnostic assays that detect antibodies to viral core or envelope proteins. The development of an effective vaccine to lentiviruses in general and EIAV in particular has been inhibited by a confounding array of immune evasion and persistence mechanisms intrinsic to these viruses, as frequently highlighted by AIDS vaccine efforts. The implementation of an effective EIAV vaccine, however, is even further complicated by present regulatory policies in certain countries that will require an ability to distinguish infected from vaccinated horses. We have previously described an experimental live attenuated EIAV vaccine, EIAVUKΔS2, which appears to achieve a high level of protection from exposure to a closely related virulent virus challenge (19). Thus, the goal of the present study was to develop a companion serological diagnostic assay that is seropositive for infected horses while being seronegative for uninfected and vaccinated horses. The data presented here indicate that a diagnostic assay based on detecting serum antibody reactivity to EIAV S2 antigen has the required specificity and sensitivity for further development for application in detecting EIAV-infected horses and distinguishing them from vaccinated horses by using a practical and simple ELISA format that can be optimized for high throughput.
The first challenge in developing a satisfactory diagnostic test was to produce adequate quantities of EIAV S2 protein for use as antigen substrate. Previous studies from our lab indicated serum antibody reactivity with small synthetic peptides derived from the S2 protein, but the level of peptide reactivity was very low, indicating the need to employ the whole protein as antigen (unpublished data). The S2 protein is not found in virus particles and is expressed at relatively low levels in infected cells. Therefore, the only practical approach to obtaining reasonable quantities was to clone the S2 gene and produce recombinant S2 protein. However, preliminary experiments in our lab indicated a frustrating inability to express S2 protein in various standard prokaryotic and eukaryotic protein expression systems. The studies described here exemplify the necessary evolution of S2 expression systems to finally achieve adequate levels of antigen production. These studies demonstrated that the lack of S2 expression was not due to deficiencies in transcription or translational efficiency, as even codon-optimized constructs failed to produce detectable protein in transfected cells, despite substantial levels of S2-specific mRNA. Deducing that the lack of S2 protein expression may be due to rapid degradation of the recombinant protein, we achieved reproducible recombinant expression by fusing the S2 protein to the carrier EGFP, a fluorescent marker that can be employed in studies to elucidate S2 function in infected cells during viral replication. It seems feasible that other carrier proteins with advantageous properties for purification or assay development may be substituted for the EGFP employed here. In addition to the fusion to a carrier protein for stability, the final HS2G protein was also engineered to contain an oligonucleotide-histidine tag for purification and for affinity ELISA. We demonstrate here that the HS2G protein can be produced in either mammalian or prokaryotic cell expression systems. Thus, the HS2G protein construct provides a reasonable system for diagnostic antigen production.
The second challenge was to identify the optimal serological assay format for developing and evaluating a diagnostic assay based on serum antibody reactivity to the HS2G antigen. Towards this goal, we compared standard and affinity ELISA formats for their sensitivity and specificity of serum antibody reactivity to the HS2G antigen. An interesting observation from these studies was that the level of serum antibody reactivity was similar in both assay formats for an S2-specific rabbit antiserum but markedly different in the level of antibody reactivity observed with immune serum from infected horses. With the equine serum, the affinity ELISA displayed substantially higher reactivity and lower backgrounds than did the standard ELISA, yielding the potential for a more sensitive and specific diagnostic assay format. Therefore, the affinity ELISA procedure was selected as the format for further evaluation. It seems reasonable that the HS2G antigen could be employed in other immunoassays, such as immunoblotting, AGID, competitive ELISA, or antigen capture (5, 11-13, 17, 23, 27).
To assess the accuracy of the affinity HS2G ELISA for detecting S2-specific serum antibodies in infected horses, we assayed a total of 65 reference serum samples from uninfected horses, from experimentally and field-infected horses, and experimentally vaccinated horses. The results of these experiments demonstrated that the affinity HS2G ELISA achieved a 92% accuracy (22 of 24 positive) in identifying EIAV-infected horses by S2-specific serum antibody reactivity. In contrast, there was no detectable S2-specific antibody reactivity in serum from uninfected horses and in horses inoculated with the attenuated EIAV vaccine. Based on these observations with this particular set of serum samples, we conclude that the affinity HS2G ELISA has the prerequisite sensitivity and specificity for further development and evaluation as a diagnostic assay that can accurately identify EIAV-infected horses and distinguish them from vaccinated horses. Future evaluations of the S2 diagnostic assay will need to determine specificity and sensitivity in large panels of horse serum samples taken from geographically diverse areas of the world, to determine the kinetics of the development and stability of S2-specific antibody responses during the course of EIAV infection, and to optimize assay formats to meet practical application.
It should be noted here that the diagnostic serological assays to detect serum antibodies to S2 may also be used to distinguish EIAV-infected horses from horses immunized with other EIAV vaccine formats, including inactivated viral particles, envelope or core protein subunit vaccines, or DNA or live vector vaccines expressing viral envelope or core proteins. None of these immunization regimens are expected to elicit host antibody responses to the S2 protein, thus providing a serological means of distinguishing infected from vaccinated horses. To increase the confidence levels of an S2 serological assay, it is possible to combine the detection of S2-specific antibodies with a complementary detection of specific antibodies to appropriate EIAV virion proteins (e.g., p26) for infected horses and detection of antibodies to an antigenic marker incorporated in the EIAV vaccine. However, all of these parameters are within the capabilities of present serological diagnostic technologies and readily adapted to the EIAV system.
Acknowledgments
We gratefully thank Kaiming Ye for his continuing advice on this study and Michelle Raabe for her expert assistance as a scientific manuscript editor. The vTF7-3 was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, from Tom Fuerst and Bernard Moss.
This project was financially supported by NIH grant RO1 AI25850, the Lucille P. Markey Charitable Trust, the Kentucky Agricultural Experiment Station, and Intervet, Inc.
REFERENCES
- 1.Archambault, D., Z. M. Wang, J. C. Lacal, A. Gazit, A. Yaniv, J. E. Dahlberg, and S. R. Tronick. 1989. Development of an enzyme-linked immunosorbent assay for equine infectious anemia virus detection using recombinant Pr55gag. J. Clin. Microbiol. 27:1167-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ball, J. M., K. E. Rushlow, C. J. Issel, and R. C. Montelaro. 1992. Detailed mapping of the antigenicity of the surface unit glycoprotein of equine infectious anemia virus by using synthetic peptide strategies. J. Virol. 66:732-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birkelt, A. J., B. Yelamos, I. Rodriguez-Crespo, F. Gavilanes, and D. L. Peterson. 1997. Cloning expression, purification, and characterization of the major core protein (p26) from equine infectious anemia virus. Biochim. Biophys. Acta 1339:62-72. [DOI] [PubMed] [Google Scholar]
- 4.Carroll, R., and D. Derse. 1993. Translation of equine infectious anemia virus bicistronic tat-rev mRNA requires leaky ribosome scanning of the tat CTG initiation codon. J. Virol. 67:1433-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coggins, L., and N. L. Norcross. 1970. Immunodiffusion reaction in equine infectious anemia. Cornell Vet. 60:330-335. [PubMed] [Google Scholar]
- 6.Cook, R. F., C. Leroux, S. J. Cook, S. L. Berger, D. L. Lichtenstein, N. N. Ghabrial, R. C. Montelaro, and C. J. Issel. 1998. Development and characterization of an in vivo pathogenic molecular clone of equine infectious anemia virus. J. Virol. 72:1383-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hammond, S. A., S. J. Cook, L. D. Falo, Jr., C. J. Issel, and R. C. Montelaro. 1999. A particulate viral protein vaccine reduces viral load and delays progression to disease in immunized ponies challenged with equine infectious anemia virus. Virology 254:37-49. [DOI] [PubMed] [Google Scholar]
- 9.Hammond, S. A., F. Li, B. M. McKeon, Sr., S. J. Cook, C. J. Issel, and R. C. Montelaro. 2000. Immune responses and viral replication in long-term inapparent carrier ponies inoculated with equine infectious anemia virus. J. Virol. 74:5968-5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henderson, L. E., R. C. Sowder, G. W. Smythers, and S. Oroszlan. 1987. Chemical and immunological characterizations of equine infectious anemia virus gag-encoded proteins. J. Virol. 61:1116-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hussain, K. A., C. J. Issel, K. L. Schnorr, P. M. Rwambo, and R. C. Montelaro. 1987. Antigenic analysis of equine infectious anemia virus (EIAV) variants by using monoclonal antibodies: epitopes of glycoprotein gp90 of EIAV stimulate neutralizing antibodies. J. Virol. 61:2956-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hussain, K. A., C. J. Issel, K. L. Schnorr, P. M. Rwambo, M. West, and R. C. Montelaro. 1988. Antigenic mapping of the envelope proteins of equine infectious anemia virus: identification of a neutralization domain and a conserved region on glycoprotein 90. Arch. Virol. 98:213-224. [DOI] [PubMed] [Google Scholar]
- 13.Issel, C. J., and W. V. Adams, Jr. 1982. Detection of equine infectious anemia virus in a horse with an equivocal agar gel immunodiffusion test reaction. J. Am. Vet. Med. Assoc. 180:276-278. [PubMed] [Google Scholar]
- 14.Issel, C. J., and L. Coggins. 1979. Equine infectious anemia: current knowledge. J. Am. Vet. Med. Assoc. 174:727-733. [PubMed] [Google Scholar]
- 15.Issel, C. J., and R. F. Cook. 1993. A review of techniques for the serologic diagnosis of equine infectious anemia. J. Vet. Diagn. Investig. 5:137-141. [DOI] [PubMed] [Google Scholar]
- 16.Issel, C. J., D. W. Horohov, D. F. Lea, W. V. Adams, Jr., S. D. Hagius, J. M. McManus, A. C. Allison, and R. C. Montelaro. 1992. Efficacy of inactivated whole-virus and subunit vaccines in preventing infection and disease caused by equine infectious anemia virus. J. Virol. 66:3398-3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knowles, D. P., Jr., and J. R. Gorham. 1993. Advances in the diagnosis of some parasitic diseases by monoclonal antibody-based enzyme-linked immunosorbent assays. Rev. Sci. Technol. 12:425-433. [DOI] [PubMed] [Google Scholar]
- 18.Kong, X. G., H. Pang, T. Sugiura, H. Sentsui, T. Onodera, Y. Matsumoto, and H. Akashi. 1997. Application of equine infectious anemia virus core proteins produced in a baculovirus expression system to serological diagnosis. Microbiol. Immunol. 41:975-980. [DOI] [PubMed] [Google Scholar]
- 19.Li, F., J. K. Craigo, L. Howe, J. D. Steckbeck, S. Cook, C. Issel, and R. C. Montelaro. 2003. A live attenuated equine infectious anemia virus proviral vaccine with a modified S2 gene provides protection from detectable infection by intravenous virulent virus challenge of experimentally inoculated horses. J. Virol. 77:7244-7253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, F., C. Leroux, J. K. Craigo, S. J. Cook, C. J. Issel, and R. C. Montelaro. 2000. The S2 gene of equine infectious anemia virus is a highly conserved determinant of viral replication and virulence properties in experimentally infected ponies. J. Virol. 74:573-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, F., B. A. Puffer, and R. C. Montelaro. 1998. The S2 gene of equine infectious anemia virus is dispensable for viral replication in vitro. J. Virol. 72:8344-8348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mia, A. S., M. M. Tierney, and D. M. Krevcak. 1982. Detection of equine infectious anemia antibody by a rapid microenzyme linked immunosorbent assay (ELISA). Proc. Annu. Meet. Am. Assoc. Vet. Lab. Diagn. 25:159-166. [Google Scholar]
- 23.Montelaro, R. C., W. G. Robey, M. D. West, C. J. Issel, and P. J. Fischinger. 1988. Characterization of the serological cross-reactivity between glycoproteins of the human immunodeficiency virus and equine infectious anaemia virus. J. Gen. Virol. 69:1711-1717. [DOI] [PubMed] [Google Scholar]
- 24.Montelaro, R. C., W. G. Robey, M. D. West, C. J. Issel, and P. J. Fischinger. 1993. Equine retroviruses, vol. 2. Plenum Press, New York, N.Y.
- 25.Raabe, M. L., C. J. Issel, S. J. Cook, R. F. Cook, B. Woodson, and R. C. Montelaro. 1998. Immunization with a recombinant envelope protein (rgp90) of EIAV produces a spectrum of vaccine efficacy ranging from lack of clinical disease to severe enhancement. Virology 245:151-162. [DOI] [PubMed] [Google Scholar]
- 26.Raabe, M. L., C. J. Issel, and R. C. Montelaro. 1999. In vitro antibody-dependent enhancement assays are insensitive indicators of in vivo vaccine enhancement of equine infectious anemia virus. Virology 259:416-427. [DOI] [PubMed] [Google Scholar]
- 27.Reischl, U. 1996. Application of molecular biology-based methods to the diagnosis of infectious diseases. Front. Biosci. 1:e72-e77. [DOI] [PubMed] [Google Scholar]
- 28.Schiltz, R. L., D. S. Shih, S. Rasty, R. C. Montelaro, and K. E. Rushlow. 1992. Equine infectious anemia virus gene expression: characterization of the RNA splicing pattern and the protein products encoded by open reading frames S1 and S2. J. Virol. 66:3455-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sellon, D. C., F. J. Fuller, and T. C. McGuire. 1994. The immunopathogenesis of equine infectious anemia virus. Virus Res. 32:111-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shane, B. S., C. J. Issel, and R. C. Montelaro. 1984. Enzyme-linked immunosorbent assay for detection of equine infectious anemia virus p26 antigen and antibody. J. Clin. Microbiol. 19:351-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soutullo, A., V. Verwimp, M. Riveros, R. Pauli, and G. Tonarelli. 2001. Design and validation of an ELISA for equine infectious anemia (EIA) diagnosis using synthetic peptides. Vet. Microbiol. 79:111-121. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki, T., S. Ueda, and T. Samejima. 1982. Enzyme-linked immunosorbent assay for diagnosis of equine infectious anemia. Vet. Microbiol. 7:307-315. [DOI] [PubMed] [Google Scholar]
- 33.Wang, S. Z., K. E. Rushlow, C. J. Issel, R. F. Cook, S. J. Cook, M. L. Raabe, Y. H. Chong, L. Costa, and R. C. Montelaro. 1994. Enhancement of EIAV replication and disease by immunization with a baculovirus-expressed recombinant envelope surface glycoprotein. Virology 199:247-251. [DOI] [PubMed] [Google Scholar]