Abstract
In the budding yeast Saccharomyces cerevisiae, selection of the bud site determines the axis of polarized cell growth and eventual oriented cell division. Bud sites are selected in specific patterns depending on cell type. These patterns appear to depend on distinct types of marker proteins in the cell cortex; in particular, the bipolar budding of diploid cells depends on persistent landmarks at the birth-scar-distal and -proximal poles that involve the proteins Bud8p and Bud9p, respectively. Rax1p and Rax2p also appear to function specifically in bipolar budding, and we report here a further characterization of these proteins and of their interactions with Bud8p and Bud9p. Rax1p and Rax2p both appear to be integral membrane proteins. Although commonly used programs predict different topologies for Rax2p, glycosylation studies indicate that it has a type I orientation, with its long N-terminal domain in the extracytoplasmic space. Analysis of rax1 and rax2 mutant budding patterns indicates that both proteins are involved in selecting bud sites at both the distal and proximal poles of daughter cells as well as near previously used division sites on mother cells. Consistent with this, GFP-tagged Rax1p and Rax2p were both observed at the distal pole as well as at the division site on both mother and daughter cells; localization to the division sites was persistent through multiple cell cycles. Localization of Rax1p and Rax2p was interdependent, and biochemical studies showed that these proteins could be copurified from yeast. Bud8p and Bud9p could also be copurified with Rax1p, and localization studies provided further evidence of interactions. Localization of Rax1p and Rax2p to the bud tip and distal pole depended on Bud8p, and normal localization of Bud8p was partially dependent on Rax1p and Rax2p. Although localization of Rax1p and Rax2p to the division site did not appear to depend on Bud9p, normal localization of Bud9p appeared largely or entirely dependent on Rax1p and Rax2p. Taken together, the results indicate that Rax1p and Rax2p interact closely with each other and with Bud8p and Bud9p in the establishment and/or maintenance of the cortical landmarks for bipolar budding.
INTRODUCTION
Cell polarization and oriented cell divisions are central to the development of most organisms. Cells of the yeast Saccharomyces cerevisiae exhibit two distinct patterns of polarization-axis selection and oriented cell division depending on their cell type (Freifelder, 1960; Hicks et al., 1977; Chant and Pringle, 1995). Thus, a and α cells (such as normal haploids) form buds in the axial pattern, in which both mother and daughter cells select new bud sites adjacent to their immediately preceding division sites, as marked by the bud scar on the mother cell and the birth scar on the daughter. In contrast, a/α cells (such as normal diploids) form buds in the bipolar pattern, in which daughter cells usually, but not always, bud first at the pole distal to the birth scar, and mother cells can choose a new bud site near either pole. Previous studies have identified many proteins that are involved in determining the cell-type-specific budding patterns. The axial pattern appears to depend on a transient cortical marker that involves Bud3p, Bud4p, and Axl2p (Chant and Herskowitz, 1991; Chant and Pringle, 1995; Chant et al., 1995; Halme et al., 1996; Roemer et al., 1996; Sanders and Herskowitz, 1996; Lord et al., 2000). Axl1p, which is expressed only in a and α cells, is also important for axial budding (Fujita et al., 1994; Adames et al., 1995; Lord et al., 2002). These proteins localize to the mother-bud neck and then are distributed to the division sites on both mother and daughter cells, where they signal to the polarity-establishment proteins through the GTPase module involving Rsr1p, Bud5p, and Bud2p (Park et al., 1993, 1999, 2002; Pringle et al., 1995; Kang et al., 2001; Marston et al., 2001). If any of the axial marker proteins is absent, a or α cells bud in the bipolar pattern, indicating that the proteins required for bipolar budding are all expressed, although not normally used, in a and α cells.
The bipolar pattern appears to depend on persistent cortical markers that are present at both poles of daughter cells and at each previously used division site on mother cells (Chant and Pringle, 1995). A screen for mutants specifically defective in bipolar budding identified bud8 mutants, which bud almost exclusively around the birth-scar-proximal pole, and bud9 mutants, which bud almost exclusively around the distal pole (Zahner et al., 1996; Harkins et al., 2001). Bud8p and Bud9p are transmembrane proteins that localize to the plasma membrane at the distal and proximal poles, respectively (Harkins et al., 2001; Schenkman et al., 2002). Taken together, the data suggest that Bud8p and Bud9p are key components of the persistent cortical markers at the distal and proximal poles of the daughter cell, respectively.
Rax1p and Rax2p are also implicated in bipolar budding. The genes encoding these proteins were originally identified by mutations that appeared to suppress the loss of axial budding in an axl1 mutant (Fujita et al., 1994; Chen et al., 2000). Subsequently, it became clear that when the bipolar-budding system is disabled, a/α cells (or axl1 mutants) can use the axial marker inefficiently despite their lack of Axl1p (see Harkins et al., 2001). Further examination of the mutant phenotypes then revealed that there is no evident effect of rax1 or rax2 mutations on axial budding but that these mutations disrupt bipolar budding, accounting for the presence of axially budding cells in axl1 rax1 and axl1 rax2 double-mutant strains (Chen et al., 2000; Ni and Snyder, 2001; this study). Observations of a Rax2p-GFP fusion protein showed that it localized to the mother-bud neck of large-budded cells and thence to the division site on both mother and daughter cells, where it persisted through multiple cell generations (Chen et al., 2000). This localization helped to explain some of the effects of rax2 mutations on bipolar budding but not the inability of rax2 mutant daughter cells to position their first buds efficiently at the distal pole (Chen et al., 2000; this study). This apparent inconsistency led us to investigate further the localization and function of Rax2p. In addition, although Rax1p has been reported to be necessary for the normal localization of Rax2p (Chen et al., 2000), there was little other information available about its structure or function. This led us to undertake parallel studies of Rax1p. A point of particular interest in these studies was the relationship between the function of Rax1p and Rax2p and that of Bud8p and Bud9p.
Here we report that Rax1p and Rax2p both localize to the distal pole as well as to the division site and that they interact both with each other and with Bud8p and Bud9p in the establishment and/or maintenance of the cortical markers for bipolar budding.
MATERIALS AND METHODS
Strains, Plasmids, Genetic Methods, and Growth Conditions
The strains and plasmids used in this study are listed in Tables 1 and 2; their construction is described below or in the tables. The sequences of the oligo-nucleotides used are listed in Supplementary Table 1. Standard methods of yeast genetics, recombinant-DNA manipulation, and growth conditions were used (Guthrie and Fink, 1991; Ausubel et al., 1999).
Table 1.
Yeast strains used in this study
| Strain | Relevant genotypea | Source/comments |
|---|---|---|
| YEF473 | a/α his3-Δ200/his3-Δ200 leu2-Δ1/leu2-Δ1 lys2-801/lys2-801 trp1-Δ63/trp1-Δ63 ura3-52/ura3-52 | Bi and Pringle (1996) |
| YEF473A | ahis3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52 | Segregant from YEF473 |
| YEF473B | α his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52 | Segregant from YEF473 |
| YHH394 | abud8-Δ1::TRP1 | Harkins et al. (2001) |
| YHH415 | a/α bud8-Δ1::TRP1/bud8-Δ1::TRP1 | Harkins et al. (2001) |
| YHH613 | abud9-Δ1::HIS3 | Harkins et al. (2001) |
| YHH615 | a/α bud9-Δ1::HIS3/bud9-Δ1::HIS3 | Harkins et al. (2001) |
| AM476 | a/α rax2Δ::HIS3/rax2Δ::HIS3 | A. McKenzie |
| HPY440 | aaxl1::URA3 | This studyb |
| HPY494 | arax1Δ::HIS3 | See text |
| HPY495 | α rax1Δ::HIS3 | See text |
| HPY496 | a/α rax1Δ::HIS3/rax1Δ::HIS3 | HPY494 × HPY495 |
| HPY543 | aaxl1:URA3 rax1Δ::HIS3 | Segregant from HPY440 × HPY495 |
| HPY584 | arax2Δ::HIS3 | See text |
| HPY591 | α rax2Δ::HIS3 | See text |
| HPY592 | a/α rax2Δ::HIS3/rax2Δ::HIS3 | HPY584 × HPY591 |
| HPY610 | arax1Δ::URA3 | See text |
| HPY612 | aGFP-RAX1-TRP1::rax1Δ::HIS3 | See text |
| HPY613 | α GFP-RAX1-TRP1::rax1Δ::HIS3 | See text |
| HPY614 | a/α GFP-RAX1-TRP1::rax1Δ::HIS3/GFP-RAX1-TRP1::rax1 Δ::HIS3 | HPY612 × HPY613 |
| HPY615 | aaxl1::URA3 GFP-RAX1-TRP1::rax1Δ::HIS3 | See text |
| HPY628 | α rax1Δ::URA3 rax2Δ::HIS3 | Segregant from HPY591 × HPY610 |
| HPY629 | arax1Δ::URA3 rax2Δ::HIS3 | Segregant from HPY591 × HPY610 |
| HPY634 | a/α rax1Δ::URA3/rax1Δ::URA3 rax2Δ::HIS3/rax2Δ::HIS3 | HPY628 × HPY629 |
| HPY635 | arax2Δ::HIS3 bud8-Δ1::TRP1 | Segregant from HPY591 × YHH394 |
| HPY636 | α rax2Δ::HIS3 bud8-Δ1::TRP1 | Segregant from HPY591 × YHH394 |
| HPY637 | arax1Δ::HIS3 bud8-Δ1::TRP1 | Segregant from HPY495 × YHH394 |
| HPY638 | α rax1Δ::HIS3 bud8-Δ1::TRP1 | Segregant from HPY495 × YHH394 |
| HPY639 | aaxl1::URA3 rax2Δ::HIS3 | Segregant from HPY440 × HPY591 |
| HPY641 | a/α rax2Δ::HIS3/rax2Δ::HIS3 bud8-Δ1::TRP1/bud8-Δ1::TRP1 | HPY635 × HPY636 |
| HPY643 | a/α rax1Δ::HIS3/rax1Δ::HIS3 bud8-Δ1::TRP1/bud8-Δ1::TRP1 | HPY637 × HPY638 |
| HPY645 | α rax2Δ::HIS3 GFP-RAX1-TRP1::rax1Δ::URA3 | See text |
| HPY646 | arax2Δ::HIS3 GFP-RAX1-TRP1::rax1Δ::URA3 | See text |
| HPY647 | a/α rax2Δ::HIS3/rax2Δ::HIS3 GFP-RAX1-TRP1::rax1Δ::URA3/GFP-RAX1-TRP1::rax1Δ::URA3 | HPY645 × HPY646 |
| HPY670 | arax2Δ::URA3 | See text |
| HPY678 | abud8-Δ1::TRP1 GFP-RAX1-LEU2::rax1Δ::HIS3 | See text |
| HPY679 | α bud8-Δ1::TRP1 GFP-RAX1-LEU2::rax1Δ::HIS3 | See text |
| HPY680 | abud8-Δ1::TRP1 RAX2-GFP-LEU2::rax2Δ::HIS3 | See text |
| HPY681 | α bud8-Δ1::TRP1 RAX2-GFP-LEU2::rax2Δ::HIS3 | See text |
| HPY682 | arax1Δ::URA3 bud9-Δ1::HIS3 | Segregant from HPY610 × YHH613 |
| HPY683 | α rax1Δ::URA3 bud9-Δ1::HIS3 | Segregant from HPY610 × YHH613 |
| HPY684 | arax2Δ::URA3 bud9-Δ1::HIS3 | Segregant from HPY670 × YHH613 |
| HPY685 | α rax2Δ::URA3 bud9-Δ1::HIS3 | Segregant from HPY670 × YHH613 |
| HPY690 | a/α bud8-Δ1::TRP1/bud8-Δ1::TRP1 GFP-RAX1-LEU2::rax1Δ::HIS3/GFP-RAX1-LEU2::rax1Δ::HIS3 | HPY678 × HPY679 |
| HPY691 | a/α bud8-Δ1::TRP1/bud8-Δ1::TRP1 RAX2-GFP-LEU2::rax2Δ::HIS3/RAX2-GFP-LEU2::rax2Δ::HIS3 | HPY680 × HPY681 |
| HPY692 | a/α rax1Δ::URA3/rax1Δ::URA3 bud9-Δ1::HIS3/bud9-Δ1::HIS3 | HPY682 × HPY683 |
| HPY693 | a/α rax2Δ::URA3/rax2Δ::URA3 bud9-Δ1::HIS3/bud9-Δ1::HIS3 | HPY684 × HPY685 |
| HPY700 | abud9-Δ1::HIS3 GFP-RAX1-LEU2::rax1Δ::URA3 | See text |
| HPY701 | α bud9-Δ1::HIS3 GFP-RAX1-LEU2::rax1Δ::URA3 | See text |
| HPY702 | abud9-Δ1::HIS3 RAX2-GFP-LEU2::rax2Δ::URA3 | See text |
| HPY703 | α bud9-Δ1::HIS3 RAX2-GFP-LEU2::rax2Δ::URA3 | See text |
| HPY704 | a/α bud9-Δ1::HIS3/bud9-Δ1::HIS3 GFP-RAX1-LEU2::rax1Δ::URA3/GFP-RAX1-LEU2::rax1Δ::URA3 | HPY700 × HPY701 |
| HPY705 | a/α bud9-Δ1::HIS3/bud9-Δ1::HIS3 RAX2-GFP-LEU2::rax2Δ::URA3/RAX2-GFP-LEU2::rax2Δ::URA3 | HPY702 × HPY703 |
| HPY729 | aaxl1::URA3 RAX2-GFP-LEU2::rax2Δ::HIS3 | See text |
| HPY730 | aaxl1::URA3 RAX2-HA3-LEU2::rax2Δ::HIS3 | See text |
| HPY793 | aaxl2Δ::HIS3 rax1Δ::URA3 | This studyc |
| HPY804 | aaxl2Δ::HIS3 rax2Δ::URA3 | This studyc |
| HPY808 | aRAX2-GFP-LEU2::rax2Δ::URA3 | See text |
| HPY814 | a/α RAX2-GFP-LEU2::rax2Δ::URA3/RAX2-GFP-LEU2::rax2Δ::URA3 | a/α conversion from HPY808 using HO |
| HPY833 | arax1Δ::URA3 RAX2-GFP-LEU2::rax2Δ::HIS3 | See text |
| HPY834 | α rax1Δ::URA3 RAX2-GFP-LEU2::rax2Δ::HIS3 | See text |
| HPY836 | a/α rax1Δ::URA3/rax1Δ::URA3 RAX2-GFP-LEU2::rax2Δ::HIS3/RAX2-GFP-LEU2::rax2Δ::HIS3 | HPY833 × HPY834 |
| HPY841 | a/α axl2Δ::HIS3/axl2Δ::HIS3 rax1Δ::URA3/rax1Δ::URA3 | a/α conversion from HPY793 using HO |
| HPY842 | a/α axl2Δ::HIS3/axl2Δ::HIS3 rax2Δ::URA3/rax2Δ::URA3 | a/α conversion from HPY804 using HO |
| HPY850 | aaxl1::URA3 RAX1-Myc13-TRP1 | See text |
| HPY16* | ahis3-Δ1 leu2 trp1-Δ63 ura3-52 prb1-1122 pep4-3 prc1-407 | Park et al. (1993) |
| HPY568* | a/α bud8-Δ1::TRP1/bud8-Δ1::TRP1 | This studyd |
| HPY570* | a/α bud9-Δ1::HIS3/bud9-Δ1::HIS3 | This studye |
| HPY601* | arax1Δ::HIS3 | See text |
| HPY599* | arax2Δ::HIS3 | See text |
| HPY644* | a/α RAX2-HA3-TRP1::rax2Δ::HIS3/RAX2-HA3-TRP1:: rax2Δ::HIS3 | See text |
| HPY798* | a/α rax1Δ::HIS3/rax1Δ::HIS3 | a/α conversion from HPY601 using HO |
| HPY843* | aRAX1-Myc13-TRP1 | See text |
All strains except those marked with asterisk (*) are congenic to YEF473 except as indicated. Strains marked with asterisk (*) are congenic to HPY16 except as indicated.
The 2.1-kb BamHI-EcoRI fragment from p98 was used to replace the wild-type AXL1 allele in strain YEF473A.
The axl2Δ::HIS3 allele was introduced into strains HPY610 and HPY670 by one-step gene replacement using a PCR-generated DNA fragment, as described by Roemer et al. (1996).
The 1.9-kb PCR fragment from a reaction using genomic DNA from strain YHH394 as template and primer pair oBUD81 and oBUD82 was used to replace the wild-type BUD8 allele in strain HPY16. The diploid strain was then created by HO-mediated mating-type conversion (Herskowitz and Jensen, 1991).
The 2.2-kb PCR fragment from a reaction using genomic DNA from strain YHH613 as template and primer pair oBUD91 and oBUD92 was used to replace the wild-type BUD9 allele in strain HPY16. The diploid strain was then created by HO-mediated mating-type conversion (Herskowitz and Jensen, 1991).
Table 2.
Plasmids used in this study
| Plasmid | Description | Source |
|---|---|---|
| pRS304 | TRP1 (integrative) | Sikorski and Hieter (1989) |
| pRS305 | LEU2 (integrative) | Sikorski and Hieter (1989) |
| pRS424 | TRP1 (high copy) | Christianson et al. (1992) |
| pRS426 | URA3 (high copy) | Christianson et al. (1992) |
| YEplac181 | LEU2 (high copy) | Gietz and Sugino (1988) |
| YEplac195 | URA3 (high copy) | Gietz and Sugino (1988) |
| p98 | axl1::URA3 (insertional knock-out construct) | Adames et al. (1995) |
| YEpHA-BUD8F | HA3-BUD8 in YEplac181 | Schenkman et al. (2002) |
| YEpGFP-BUD8F | GFP-BUD8 in YEplac181 | Schenkman et al. (2002) |
| pCA5015 | 1.7-kb BamHI fragment containing the HIS3 gene in pBluescript-KS(+) BamHI site | T.-H. Chang |
| pHP835 | BUD5-HA6 in YCp50 | Kang et al. (2001) |
| pHP896 | 1.9-kb RAX1 in pBluescript-SK(+) | See text |
| pHP898 | rax1Δ::HIS3 in pBluescript-SK(+) | See text |
| pHP899 | 2.6-kb RAX1 in pBluescript-SK(+) | See text |
| pHP924 | rax1Δ::URA3 in pBluescript-SK(+) | See text |
| pHP926 | pBluescript-SK(+) with NotI site destroyed | This study |
| pHP933 | RAX1 with N-terminal NotI site in pHP926 | See text |
| pHP995 | rax2Δ::HIS3 in pBluescript-SK(+) | See text |
| pHP996 | 4.2-kb RAX2 in pBluescript-SK(+) | See text |
| pHP1012 | RAX2 with C-terminal NotI site in pBluescript-SK(+) | See text |
| pHP1013 | RAX2 with C-terminal NotI site in pRS304 | See text |
| pHP1016 | RAX2-HA3 in pRS304 | See text |
| pHP1017 | RAX2-GFP in pRS304 | See text |
| pHP1061 | GFP-RAX1 in pRS304 | See text |
| pHP1090 | RAX2-HA3 in pRS305 | See text |
| pHP1091 | RAX2-GFP in pRS305 | See text |
| pHP1102 | RAX2-HA3 with additional 5′ sequences in pRS305 | This studya |
| pHP1109 | GFP-RAX1 in pRS305 | See text |
| pHP1110 | rax2Δ::URA3 in pBluescript-SK(+) | See text |
| pHP1156 | pGAL1p-GST-HIS6-RAX1 | Zhu et al. (2001) |
| pHP1158 | pGAL1p-GST-HIS6-RHO5 | Zhu et al. (2001) |
| pHP1159 | pEGFP-C3 with NotI sites | See text |
| pHP1169 | AXL2-Protein C (C-terminus) in pRS424 | This studyb |
| pHP1183 | RAX2-HA3 in pRS426 | This studyc |
| pHP1202 | GFP3-BUD9 in YEplac195 | See text |
| pHP1319 | HA3-BUD9 in YEplac181 | This studyd |
Contains 1350 bp of RAX2 upstream sequences.
The 4-kb NsiI fragment from YEp24 AXL2-Protein C (Kang et al., 2001) was ligated into the PstI site of pRS424.
The 4.8-kb HindIII-XhoI fragment from pHP1102 was ligated into the HindIII and XhoI sites of pRS426.
The 2.7-kb SphI fragment from YEpHA-BUD9 (Harkins et al., 2001) was ligated into the SphI site of YEplac181.
Cloning, Deletion, and Tagging of RAX1 and RAX2
RAX1 was cloned by PCR using genomic DNA from strain YEF473A as template and primer pair oRAX11 and oRAX12. The 2.6-kb product (containing the RAX1 ORF plus ∼680 base pairs of upstream and 650 base pairs of downstream sequence) was digested with HindIII (sites included in the primers) and cloned into the HindIII site of pBluescript SK(+) (Stratagene, La Jolla, CA), yielding plasmid pHP899. RAX2 was cloned similarly using primers oRAX21 and oRAX22 and digestion with SacII to clone the 4.2-kb product (containing the RAX2 ORF plus ∼327 base pairs of upstream and 180 base pairs of downstream sequence) into pBluescript SK(+), yielding plasmid pHP996. DNA sequencing confirmed that the wild-type genes had been cloned.
To construct RAX1 deletion plasmids pHP898 and pHP924, the 1.9-kb DraI fragment from pHP899 was first subcloned into EcoRV/HincII-digested pBluescript SK(+), yielding plasmid pHP896. The complete RAX1 coding sequence in pHP896 was then replaced with a 1.7-kb BamHI fragment carrying the HIS3 marker from plasmid pCA5015 (kindly provided by T.-H. Chang, The Ohio State University) or a 1.1-kb BglII fragment carrying the URA3 marker from plasmid p98 (Table 2). PCR was carried out using pHP896 as template and primers oRAX13 and oRAX14. The product was digested with BamHI and then ligated to the HIS3 and URA3 fragments, yielding plasmids pHP898 and pHP924, respectively. The chromosomal RAX1 gene was then deleted in strains YEF473 and HPY16 by one-step gene disruption (Rothstein, 1991) using the 2.3- and 1.7-kb EcoRI-XhoI fragments from pHP898 and pHP924, respectively. The presence of the desired deletions was confirmed by colony PCR, and appropriate segregants from a YEF473 transformant were mated to obtain the homozygous rax1Δ::HIS3 diploid strain HPY496. RAX2 deletion plasmids pHP995 and pHP1110 were constructed similarly using plasmid pHP996 and primers oRAX23 and oRAX24. The chromosomal RAX2 gene was then deleted in strains YEF473 and HPY16 using the 2.2- and 1.6-kb SacII fragments from pHP995 and pHP1110, respectively, and appropriate segregants from a YEF473 transformant were mated to obtain the homozygous rax2Δ::HIS3 diploid strain HPY592.
To express Rax1p with green fluorescent protein (GFP) fused to its N-terminus, a NotI site was first introduced just after the RAX1 start codon by a two-step PCR strategy. First, with pHP899 as template, separate reactions were run using primers oRAX11 and oRAX15 and primers oRAX16 and oRAX12. The products from these reactions were ligated together after NotI digestion and used as template in a second PCR with primers oRAX11 and oRAX12. The resulting product was digested with HindIII and cloned into the HindIII site of pHP926 (Table 2), yielding pHP933. Sequencing of the entire HindIII fragment confirmed the presence of wild-type RAX1 with the desired in-frame NotI site. The 2.6-kb SacII-ApaI fragment of pHP933 was then subcloned into pRS304, and a 720-base pairs NotI fragment encoding GFPS65T,V163A,S175G (Straight et al., 1998) was inserted into the NotI site, yielding pHP1061. The 3.3-kb SacII-ApaI fragment from pHP1061 was also subcloned into pRS305, yielding pHP1109.
To construct strains expressing GFP-Rax1p from its chromosomal locus, rax1 deletion strains were transformed with pHP1061 or pHP1109 after digestion with NsiI (site 297 base pairs upstream of the RAX1 start codon). To determine whether GFP-Rax1p is functional, pHP1061 was integrated into strain HPY543 (a axl1 rax1), yielding strain HPY615. As expected (Fujita et al., 1994), strain HPY440 (a axl1) budded in a bipolar pattern, whereas HPY543 budded in a partially axial pattern. In contrast, both HPY615 and a diploid strain homozygous for the GFP-RAX1 allele (HPY614) budded in a bipolar pattern (see Supplementary Table 2), indicating that GFP-Rax1p is functional.
To construct strains expressing Rax1p with a C-terminal Myc13 tag, the PCR-based one-step-replacement method (Longtine et al., 1998) was used with primers oRAX111 and oRAX112 and plasmid pFA6a-13Myc-TRP1 as template. The PCR product was used to transform strains HPY440 and HPY16, yielding strains HPY850 and HPY843, respectively. Successful tagging was confirmed by genomic PCR and by immunoblotting using anti-Myc antibody. Rax1p-Myc13 was confirmed to be fully functional by examining the budding pattern of strain HPY850 (see Supplementary Table 2), as described above.
To express Rax2p with GFP or a triple hemagglutinin epitope (HA3) fused to its C-terminus, a NotI site was first introduced just before the RAX2 stop codon by two-step PCR. First, with pHP996 as template, separate reactions were run using primers oRAX25 and oRAX26 and primers oRAX27 and oRAX28. The products were ligated together after NotI digestion and used as template in a second PCR with primers oRAX25 and oRAX28. The resulting 777-base pair product was digested with NheI and XhoI and used to replace the 748-base pairs NheI-XhoI fragment of pHP996, yielding pHP1012. Sequencing of the NheI-XhoI region confirmed the wild-type sequence with the desired in-frame NotI site. The 4.2-kb SacII-XhoI fragment of pHP1012 was then subcloned into pRS304, yielding pHP1013. For tagging with HA3, the 111-base pair NotI HA3 fragment from plasmid pHP835 (Table 2) was inserted at the NotI site of pHP1013, yielding pHP1016. For tagging with GFP, the 720-base pair NotI fragment encoding GFPS65T,V163A,S175G (Straight et al., 1998) was inserted into the NotI site of pHP1013, yielding pHP1017. The SacII-XhoI fragments from pHP1016 and pHP1017 were then subcloned into pRS305, yielding pHP1090 and pHP1091. To construct strains expressing Rax2p-HA3 or Rax2p-GFP from its chromosomal locus, rax2 deletion strains were transformed with pHP1016, pHP1090, or pHP1091 after digestion with NruI (site 216 base pairs upstream of the RAX2 start codon). Rax2p-GFP and Rax2p-HA3 were confirmed to be fully functional essentially as described for Rax1p (see Supplementary Table 2).
Tagging of BUD9
To construct a plasmid expressing Bud9p with a triple-GFP tag, we first constructed a DNA fragment containing three tandem copies of GFP sequences flanked by NotI sites, as follows. Plasmid pEGFP-C3 (a gift from B. Glick, University of Chicago; see Rossanese et al., 2001) was digested with EcoRI, filled in with Klenow enzyme, and ligated to NotI linkers (New England Biolabs, Beverly, MA), and the resulting plasmid was digested with AgeI, filled in with Klenow enzyme, and ligated to NotI linkers, yielding pHP1159. Plasmid YEpHA-BUD9 (Harkins et al., 2001) was then digested with NotI, removing the HA3 sequences at the N-terminus of BUD9, and ligated to the 2.2-kb NotI fragment from pHP1159, yielding plasmid pHP1202. The correct orientation of the GFP3 insert in pHP1202 was confirmed by digestion with BglII. To determine whether GFP3-Bud9p is functional, pHP1202 was transformed into bud9Δ diploid strain YHH615. The transformed cells budded in a bipolar pattern with minor abnormalities, as observed previously for cells expressing GFP-Bud9p from a multicopy plasmid (Harkins et al., 2001).
Protein Analyses
Except as noted, yeast cells were grown to mid-log phase (OD600 ∼ 1.0) in SC medium lacking nutrients as needed to maintain various plasmids, and lysate preparation and glutathione-S-transferase (GST) pull-down experiments were carried out essentially as described previously (Park et al., 1997; Kozminski et al., 2003). The HA3-tagged Bud8p and Bud9p used in these experiments were shown previously to be largely, but not completely, functional by examining their abilities to support normal bipolar budding (Harkins et al., 2001). Similar assays indicated that the GST-Rax1p (Zhu et al., 2001) and protein C-tagged Axl2p (Kang et al., 2001) were also partly, but not completely, functional (our unpublished results). Before performing the pull-down experiments, the membrane fraction was prepared using a lysis buffer (50 mM HEPES, pH 7.6, 50 mM KCl, 1 mM EGTA, 1 mM MgCl2, 1 mM dithiothreitol, 10% glycerol) with a cocktail of various protease inhibitors, and proteins were then solubilized with 1% Triton X-100. Because Rax1p appeared to aggregate when incubated in SDS solution at temperatures ≥65°C, protein samples for most experiments involving this protein were mixed with SDS-loading buffer (Laemmli, 1970) and loaded directly onto the SDS polyacrylamide gel without heating. The HA epitope-tagged proteins, Myc epitope-tagged protein, protein C-tagged protein, GST-fusion proteins, and Nap1p were detected using monoclonal anti-HA antibody HA11 (Covance Research Products, Denver, PA) or 3F10 (Roche Molecular Biochemicals, Indianapolis, IN), monoclonal anti-Myc antibody 9E10 (gift from M. Bishop, University of California-San Francisco), monoclonal anti-protein C antibody (Roche Molecular Biochemicals), polyclonal anti-GST antibodies (Santa Cruz Biotechnology, Santa Cruz, CA), and polyclonal anti-Nap1p antibodies (gift from D. Kellogg, University of California-Santa Cruz), respectively. Protein bands were then detected using HRP-conjugated anti-mouse IgG, anti-rat IgG, or anti-rabbit IgG secondary antibodies and the ECL chemiluminescence system (Amersham Pharmacia Biotech, Piscataway, NJ).
Possible N-linked glycosylation was determined by digestion of proteins in total cell lysates either with a recombinant endo-β-N-acetylglucosaminidase H/maltose-binding protein fusion protein (EndoH; New England Biolabs) or with peptide-N-glycosidase F (PNGase F; New England Biolabs). Cell pellet, 0.1 ml, was washed once with 0.5 ml of lysis buffer (10 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 100 mM NaCl, 300 mM sorbitol, containing one proteinase inhibitor cocktail tablet [Roche Molecular Biochemicals] per 25 ml) and re-suspended in 0.2 ml of this buffer. Cells were broken by vortexing with 0.2 ml of glass beads, the crude lysate was centrifuged at 1000 × g for 5 min, and the supernatant was collected as the total cell lysate. EndoH and PNGase F treatments essentially followed the manufacturer's protocols. Total cell lysate, 100 μl, and 10 μl of 10× denaturing buffer (5% SDS, 10% β-mercaptoethanol) were mixed and treated at 100°C for 10 min. For Endo H treatment, 55 μl of denatured sample, 7 μl of 10× G5 buffer (500 mM Na citrate, pH 5.5), and 7 μl of water were mixed. For PNGase F treatment, 55 μl of denatured sample, 7 μl of 10× G7 buffer (500 mM Na phosphate, pH 7.5), and 7 μl of 10% NP-40 solution (to prevent inhibition of PNGase F by SDS) were mixed. Reactions were initiated by addition of 2 μl of enzyme solution (1000 U) or water and incubated at 37°C for 90 min. SDS sample buffer, 8 μl at 5× (Laemmli, 1970), was then added to each reaction, the mixtures were applied to 6% SDS-polyacrylamide gels, and immunoblotting was performed as described above.
Staining and Microscopy
To visualize bud scars and birth scars for determination of budding patterns, cells were stained with Calcofluor (Pringle, 1991). To visualize GFP-fusion proteins, exponentially growing cells were observed using a Nikon E800 microscope (Nikon, Tokyo, Japan) fitted with a 100× oil-immersion objective (N.A. = 1.30) as previously described (Kang et al., 2001) using filters from Chroma (Brattleboro, VT). Images were collected using IPLab software (Signal Analytics Corporation, Vienna, VA) with a Hamamatsu ORCA-2 CCD camera (Hamamatsu Photonics, Bridgewater, NJ).
RESULTS
Effects of rax1 and rax2 Mutations on Bipolar Budding
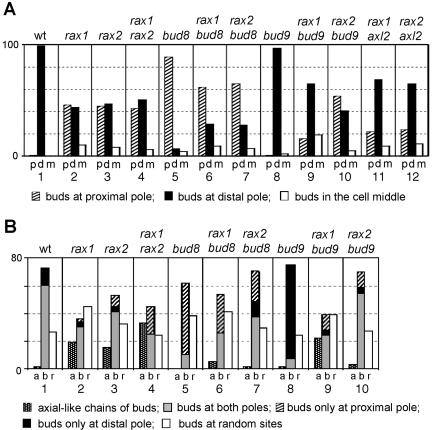
Previous studies had indicated that Rax1p and Rax2p are involved in bipolar budding (see Introduction) but had left considerable uncertainty as to the nature of their roles and how these might relate to the roles of Bud8p and Bud9p. To explore these issues further, we first examined the budding patterns of a set of congenic diploid strains that were homozygous for mutations in RAX1, RAX2, BUD8, or BUD9 as well as in the related double mutants. In the wild-type strain, as observed previously (Chant and Pringle, 1995; Harkins et al., 2001), nearly all first buds (>98%) were at the distal pole of the daughter cell, and mother cells that had budded multiple times typically had most or all of the bud scars clustered around the two poles of the cell (Figure 1, A and B, column 1). The bud8 and bud9 mutants also had the phenotypes reported previously: in the bud8 mutant, nearly all first buds, and most subsequent buds, were at the proximal pole, whereas in the bud9 mutant, nearly all first buds, and most subsequent buds, were at the distal pole (Zahner et al., 1996; Harkins et al., 2001; Figure 1, A and B, columns 5 and 8). The rax1 and rax2 mutants also displayed a severe disruption of bipolar budding, but their phenotypes were strikingly different from those of the bud8 and bud9 mutants. Only about half the first buds were at the distal pole, and mother cells showed a wide variety of bud-scar patterns (Figure 1, A and B, columns 2 and 3). The phenotype of the rax1 rax2 double mutant was very similar to those of the single mutants (Figure 1, A and B, column 4), suggesting that Rax1p and Rax2p may function in the same pathway and perhaps be interdependent for function.
Figure 1.
Budding patterns of diploid strains homozygous for various mutations. At least 300 cells were scored for each type of count on each strain, and percentages are indicated. Strains used were wild-type (YEF473), rax1 (HPY496), rax2 (HPY592), rax1 rax2 (HPY634), bud8 (YHH415), rax1 bud8 (HPY643), rax2 bud8 (HPY641), bud9 (YHH615), rax1 bud9 (HPY692), rax2 bud9 (HPY693), rax1 axl2 (HPY841), and rax2 axl2 (HPY842). (A) The positions of the first buds on daughter cells were scored as being at the pole proximal to the birth scar (p), at the pole distal to the birth scar (d), or in the middle region (m) of the cell. (B) The budding patterns of the strains were determined by counting cells that had >3 bud scars. (a) One or more chains of bud scars similar to those of the axial pattern; (b) bud scars concentrated at one or both poles of the cell; (r) one or more bud scars at seemingly random sites. Within category b, the gray, black, and hatched boxes indicate cells with bud scars at both poles, exclusively at the distal pole, or exclusively at the proximal pole, respectively.
The partial loss of distal-pole first buds in the rax1 and rax2 mutants suggests that Rax1p and Rax2p contribute to, but are not absolutely essential for, the establishment and/or maintenance of the Bud8p-based distal-pole marker. Several other observations support this interpretation. First, deletion of BUD8 in a rax1 or rax2 mutant background further reduced the fraction of distal-pole first buds (Figure 1A, compare columns 6 and 7 to columns 2 and 3). Second, the fraction of distal-pole first buds was reduced in rax1 bud9 and rax2 bud9 double mutants in comparison to a bud9 single mutant (Figure 1A, compare columns 9 and 10 to column 8). Because the distal-pole buds in a bud9 mutant appear to be largely dependent on Bud8p (Harkins et al., 2001), these observations support the hypothesis that Bud8p function is reduced but not eliminated by loss of Rax1p or Rax2p. Finally, introduction of a high-copy BUD8 plasmid largely restored the distal-pole first buds in a rax1 or rax2 mutant strain (Nakashima and Pringle, unpublished observations).
Of the first buds that are not at the distal pole in the rax1 and rax2 mutants, most are at the proximal pole (Figure 1A, columns 2 and 3). At first glance, this might suggest that the Bud9p-based proximal-pole marker is not affected (or is even enhanced) by the loss of Rax1p or Rax2p. However, these strains possess an intact axial budding system, which also provides a marker at the proximal pole that can be utilized to a significant extent in a diploid strain if the bipolar system is sufficiently compromised (Harkins et al., 2001; see Introduction). Moreover, if there is no effective marking of potential bud sites, some first buds should be near the proximal pole by chance alone. Indeed, several observations suggest that the establishment and/or maintenance of the Bud9p-based marker is strongly compromised by the loss of Rax1p or Rax2p. First, significant fractions of rax1, rax2, and rax1 rax2 mother cells show axial-like chains of bud scars (Figure 1B, columns 2-4), suggesting that the axial system is indeed playing a role in determining bud position in these strains. Indeed, deletion of AXL2 (which encodes an essential component of the axial marker; see Introduction) in a rax1 or rax2 mutant background led to a substantial reduction in the fraction of proximal-pole first buds (Figure 1A, compare columns 11 and 12 to columns 2 and 3). Second, deletion of RAX1 or RAX2 in a bud8 mutant background caused a significant decrease in the fraction of proximal-pole first buds (Figure 1A, compare columns 6 and 7 to column 5). Because Bud9p is important for the positioning of buds in a bud8 mutant (Harkins et al., 2001) and Rax1p and Rax2p have no detectable role in the axial pattern (see Introduction), these observations suggest that deletion of RAX1 or RAX2 affects Bud9p function. Third, deletion of BUD9 in a rax2 mutant background produced no detectable decrease in the fraction of proximal-pole first buds (Figure 1A, compare columns 3 and 10), supporting the hypothesis that these buds may mostly be positioned by the axial marker. On the other hand, deletion of BUD9 in a rax1 mutant background did appear to produce a significant decrease in the fraction of proximal-pole first buds (Figure 1A, compare columns 2 and 9), suggesting that the Bud9p-based proximal-pole marker, like the Bud8p-based distal-pole marker, may remain partially functional in the absence of Rax1p.
Interdependent Localization of Rax1p and Rax2p to Both the Division Site and the Distal Pole
It has been reported previously that Rax2p localizes to the division site on mother and daughter cells and that Rax1p is necessary for this localization (Chen et al., 2000). These observations are consistent with the apparent effects of rax1 and rax2 mutations on the ability of cells to establish and/or maintain a bipolar marker at the proximal pole and to maintain a bipolar pattern through multiple cell cycles (Chen et al., 2000; and see above). However, the effects of rax1 and rax2 mutations on the ability of daughter cells to bud at the distal pole (see above) were difficult to explain on the basis of these previous observations. To investigate this matter further and to explore further the interrelationship between Rax1p and Rax2p function, we reexamined the localization of Rax2p and examined the localization of Rax1p using functional GFP-tagged proteins that were expressed from their normal promoters at their normal chromosomal loci (see Materials and Methods).
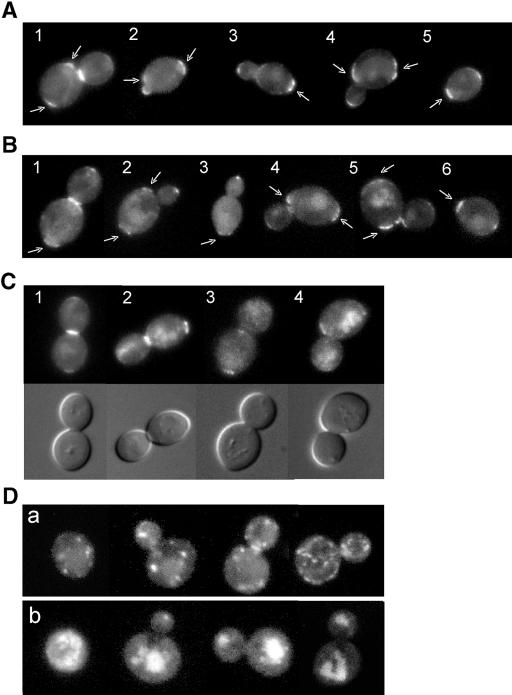
As expected (Chen et al., 2000), Rax2p-GFP was observed at the mother-bud neck in many large-budded cells (Figure 2A, cell 1) and at previous division sites on both mother and daughter cells (Figure 2A, cells 1-5), suggesting that Rax2p may be a stable component of the bipolar landmark at these sites. In addition, however, Rax2p-GFP was observed clearly at the tips of >50% of buds (Figure 2A, cells 2-4) and at the distal poles of most newborn daughter cells (Figure 2A, cell 5).
Figure 2.
Interdependent localization of Rax1p and Rax2p to both the division site and the distal pole. (A-C) Wild-type diploid cells expressing Rax2p-GFP (A; strain HPY814) or GFP-Rax1p (B and C; strain HPY614) were examined for the localization of GFP fluorescence at various stages in the cell cycle. Cells are numbered for reference in the text; arrows indicate signal at previous division sites. In C, the companion DIC images show the presence (cells 1 and 2) or absence (cells 3 and 4) of a fully formed septum. (D) Diploid cells expressing Rax2p-GFP in a rax1Δ background (a; strain HPY836) or GFP-Rax1p in a rax2Δ background (b; strain HPY647) were examined for the localization of GFP fluorescence.
GFP-Rax1p displayed essentially the same localization as Rax2p-GFP: it was observed at the mother-bud neck in many large-budded cells (Figure 2B, cell 1; Figure 2C, cells 1 and 2) and at previous division sites on both mother and daughter cells (Figure 2B, cells 1-6; Figure 2C, cells 1-4). Thus, Rax1p may also be a stable component of the bipolar landmark at previous division sites. Consistent with this idea, we found that GST-Rax1p, like Rax2p (Chen et al., 2000), remained stable as judged by Western blotting for up to 10 h after turning off its expression from the GAL1 promoter (Kang and Park, unpublished observations). Interestingly, examination of large-budded cells by DIC microscopy revealed that GFP-Rax1p signal was only observed clearly at the necks of cells that had fully formed septa, and it was present in nearly all such cells (Figure 2C, compare cells 1 and 2 with cells 3 and 4). In addition, like Rax2p-GFP, GFP-Rax1p was also observed clearly at the tips of ∼50% of buds (Figure 2B, cells 1-4) and at the distal poles of newborn daughter cells (Figure 2B, cell 6).
We next examined the relationships between Rax1p and Rax2p localization. As reported previously (Chen et al., 2000), when Rax2p-GFP was expressed in a rax1 mutant strain, it failed to localize normally. Instead, the GFP signal was observed in small patches or vesicles (Figure 2Da) or in structures that appeared to be the vacuoles (our unpublished results). In addition, when GFP-Rax1p was expressed in a rax2 mutant strain, it also failed to localize normally, and the GFP signal was observed primarily in structures that appeared to be the vacuoles (Figure 2Db). Thus, Rax1p and Rax2p appear to be interdependent for localization (and thus, presumably, for function), consistent with the very similar phenotypes of rax1, rax2, and rax1 rax2 mutant strains.
Biochemical Properties and Interaction of Rax1p and Rax2p
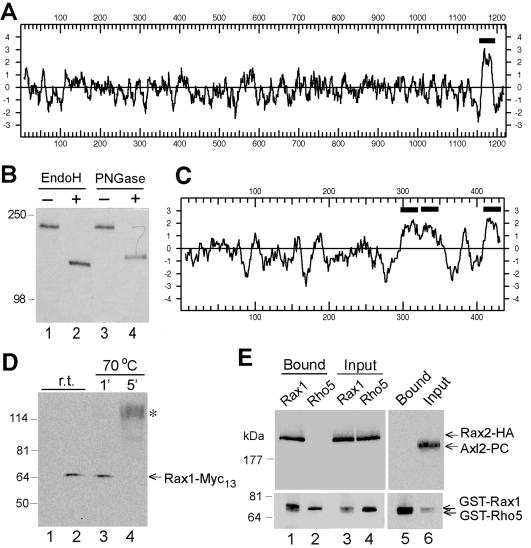
Rax2p contains an apparent transmembrane domain near its C-terminus (Figure 3A) and has been proposed to be a type II membrane protein, with its long N-terminal region in the cytoplasm (Chen et al., 2000). However, the N-terminal region is very S/T rich (21%), suggesting possible O-linked glycosylation, and also contains 52 potential sites for N-linked glycosylation. These observations suggested that Rax2p might instead have its N-terminal region in the extracytoplasmic space and only its short C-terminal region in the cytoplasm. This topology would resemble those of the putative landmark proteins Axl2p (Roemer et al., 1996), Bud8p, and Bud9p (Harkins et al., 2001), except that the latter two proteins appear to have two transmembrane domains surrounding a short cytoplasmic loop. Interestingly, programs for predicting membrane topology differ in their predictions for Rax2p. The programs of von Heijne (Nielsen et al., 1997) and Hartmann et al. (1989) predict that there is no N-terminal signal sequence and that the long N-terminal region is cytoplasmic, whereas TM-Pred (Hofmann and Stoffel, 1993) predicts that the short hydrophobic region at the N-terminus may be a signal sequence and thus that the long N-terminal domain is extracytoplasmic. To resolve this discrepancy, we examined the mobility during SDS-PAGE of HA-epitope-tagged Rax2p with and without treatment with enzymes that remove N-linked glycosyl moieties. In the absence of enzyme treatment, Rax2p-HA3 migrated with an apparent molecular mass of ∼200 kDa, much higher than the 134 kDa predicted from the sequence of its polypeptide chain (Figure 3B). After enzyme treatment, the apparent molecular mass was reduced to ∼130 kDa, closer to that predicted for the polypeptide chain (Figure 3B). Both before and after enzyme treatment, Rax2p-HA3 formed bands that were sharper than usual for a glycoprotein. We currently have no explanation for this finding. Nonetheless, because the short C-terminal region has no potential N-linked glycosylation sites, the clear evidence for glycosylation suggests that Rax2p is oriented in the membrane with its long N-terminal region in the extracytoplasmic space and therefore with its short C-terminal region in the cytoplasm.
Figure 3.
Biochemical properties and interaction of Rax1p and Rax2p. (A) Hydropathy plot of Rax2p generated by the method of Kyte and Doolittle (1982) with a window size of 11. The region N-terminal to the predicted transmembrane domain (black bar) contains 21% S + T and 52 potential N-linked glycosylation sites (N-X-S/T, where X is any amino acid other than P; Tanner and Lehle, 1987). (B) N-linked glycosylation of Rax2p. Total cell lysates from rax2Δ strain AM476 containing RAX2-HA3 plasmid pHP1183 were either mock-digested (-) or digested with Endo H or PNGase (+) before SDS-PAGE and immunoblotting with anti-HA antibody 3F10. (C) Hydropathy plot of Rax1p generated as described in A. Black bars, possible transmembrane domains. (D) Mobility of Rax1p-Myc13 in SDS-PAGE and its aggregation upon heating in SDS solution. Immunoblotting was performed using anti-Myc antibody 9E10 (see Materials and Methods). Lanes 1 and 2, proteins from total cell lysates of wild-type strain HPY16 (lane 1) or RAX1-MYC13 strain HPY843 (lane 2) were mixed with SDS-PAGE sample buffer and loaded onto a 6% gel after a 5-min incubation at room temperature (r.t.). Lanes 3 and 4, total proteins from HPY843 were mixed with SDS-PAGE sample buffer and heated at 70°C for 1 (lane 3) or 5 (lane 4) min before loading onto the gel. The asterisk indicates the aggregated Rax1p-Myc13. (E) Copurification of Rax1p and Rax2p from yeast. Cells of strain HPY644 (RAX2-HA3) carrying GAL1p-GST-HIS6-RAX1 plasmid pHP1156 (lanes 1 and 3) or GAL1p-GST-HIS6-RHO5 plasmid pHP1158 (lanes 2 and 4) were grown in SC-Ura medium with 2% sucrose as carbon source and then induced for 4 h by adding galactose to 2%. The membrane fractions were isolated, solubilized with 1% Triton X-100, and subjected to pull-down of the GST-tagged protein (see Materials and Methods). Samples of the proteins eluted from the glutathione Sepharose (lanes 1 and 2) and of the input material (lanes 3 and 4) were analyzed by SDS-PAGE and immunoblotting with anti-HA antibody HA11 (top panel) and anti-GST antibody (bottom panel; see Materials and Methods). As a control (lanes 5 and 6), the membrane fraction prepared from cells of strain HPY798 (rax1Δ) containing plasmids pHP1169 (AXL2-PC) and pHP1156 was analyzed similarly using antibodies against protein C (top panel) and GST (bottom panel).
The hydropathy plot of Rax1p shows that it also has one or more potential transmembrane domains (Figure 3C), and application of the “positive inside” rule (von Heijne and Gavel, 1988; Hartmann et al., 1989) predicts a type IIIa membrane topology with the N-terminus oriented toward the cytoplasm. Crude fractionation experiments suggest that Rax1p is enriched in the membrane fraction but can be solubilized with nonionic detergents such as 1% Triton X-100 (Angerman and Park, unpublished data). Like some membrane proteins, particularly those that have multiple hydrophobic domains (Franzusoff et al., 1991; Harsay and Bretscher, 1995), Rax1p tagged with either Myc13 or GST appeared to aggregate with itself and/or other proteins upon heating in SDS solution. Surprisingly, this aggregation was observed even after short incubations at temperatures as low as 70°C (Figure 3D, lane 4). When aggregation was avoided, Rax1p-Myc13 and GST-Rax1p migrated with apparent molecular masses of ∼65 and ∼76 kDa, respectively (Figure 3, D, lanes 2 and 3, and 3E, lanes 1 and 3), close to the values predicted for their polypeptide chains, suggesting that Rax1p is not extensively modified either by glycosylation or in other ways.
Given the similar phenotypes of the rax1 and rax2 mutants and the interdependent localization of Rax1p and Rax2p, we explored the possible physical association between these proteins by expressing either GST-Rax1p or GST-Rho5p (as a control) in yeast cells that also expressed Rax2p-HA3. These proteins were all solubilized with 1% Triton X-100, as judged by SDS-PAGE and immunoblotting of samples of the supernatants obtained after Triton treatment (Figure 3E, lanes 3 and 4). When the GST-fusion proteins were pulled down with glutathione Sepharose, Rax2p-HA3 was copurified with GST-Rax1p but not with GST-Rho5p (Figure 3E, compare lane 1 with lane 2). In a second control experiment, the axial landmark component Axl2p was expressed as a protein C-epitope-tagged protein (Axl2p-PC) in cells that also expressed GST-Rax1p (Figure 3E, lane 6). Like Rax2p, Axl2p is an N-glycosylated transmembrane protein with type I membrane topology (Roemer et al., 1996), but it did not associate with GST-Rax1p after being subjected to the same procedure (Figure 3E, lane 5). Thus, it appears that Rax1p interacts specifically with Rax2p.
Interaction between Rax1p/Rax2p and Bud8p
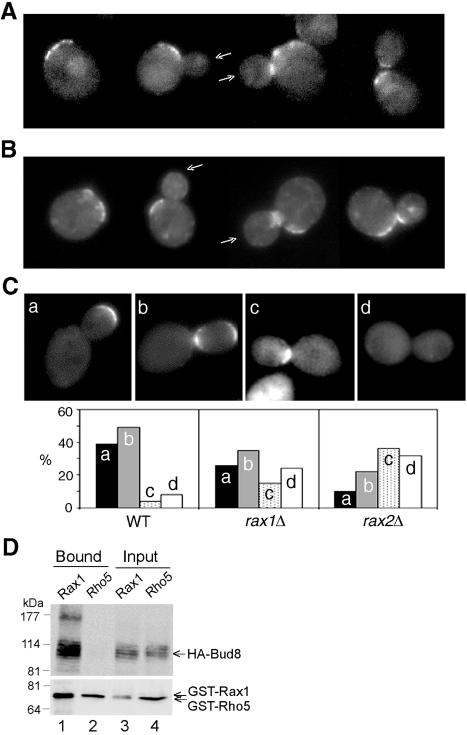
As described above, Rax1p and Rax2p localize interdependently to the bud tip and the distal pole of the daughter cell, and rax1 or rax2 mutant daughter cells do not bud efficiently at their distal poles. These observations suggested that Rax1p and/or Rax2p might interact with Bud8p as well as with each other, a hypothesis that is supported by several additional observations. First, when GFP-Rax1p or Rax2p-GFP was expressed in cells lacking Bud8p, localization to the bud tips and the distal poles of daughter cells was largely or entirely lost (>90%; n = 60∼80), while localization to the mother-bud neck, proximal poles, and previous division sites seemed unaffected (Figure 4, A and B). Second, when GFP-Bud8p was expressed in cells lacking Rax1p or Rax2p, some cells showed seemingly normal localization of GFP-Bud8p to the bud tips and distal poles, but many other cells had apparently lost such localization; the rax2 mutation appeared to have a somewhat stronger effect than did the rax1 mutation (Figure 4C). Finally, HA-Bud8p could be copurified with GST-Rax1p from yeast cells (Figure 4D). Taken together, these results suggest that Rax1p (and hence, at least indirectly, Rax2p) interacts with Bud8p; that this interaction is essential for the localization of Rax1p and Rax2p to the bud tip and distal pole; and that this interaction is also important, although apparently not essential, for the normal localization of Bud8p to the distal pole.
Figure 4.
Interaction between Rax1p/Rax2p and Bud8p. (A and B) Localization of Rax1p and Rax2p to the bud tip depends on Bud8p. Cells of GFP-RAX1 bud8Δ strain HPY690 (A) and RAX2-GFP bud8Δ strain HPY691 (B) were examined for the localization of GFP fluorescence. Arrows indicate bud tips lacking GFP-Rax1p or Rax2p-GFP; localization to mother-bud necks and previous division sites appears normal. (C) Efficient localization of Bud8p to the bud tip depends on Rax1p and Rax2p. The bud8Δ strain YHH415 (WT), rax1Δ bud8Δ strain HPY643 (rax1Δ), and rax2Δ bud8Δ strain HPY641 (rax2Δ) were transformed with GFP-BUD8 plasmid YEpGFP-BUD8F, and ∼100 large-budded cells were examined for the localization of GFP fluorescence. Four patterns of GFP-Bud8p localization were observed, as shown in the representative images: (a) fluorescence exclusively at or near the bud tip; (b) fluorescence at both the bud tip and neck; (c) fluorescence exclusively at the neck (often specifically on the daughter side); and (d) no localized fluorescence. The percentage of each pattern of each strain is shown in the histograms. Note that the high perentage of wild-type cells with GFP-Bud8p at the neck as well as the distal pole probably reflects the overexpression of GFP-Bud8p in these cells (Harkins et al., 2001; Schenkman et al., 2002). (D) Copurification of Rax1p and Bud8p from yeast. Cells of bud8Δ strain HPY568 carrying HA3-BUD8 plasmid YEpHA-BUD8F and either GAL1p-GST-HIS6-RAX1 plasmid pHP1156 (lanes 1 and 3) or GAL1p-GST-HIS6-RHO5 plasmid pHP1158 (lanes 2 and 4) were grown and analyzed as described for Figure 3E.
Interaction between Rax1p/Rax2p and Bud9p
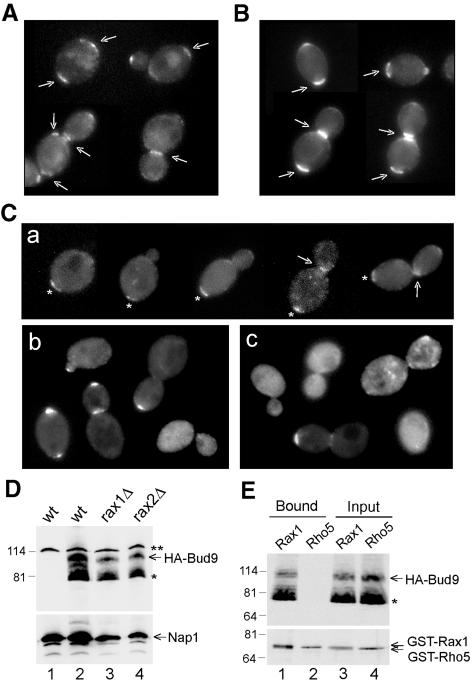
As described above, Rax1p and Rax2p localize interdependently to the necks of large-budded cells and to the division sites on both mother and daughter cells, and rax1 or rax2 mutant daughter cells appear largely or entirely unable to bud at their proximal poles using a bipolar (Bud9p-dependent) marker. These observations suggested that Rax1p and/or Rax2p might interact with Bud9p as well as with each other. Surprisingly, the localization of GFP-Rax1p and Rax2p-GFP did not appear to be affected by an absence of Bud9p (Figure 5, A and B). GFP-Rax1p or Rax2p-GFP was observed at the previous division sites in >90% of the bud9 mutant cells (n = 100) and at the mother-bud neck in >60% of the bud9 mutant cells with large-sized buds (n = 80). In contrast, however, the normal localization of Bud9p to the daughter side of the neck in large-budded cells and to the proximal pole of the daughter cell (Figure 5Ca) was largely lost in the rax1 and rax2 mutants. In the rax1 mutant, many cells had GFP3-Bud9p localized to the bud tip, some cells had little or no detectable GFP signal, but few cells showed localization to the normal site (Figure 5Cb). The rax2 mutation appeared to have a similar effect on Bud9p localization, but fewer cells had GFP3-Bud9p localized to the bud tip than in the rax1 mutant (Figure 5Cc). Because the rax1 and rax2 mutations had no obvious effect on the levels of HA-tagged Bud9p (Figure 5D), the absence of Rax1p or Rax2p appears to affect the localization of Bud9p rather than its synthesis or stability. Finally, HA-Bud9p could be copurified with GST-Rax1p from yeast cells (Figure 5E). Taken together, these results suggest that Rax1p (and hence, at least indirectly, Rax2p) interacts with Bud9p; that this interaction is important, although perhaps not absolutely essential, for the normal localization of Bud9p to the division site and the proximal pole; but that this interaction is not essential for the localization of Rax1p and Rax2p to the division site and the proximal pole.
Figure 5.
Interaction between Rax1p/Rax2p and Bud9p. (A and B) Localization of Rax1p and Rax2p does not appear to depend on Bud9p. Cells of GFP-RAX1 bud9Δ strain HPY704 (A) and RAX2-GFP bud9Δ strain HPY705 (B) were examined for the localization of GFP fluorescence. Arrows indicate seemingly normal localization of GFP-Rax1p and Rax2p-GFP to mother-bud necks and previous division sites. (C) Localization of Bud9p depends on Rax1p and Rax2p. Wild-type strain YEF473 (a), rax1Δ strain HPY496 (b), and rax2Δ strain HPY592 (c) were transformed with GFP3-BUD9 plasmid pHP1202, and cells were examined for the localization of GFP fluorescence. In panel a, asterisks indicate the proximal poles as judged by Calcofluor staining, and arrows indicate GFP3-Bud9p localized normally to the daughter side of the neck. (D) Bud9p levels are approximately normal in the absence of Rax1p or Rax2p. Wild-type strain YEF473 (lane 2), rax1Δ strain HPY496 (lane 3), and rax2Δ strain HPY592 (lane 4) were transformed with HA3-BUD9 plasmid pHP1319, and total cell lysates from the same OD units of cells were analyzed by immunoblotting using anti-HA antibody HA11 (top panel) or anti-Nap1p antibodies as a loading control (bottom panel). As a control, total cell lysate was prepared from YEF473 without plasmid and subjected to the same procedure (lane 1). The asterisk indicates a presumably degraded or undermodified Bud9p; the double asterisk indicates a protein cross-reacting nonspecifically with the anti-HA antibody. (E) Copurification of Rax1p and Bud9p from yeast. Cells of bud9Δ strain HPY570 carrying HA3-BUD9 plasmid pHP1319 and either GAL1p-GST-HIS6-RAX1 plasmid pHP1156 (lanes 1 and 3) or GAL1p-GST-HIS6-RHO5 plasmid pHP1158 (lanes 2 and 4) were grown and analyzed as described for Figure 3E. The asterisk indicates a presumably degraded or undermodified Bud9p.
DISCUSSION
RAX1 and RAX2 were originally identified by mutations that appeared to suppress the axl1 mutant defect in axial budding (Fujita et al., 1994; Chen et al., 2000). However, it now seems clear that this phenotype reflects roles of Rax1p and Rax2p in bipolar rather than axial budding (see Introduction). Chen et al. (2000) observed that Rax2p localizes to the division site on both mother and daughter cells in a Rax1p-dependent manner and that this localization persists through multiple cell cycles, and they hypothesized that Rax2p is a component of the persistent cortical landmarks that had been predicted to provide the spatial information for bipolar budding (Chant and Pringle, 1995). However, many details of the structure and function of Rax1p and Rax2p remained unclear, including the relationship between their function and that of the putative bipolar landmark proteins Bud8p and Bud9p (Zahner et al., 1996; Harkins et al., 2001; Schenkman et al., 2002). The studies reported here have now clarified several of the outstanding issues.
First, some aspects of the structure and membrane association of Rax2p and Rax1p have been clarified. Rax2p was previously suggested to be an integral membrane protein with type II orientation (Chen et al., 2000), apparently on the basis of predictions using the programs of von Heijne (Nielsen et al., 1997) and/or Hartmann et al. (1989). However, the program TM-pred (Hofmann and Stoffel, 1993) predicts a type I orientation, with the long N-terminal domain in the extracyotplasmic space. This prediction is consistent with the S/T richness and mulitple potential N-linked glycosylation sites of the N-terminal domain and is strongly supported by the evidence that Rax2p is indeed N-glycosylated. The type I orientation presumably implies that the short hydrophobic region at the extreme N-terminus serves as a signal sequence for membrane insertion, but this point requires further investigation. It is of interest that Rax2p now appears to resemble the other putative landmark proteins Axl2p, Bud8p, and Bud9p in having a long, glycosylated N-terminal domain in the extracytoplasmic space and only a relatively short C-terminal domain in the cytoplasm. The bulky N-terminal domains of these proteins may serve to anchor them in the cell wall so that they do not diffuse in the plane of the plasma membrane, to allow interactions with other proteins of similar structure, or both. To distinguish between these and other possible models will require further investigation.
Rax1p also appears to be an integral membrane protein. Its hydropathy plot predicts three possible transmembrane domains, and, in crude fractionation experiments, it is found in the pellet but is effectively solubilized by a nonionic detergent. It also appears to aggregate with remarkable avidity upon mild heating in the presence of SDS, presumably by association of exposed hydrophobic domains. There is no indication that Rax1p is glycosylated or subject to other bulky posttranslational modification. However, the biochemical properties of Rax1p require considerable further investigation.
After solubilization with Triton X-100, Rax1p and Rax2p can be copurified from yeast cell extracts. The apparent physical association of the proteins presumably helps to explain their interdependent localization to specific sites in the plasma membrane (Chen et al., 2000; this study). It is not yet clear whether the Rax1p-Rax2p association is direct or mediated by other proteins or whether it involves the extracytoplasmic, transmembrane, or cytoplasmic domains of the proteins. Additional biochemical studies will be required to clarify these points.
Careful analysis of the rax1, rax2, and rax1 rax2 mutant phenotypes (see Results) suggests that Rax1p and Rax2p function together in helping to mark all of the sites that are thought to possess landmarks used in bipolar budding, namely the distal pole of daughter cells, the proximal pole (division site) of daughter cells, and each previous division site on mother cells (Chant and Pringle, 1995). Several additional observations are consistent with this model. First, Rax1p and Rax2p localize interdependently to all of the relevant sites: functional GFP-tagged proteins were observed at the tips of buds and the distal poles of daughter cells as well as in persistent patches or rings at the previous division sites on both mother and daughter cells. Second, it seems clear that Rax1p and Rax2p interact both with Bud8p at the bud tip and distal pole and with Bud9p at the proximal pole. The existence of these interactions is supported by biochemical experiments indicating that both Bud8p and Bud9p can be copurified with Rax1p (and thus, presumably, also with Rax2p). We also found that Rax1p and Rax2p were almost totally unable to localize to the bud tip or distal pole in the absence of Bud8p, whereas Bud8p suffered a partial loss of ability to localize and function to direct budding events in the absence of Rax1p and Rax2p. Thus, Bud8p appears to be able to provide a spatial signal without Rax1p and Rax2p, but does so only inefficiently.
In contrast, Rax1p and Rax2p appear to be able to localize normally to the proximal poles of daughter cells in the absence of Bud9p, whereas Bud9p was largely unable to localize normally and appeared to retain little if any function to direct budding events, in the absence of Rax1p or Rax2p. However, Rax1p and Rax2p appear to have no significant ability to provide a spatial signal at the proximal pole in the absence of Bud9p, because a bud9 null mutant almost never buds at the proximal pole (Zahner et al., 1996; Harkins et al., 2001; Figure 1). Thus, it appears that Rax1p, Rax2p, and Bud9p must all function together to provide a spatial signal at this site.
The above observations highlight the interesting problem of what provides the spatial signal for bipolar budding at the previous division sites on mother cells. Rax1p and Rax2p both localize persistently to these sites (Chen et al., 2000; Figure 1), but neither Bud8p nor Bud9p appears to be present at these sites (Harkins et al., 2001; Schenkman et al., 2002). If Rax1p and Rax2p by themselves can provide the spatial signal at these sites, why do they seem unable to do so at the proximal pole of daughter cells? Perhaps there is some other partner protein that has not yet been identified. If so, however, it is one that is not readily recognized on the basis of sequence similarity to Bud8p or Bud9p (our unpublished results). The situation is puzzling, because the similarity in sequence between the cytoplasmic domains of Bud8p and Bud9p is one of the observations suggesting that these domains might provide the proximal recognition signal for the Rsr1p/Bud5p/Bud2p module.
Normally, BUD9 is expressed early in the cell cycle, but Bud9p is not delivered to the plasma membrane until late in the cell cycle, when it arrives at the mother-bud neck along with other cell-surface materials (Schenkman et al., 2002). In contrast, in the rax1 mutant, many cells showed a strong GFP3-Bud9p signal at the bud tip beginning early in the cell cycle. Although there is a significant caveat arising from the possible promotion of mislocalization by overexpression of the plasmid-borne GFP3-BUD9, this observation suggests that in the absence of Rax1p, the mechanism that normally delays delivery of Bud9p to the surface may be inoperative, so that the newly made Bud9p is delivered immediately to the cell surface at the growth zone active at that time in the cell cycle.
Despite the various clues discussed above, many mysteries remain about the interactions and specific functions of Rax1p, Rax2p, Bud8p, and Bud9p. What are the precise functions of the extracytoplasmic, transmembrane, and cytoplasmic domains of each protein? Which protein or proteins provide the proximal signal that is recognized by the Rsr1p/Bud5p/Bud2p module? Do some of these proteins function to facilitate the passage of the others through the secretory system to the correct locations? Do some of these proteins function to anchor and/or stabilize the other proteins in these correct locations? Our future studies will address these challenging problems.
Supplementary Material
Acknowledgments
We thank A. McKenzie, L. Schenkman, B. Glick, S. Emr, G. Petsko, and T. J. Proszynski for helpful suggestions, stimulating discussions, and help with some experiments. We also thank M. Snyder, C. Boone, B. Glick, T.-H. Chang, M. Longtine, and B. Lee for plasmids and M. Bishop and D. Kellogg for antibodies. This work was supported by National Institutes of Health grants GM56997 (to H.-O.P.) and GM31006 (to J.R.P.).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04-07-0600. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-07-0600.
The online version of this article contains supplementary material accessible through http://www.molbiolcell.org.
References
- Adames, N., Blundell, K., Ashby, M.N., and Boone, C. (1995). Role of yeast insulin-degrading enzyme homologs in propheromone processing and bud site selection. Science 270, 464-467. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., and Struhl, K. (1999). Current Protocols in Molecular Biology, New York: John Wiley & Sons.
- Bi, E., and Pringle, J.R. (1996). ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae. Mol. Cell. Biol. 16, 5264-5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chant, J., and Herskowitz, I. (1991). Genetic control of bud site selection in yeast by a set of gene products that constitute a morphogenetic pathway. Cell 65, 1203-1212. [DOI] [PubMed] [Google Scholar]
- Chant, J., Mischke, M., Mitchell, E., Herskowitz, I., and Pringle, J.R. (1995). Role of Bud3p in producing the axial budding pattern of yeast. J. Cell Biol. 129, 767-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chant, J., and Pringle, J.R. (1995). Patterns of bud-site selection in the yeast Saccharomyces cerevisiae. J. Cell Biol. 129, 751-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, T., Hiroko, T., Chaudhuri, A., Inose, F., Lord, M., Tanaka, S., Chant, J., and Fujita, A. (2000). Multigenerational cortical inheritance of the Rax2 protein in orienting polarity and division in yeast. Science 290, 1975-1978. [DOI] [PubMed] [Google Scholar]
- Christianson, T.W., Sikorski, R.S., Dante, M., Shero, J.H., and Hieter, P. (1992). Multifunctional yeast high-copy-number shuttle vectors. Gene 110, 119-122. [DOI] [PubMed] [Google Scholar]
- Franzusoff, A., Rothblatt, J., and Schekman, R. (1991). Analysis of polypeptide transit through yeast secretory pathway. Methods Enzymol. 194, 662-674. [DOI] [PubMed] [Google Scholar]
- Freifelder, D. (1960). Bud position in Saccharomyces cerevisiae. J. Bacteriol. 80, 567-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, A., Oka, C., Arikawa, Y., Katagai, T., Tonouchi, A., Kuhara, S., and Misumi, Y. (1994). A yeast gene necessary for bud-site selection encodes a protein similar to insulin-degrading enzymes. Nature 372, 567-570. [DOI] [PubMed] [Google Scholar]
- Gietz, R.D., and Sugino, A. (1988). New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74, 527-534. [DOI] [PubMed] [Google Scholar]
- Guthrie, C., and Fink, G.R. (1991). Guide to Yeast Genetics and Molecular Biology. Methods in Enzymology, Vol. 194, San Diego: Academic Press. [PubMed]
- Halme, A., Michelitch, M., Mitchell, E.L., and Chant, J. (1996). Bud10p directs axial cell polarization in budding yeast and resembles a transmembrane receptor. Curr. Biol. 6, 570-579. [DOI] [PubMed] [Google Scholar]
- Harkins, H.A., Pagé, N., Schenkman, L.R., De Virgilio, C., Shaw, S., Bussey, H., and Pringle, J.R. (2001). Bud8p and Bud9p, proteins that may mark the sites for bipolar budding in yeast. Mol. Biol. Cell 12, 2497-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harsay, E., and Bretscher, A. (1995). Parallel secretory pathways to the cell surface in yeast. J. Cell Biol. 131, 297-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann, E., Rapoport, T.A., and Lodish, H.F. (1989). Predicting the orientation of eukaryotic membrane-spanning proteins. Proc. Natl. Acad. Sci. USA 86, 5786-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz, I., and Jensen, R.E. (1991). Putting the HO gene to work: practical uses for mating-type switching. Methods Enzymol. 194, 132-146. [DOI] [PubMed] [Google Scholar]
- Hicks, J.B., Strathern, J.N., and Herskowitz, I. (1977). Interconversion of yeast mating types. III. Action of the homothallism (HO) gene in cells homozygous for the mating type locus. Genetics 85, 395-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann, K., and Stoffel, W. (1993). A database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 374, 166 [Google Scholar]
- Kang, P.J., Sanson, A., Lee, B., and Park, H.-O. (2001). A GDP/GTP exchange factor involved in linking a spatial landmark to cell polarity. Science 292, 1376-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski, K.G., Beven, L., Angerman, E., Tong, A.H.Y., Boone, C., and Park, H.-O. (2003). Interaction between a Ras and a Rho GTPase couples selection of a growth site to the development of cell polarity in yeast. Mol. Biol. Cell 14, 4958-4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte, J., and Doolittle, R.F. (1982). A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105-132. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680-685. [DOI] [PubMed] [Google Scholar]
- Longtine, M.S., McKenzie, A., III, Demarini, D.J., Shah, N.G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J.R. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953-961. [DOI] [PubMed] [Google Scholar]
- Lord, M., Inose, F., Hiroko, T., Hata, T., Fujita, A., and Chant, J. (2002). Subcellular localization of Axl1, the cell type-specific regulator of polarity. Curr. Biol. 12, 1347-1352. [DOI] [PubMed] [Google Scholar]
- Lord, M., Yang, M.C., Mischke, M., and Chant, J. (2000). Cell cycle programs of gene expression control morphogenetic protein localization. J. Cell Biol. 151, 1501-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston, A.L., Chen, T., Yang, M.C., Belhumeur, P., and Chant, J. (2001). A localized GTPase exchange factor, Bud5, determines the orientation of division axes in yeast. Curr. Biol. 11, 803-807. [DOI] [PubMed] [Google Scholar]
- Ni, L., and Snyder, M. (2001). A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae. Mol. Biol. Cell 12, 2147-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, H., Engelbrecht, J., Brunak, S., and von Heijne, G. (1997). Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10, 1-6. [DOI] [PubMed] [Google Scholar]
- Park, H.-O., Bi, E., Pringle, J.R., and Herskowitz, I. (1997). Two active states of the Ras-related Bud1/Rsr1 protein bind to different effectors to determine yeast cell polarity. Proc. Natl. Acad. Sci. USA 94, 4463-4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, H.-O., Chant, J., and Herskowitz, I. (1993). BUD2 encodes a GTPase-activating protein for Bud1/Rsr1 necessary for proper bud-site selection in yeast. Nature 365, 269-274. [DOI] [PubMed] [Google Scholar]
- Park, H.-O., Kang, P.J., and Rachfal, A.W. (2002). Localization of the Rsr1/Bud1 GTPase involved in selection of a proper growth site in yeast. J. Biol. Chem. 277, 26721-26724. [DOI] [PubMed] [Google Scholar]
- Park, H.-O., Sanson, A., and Herskowitz, I. (1999). Localization of Bud2p, a GTPase-activating protein necessary for programming cell polarity in yeast to the presumptive bud site. Genes Dev. 13, 1912-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle, J.R. (1991). Staining of bud scars and other cell wall chitin with Calcofluor. Methods Enzymol. 194, 732-735. [DOI] [PubMed] [Google Scholar]
- Pringle, J.R., Bi, E., Harkins, H.A., Zahner, J.E., De Virgilio, C., Chant, J., Corrado, K., and Fares, H. (1995). Establishment of cell polarity in yeast. Cold Spring Harbor Symp. Quant. Biol. 60, 729-744. [DOI] [PubMed] [Google Scholar]
- Roemer, T., Madden, K., Chang, J., and Snyder, M. (1996). Selection of axial growth sites in yeast requires Axl2p, a novel plasma membrane glycoprotein. Genes Dev. 10, 777-793. [DOI] [PubMed] [Google Scholar]
- Rossanese, O.W., Reinke, C.A., Bevis, B.J., Hammond, A.T., Sears, I.B., O'Connor, J., and Glick, B.S. (2001). A role for actin, Cdc1p, and Myo2p in the inheritance of late Golgi elements in Saccharomyces cerevisiae. J. Cell Biol. 153, 47-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein, R. (1991). Targeting, disruption, replacement, and allele rescue: Integrative DNA transformation in yeast. Methods Enzymol. 194, 281-301. [DOI] [PubMed] [Google Scholar]
- Sanders, S.L., and Herskowitz, I. (1996). The Bud4 protein of yeast, required for axial budding, is localized to the mother/bud neck in a cell cycle-dependent manner. J. Cell Biol. 134, 413-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkman, L.R., Caruso, C., Pagé, N., and Pringle, J.R. (2002). The role of cell cycle-regulated expression in the localization of spatial landmark proteins in yeast. J. Cell Biol. 156, 829-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R.S., and Hieter, P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight, A.F., Sedat, J.W., and Murray, A.W. (1998). Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J. Cell Biol. 143, 687-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner, W., and Lehle, L. (1987). Protein glycosylation in yeast. Biochim. Biophys. Acta 906, 81-99. [DOI] [PubMed] [Google Scholar]
- von Heijne, G., and Gavel, Y. (1988). Topogenic signals in integral membrane proteins. Eur. J. Biochem. 174, 671-678. [DOI] [PubMed] [Google Scholar]
- Zahner, J., Harkins, H.A., and Pringle, J.R. (1996). Genetic analysis of the bipolar pattern of bud site selection in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 16, 1857-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, H. et al. (2001). Global analysis of protein activities using proteome chips. Science 293, 2101-2105. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.