Abstract
Previous studies of the kinetochore in mammalian systems have demonstrated that this structure undergoes reorganizations after microtubule attachment or in response to activation of the spindle checkpoint. Here, we show that the Caenorhabditis elegans kinetochore displays analogous rearrangements at prometaphase, when microtubule/chromosome interactions are being established, and after exposure to checkpoint stimuli such as nocodazole or anoxia. These reorganizations are characterized by a dissociation of several kinetochore proteins, including HCP-1/CeCENP-F, HIM-10/CeNuf2, SAN-1/CeMad3, and CeBUB-1, from the centromere. We further demonstrate that at metaphase, despite having dissociated from the centromere, these reorganized kinetochore proteins maintain their associations with the metaphase plate. After checkpoint activation, these proteins are detectable as large “flares” that project out laterally from the metaphase plate. Disrupting these gene products via RNA interference results in sensitivity to checkpoint stimuli, as well as defects in the organization of chromosomes at metaphase. These phenotypes suggest that these proteins, and by extension their reorganization during mitosis, are important for mediating the checkpoint response as well as directing the assembly of the metaphase plate.
INTRODUCTION
The kinetochore is a dynamic structure that assembles onto mitotic chromosomes and plays multiple roles during mitosis (reviewed in Rieder and Salmon, 1998; Cleveland et al., 2003). It functions initially during prometaphase, providing an attachment site for kinetochore microtubules and serving as the connection between the chromosomes and the mitotic spindle. After attachment, the kinetochore interacts with microtubules to generate the force required to direct chromosome movement (Maney et al., 1998; Pearson et al., 2003). Finally, kinetochores help ensure accurate chromosome segregation by monitoring the alignment of metaphase chromosomes. If chromosomes have not established proper connections to the spindle or are misaligned within the metaphase plate, the kinetochore is able to transmit a signal delaying the onset of anaphase (reviewed in Musacchio and Hardwick, 2002). Consistent with this idea, a number of spindle checkpoint proteins localize to the kinetochore (Chen et al., 1996; Li and Benezra, 1996; Taylor et al., 1998; Kitagawa and Rose, 1999; Sharp-Baker and Chen, 2001; Nystul et al., 2003).
Studies of the kinetochore in mammalian systems show that this structure is dynamic and undergoes rearrangements at various points throughout mitosis and in response to external stimuli (reviewed in Rieder and Salmon, 1998). These alterations were first detected as variations in the size of the kinetochore during the transition between prometaphase and metaphase (Cassimeris et al., 1990), coinciding with the time when interactions between chromosomes and microtubules are being established. Using immunofluorescence microscopy, it has been demonstrated that the association of many kinetochore proteins varies considerably after microtubule attachment (Yao et al., 1997; Jablonski et al., 1998; Waters et al., 1998; Martinez-Exposito et al., 1999; King et al., 2000; Hoffman et al., 2001). In addition, treatment of mammalian cells with the microtubule destabilizing drug nocodazole induces dramatic changes in kinetochores, causing them to adopt crescent or ring-shaped morphologies (Thrower et al., 1996; Martinez-Exposito et al., 1999; Hoffman et al., 2001). Although the purpose of these rearrangements remains unclear, it is possible that they are involved in regulating the diverse functions of the kintochore.
In an effort to expand upon these observations, we have begun to examine kinetochore reorganization in the holocentric organism Caenorhabditis elegans. The C. elegans kinetochore extends the length of the chromosome (Albertson and Thomson, 1982; reviewed in Dernburg, 2001), offering an increased capacity for cytological examination of this structure and any rearrangements it may undergo. And although holocentric chromosomes seem distinct from monocentric chromosomes on a superficial level, there are many parallels between these systems. For example, in mammalian cells, the centromeres line up in the middle of the cell at metaphase, whereas the chromosome arms freely oscillate within the cell. In C. elegans, the centromere, and thus microtubule binding sites, are evenly distributed along the chromosome; therefore, the entire chromosome migrates to the center of the cell (Albertson and Thomson, 1982; reviewed in Dernburg, 2001). We suggest that the alignment of C. elegans chromosomes presents a magnified view of what must happen for mammalian centromeres to align at metaphase. In addition, it has been proposed (Zinkowski et al., 1991) that the mammalian kinetochore is composed of discrete centromere subunits that associate during mitosis to form a functional mammalian kinetochore. This situation is analogous to what is seen in C. elegans, where the punctate pattern of centromere staining present in the interphase nucleus coalesces into lines on prophase chromosomes (Buchwitz et al., 1999). These results suggest that similar dynamic mechanisms may regulate the assembly of the both monocentric and holocentric centromeres.
Here, we demonstrate that, analogous to what occurs in mammalian systems, several C. elegans kinetochore proteins reorganize at prometaphase, as well as in response to checkpoint stimuli. We find that these kinetochore proteins dissociate from the centromere yet remain associated with the poleward faces of metaphase chromosomes. To better understand the function of the gene products involved in these rearrangements, we have used RNA interference (RNAi) to disrupt the expression of this set of kinetochore proteins. The resulting RNAi phenotypes suggest that these kinetochore proteins, and by extension their reorganization during mitosis, are important for regulating chromosome segregation.
MATERIALS AND METHODS
RNA Interference
We generated a polymerase chain reaction template for the production of double-stranded RNA as described previously (Moore et al., 1999), by using primer sets specific for hcp-1, hcp-2, him-10, and san-1. RNAi embryos were prepared by soaking wild-type young adults in concentrated RNA solutions or by injecting RNA directly into the gonad of adults (Fire et al., 1998; Moore and Roth, 2001). In either case, the worms were allowed to purge for 12-18 h after RNAi treatment before embryos were examined for RNAi phenotypes.
Antibody Staining and Microscopy
In general, embryos were fixed with N,N-dimthylformamide and stained as described previously (Moore et al., 1999; Moore and Roth, 2001). To help ensure that our visualization of the kinetochore reorganization was not an artifact of the fixation process, we also prepared samples by using methanol (Moore et al., 1999) or paraformaldehyde (Howe et al., 2001), with the slight variation that after freeze-cracking, slides were placed into cold methanol. The following primary antibodies were used: α-HCP-3 (Buchwitz et al., 1999), α-HCP-1 (Moore et al., 1999), and α-SAN-1 and YL1/2, an α-tubulin antibody (Amersham Biosciences, Piscataway, NJ). Tony Hyman generously provided α-MCAK and α-BUB-1 antibodies (Oegema et al., 2001), and Barbara Meyer made the α-HIM-10 antibody available (Howe et al., 2001). Staining was detected with Alexa Fluor 488 and 546 secondary antibodies (Molecular Probes, Eugene, OR), and samples were mounted in a solution of 50% glycerol, 2 mM MgCl2, and 2 μg/ml 4,6-diamidino-2-phenylindole (DAPI) in phosphate-buffered saline. Microscopy on fixed samples, as well as green fluorescent protein (GFP)-labeled chromosomes was performed using an Olympus IX70 Deltavision microscope (Applied Precision, Issaquah, WA) with a 100× UPLANO 1.35 numerical aperture oil immersion lens. Images were collected a Photometrics CH350 digital camera, subsequently deconvolved using the Softworx program (Applied Precision), and processed using Photoshop (Adobe Systems, Mountain View, CA).
Cytological Examination of Anoxic and Nocodazole-treated Embryos
Anoxic embryos were prepared for cytological analysis in a manner identical to what was described previously (Padilla et al., 2002). Briefly, large populations of embryos were isolated from adults, exposed to anoxia for 24 h, and then immediately frozen on dry ice. For nocodazole treatment, embryos were obtained from hypochlorite-treated adults and added to drops of nocodazole solution on slides. Coverslips were placed on the embryos and pressed down to crush the eggshell and facilitate the entry of nocodazole into the embryo. Embryos were exposed to 40 μg/ml nocodazole for 5-20 min and then frozen on dry ice. Both anoxia and nocodazole-treated embryos were subsequently processed for immunostaining as described above.
RESULTS
Components of the C. elegans Kinetochore Dissociate from the Centromere at Prometaphase
To investigate whether the C. elegans kinetochore undergoes a structural reorganization during mitosis, we examined the localization of centromere and kinetochore components by immunofluorescence microscopy. For this work, we define proteins as being centromere components if they are constitutively associated with the chromosomes. This includes the centromeric histone HCP-3/CeCENP-A (Buchwitz et al., 1999), and HCP-6, a protein with homology to the condensin subunit CAP-D2 (Stear and Roth, 2002). In contrast, kinetochore proteins assemble onto the chromosomes, at the centromere, during prophase. These antigens include HCP-4/CeCENP-C (Moore and Roth, 2001; Oegema et al., 2001), HIM-10/CeNuf2 (Howe et al., 2001; Desai et al., 2003), SAN-1/CeMAD3 (Nystul et al., 2003), CeMCAK, CeBUB-1 (Oegema et al., 2001), as well as HCP-1, a protein related to the mammalian kinetochore protein CENP-F (Moore et al., 1999).
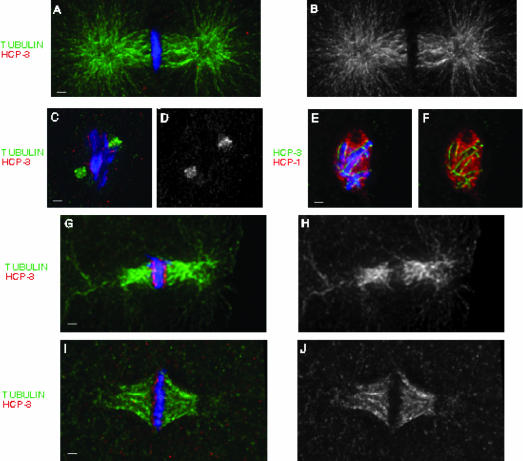
Due to the large number of proteins we examined, as well as similarities between their staining patterns, we provide the localization of the kinetochore protein HCP-1/CeCENP-F and the centromere component HCP-3/CeCENP-A as representative examples for the appearance of the C. elegans kinetochore and centromere (Figure 1). Consistent with previous reports, all of these antigens were present in two parallel lines on prophase chromosomes (Figure 1, a-c; our unpublished data), and a high degree of colocalization could be detected between centromere and kinetochore proteins (Buchwitz et al., 1999; Moore et al., 1999; Howe et al., 2001; Moore and Roth, 2001; Oegema et al., 2001; Stear and Roth, 2002; Desai et al., 2003). Furthermore, when metaphase plates were examined from a view perpendicular to the spindle axis (Supplemental Figure 1), all of the centromere and kinetochore components were present on the poleward faces of the metaphase plate, as had been previously shown (Figure 1, d-f; our unpublished data) (Buchwitz et al., 1999; Moore et al., 1999; Hoffman et al., 2001; Howe et al., 2001; Moore and Roth, 2001; Oegema et al., 2001; Stear and Roth, 2002; Desai et al., 2003; Nystul et al., 2003). Although these data indicate that components of the centromere and kinetochore are present within the same planes on the metaphase plate, this view does not allow us to visualize the distribution of centromere and kinetochore proteins on individual chromosomes.
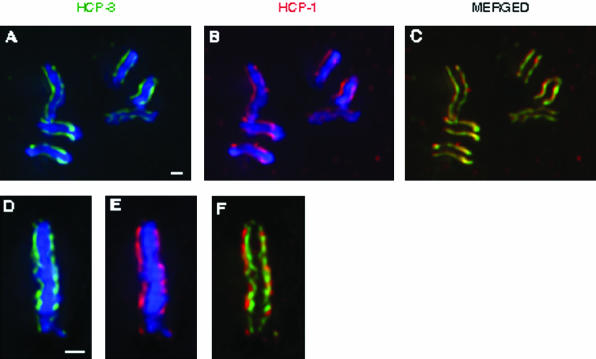
Figure 1.
Localization of centromere and kinetochore components at prophase and metaphase. Embryos were stained with antibodies against HCP-3/CeCENP-A (green) and HCP-1/CeCENP-F (red). DNA was visualized using DAPI (blue). These images were collected from one-cell embryos. At prophase, centromere and kinetochore proteins colocalize to parallel lines running the length of each chromosome (a-c). At metaphase, all known components of the centromere and kinetochore are present on the poleward faces of the metaphase plate, as seen from the perpendicular perspective (d-f). Bar, 1 μm.
To determine whether the colocalization that is observed between centromere and kinetochore proteins at prophase is maintained at metaphase, we examined metaphase plates from the centrosomal perspective (Supplemental Figure 1). This view enabled us to visualize chromosomes within the plane perpendicular to the spindle axis and directly observe the patterns of centromere and kinetochore staining on individual metaphase chromosomes. Consistent with past observations (Stear and Roth, 2002), centromere components such as HCP-3/CeCENP-A and HCP-6, as well as the kinetochore proteins HCP-4/CeCENP-C and MCAK, were all detectable as lines of reactivity that lay coincident with the metaphase chromosomes (Figure 2, a and d; our unpublished data). The fact that the localization of these proteins at metaphase is indistinguishable from their distribution on prophase chromosomes indicates that these centromere and kinetochore proteins do not undergo a detectable reorganization before metaphase.
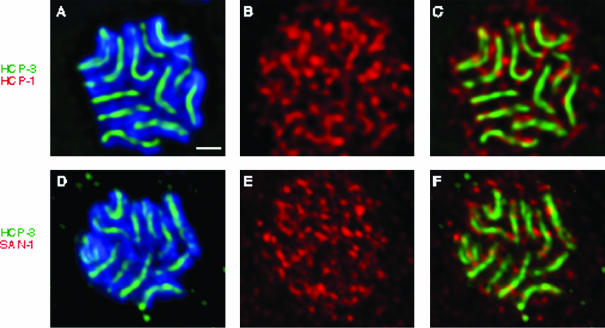
Figure 2.
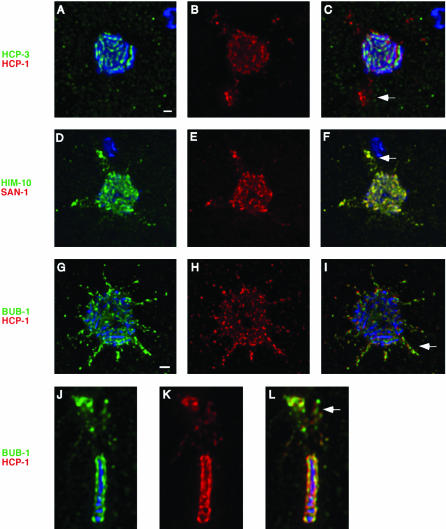
HCP-1/CeCENP-F and SAN-1/CeMAD3 dissociate from the centromere before metaphase. After staining with DAPI (blue), and antibodies against HCP-3/CeCENP-A (green), and either HCP-1/CeCENP-F (a-c) or SAN-1/CeMAD3 (d-f) (red), metaphase plates in four cell embryos were examined from the centrosomal perspective. Each image represents a 0.4-μm section through the metaphase plate in which both the centromere and the kinetochore are in focus. HCP-3/CeCENP-A is detectable as lines of staining coincident with the chromosomes (a and d), whereas HCP-1/CeCENP-F and SAN-1/CeMAD3 are present in a mesh-like network that is not coincident with the centromere (b and c, e and f). Bar, 1 μm.
If additional components of the C. elegans kinetochore undergo a structural reorganization during mitosis, it could be detectable as an alteration in the distribution of these kinetochore proteins relative to the centromere. Thus, using HCP-3/CeCENP-A as a marker for the centromere, we investigated whether any kinetochore proteins exhibited a staining pattern at metaphase that was not consistent with centromere localization. We found that two previously identified kinetochore components, HCP-1/CeCENP-F and SAN-1/CeMAD3 (Moore et al., 1999; Nystul et al., 2003), exhibited a staining pattern that did not coincide with the centromere (Figure 2, b and c, e and f). These proteins, both of which localize to the centromere at prophase, were detectable as a mesh-like network that lay on each face of the metaphase plate. It seems probable that this reorganization occurs during prometaphase, similar to what has previously been observed in mammalian cells. Consistent with this idea, when we examined prometaphase chromosomes stained with α-HCP-3 and α-HCP-1 antibodies (Figure 3, a-c), we observed several chromosomes in which lines of HCP-1/CeCENP-F were associated with the centromeres (Figure 3c, arrows). However, a great deal of HCP-1/CeCENP-F staining also had dissociated from the centromeres and was detectable as a diffuse cloud surrounding the mitotic chromosomes (Figure 3b). These pictures are likely to represent an intermediate stage in the dissociation of HCP-1/CeCENP-F from the centromere, suggesting that the reorganization of the kinetochore occurs during prometaphase, before the assembly of a metaphase plate.
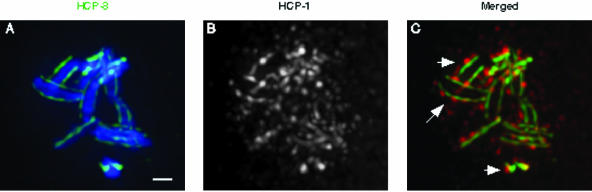
Figure 3.
HCP-1/CeCENP-F dissociates from the centromere during prometaphase. A prometaphase nucleus from a four-cell embryo was stained with DAPI (blue) and α-HCP-3 (green) and α-HCP-1 (white [b] or red [c]) antibodies. At this stage in mitosis, fully resolved centromeres can be observed on all the chromosomes (a). HCP-1 has maintained associations with some of the centromeres (arrows in c), but it is also present in a diffuse sphere around the chromosomes (b and c), indicative of an intermediate stage in its reorganization away from the centromere. Bar, 1 μm.
To determine whether other kinetochore proteins display similar patterns of localization at metaphase, we examined the localization of HIM-10/CeNUF2 or BUB-1 (Howe et al., 2001; Oegema et al., 2001; Desai et al., 2003), both of which had been previously characterized as components of the C. elegans kinetochore. The staining pattern of these antigens on the metaphase plate was very similar to that of both HCP-1/CeCENP-F and SAN-1/CeMAD3 (Figure 4, a-f). Instead of the tight, nonoverlapping lines observed for centromere proteins, HIM-10/CeNUF2 and BUB-1 seemed to assemble into a more diffuse net (Figure 4, a and d) and exhibited partial colocalization with HCP-1/CeCENP-F (Figure 4, c and f). These data lead us to conclude that like HCP-1/CeCENP-F and SAN-1/CeMAD3, HIM-10/CeNUF2 and BUB-1 also dissociate from the centromere before metaphase, yet remain associated with the metaphase plate.
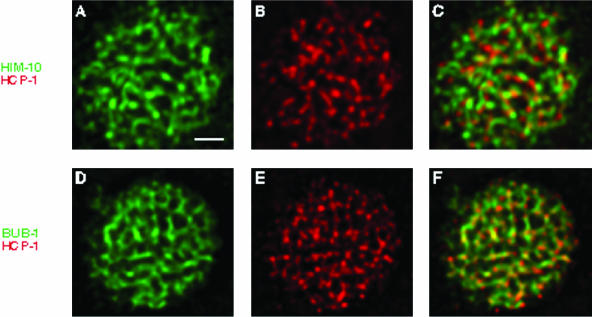
Figure 4.
HIM-10/CeNUF2 and BUB-1 exhibit localization patterns similar to HCP-1/CeCENP-F. Metaphase plates from four cell embryos were stained with antibodies against HCP-1/CeCENP-F (b and c and e and f) (red), and either HIM-10/CeNUF2 (a-c) or BUB-1 (d-f) (green). The staining patterns observed for HIM-10/CeNUF2 and BUB-1 are similar to those described for HCP-1/CeCENP-F and SAN-1/CeMAD3. Furthermore, costaining experiments demonstrate that HIM-10/CeNUF2 and BUB-1 exhibit partial colocalization with the HCP-1/CeCENP-F protein. Bar, 1 μm.
The Role of Microtubules in Mediating Kinetochore Reorganization
Because the dissociation of these kinetochore proteins from the C. elegans centromere coincides with the establishment of chromosome/microtubule attachments, we wanted to investigate whether spindle microtubules are required for this reorganization. To address this question, we treated C. elegans with the microtubule depolymerizing agent nocodazole and asked whether the kinetochore remained associated with the centromere in the absence of spindle microtubules. After a prolonged exposure of embryos to nocodazole (20 min at 40 μg/ml), we were able to identify mitotic blastomeres with no polymerized microtubules (Figure 5, a-d). We demonstrated that these cells had reached later stages of mitosis by the absence of staining with mAb414, a marker for the nuclear envelope (our unpublished data). Lacking spindle microtubules, the chromosomes aggregated in a disorganized clump in the center of the cell, but still exhibited a normal distribution of HCP-3/CeCENP-A (Figure 5, c-f). To examine whether kinetochore proteins are able to reorganize in the absence of microtubule interactions, we stained with antibodies against HCP-1/CeCENP-F. This antigen was present in a mesh-like network that was tightly associated with the chromosomes, but very distinct from the centromere (Figure 5, e and f). These results lead us to conclude that the reorganization of the C. elegans kinetochore away from the centromere at prometaphase is not dependent on the presence of microtubules, and does not merely represent the migration of these kinetchore proteins along the mitotic spindle.
Figure 5.
The response of C. elegans embryos to nocodazole and anoxia treatment. Wild-type embryos were stained with DAPI (blue), and α-tubulin (green) and α-HCP-3 (red) antibodies before (a and b) or after (c and d) a 20-min exposure to 40 μg/ml nocodazole. This treatment completely disrupted the microtubule network within the cell and enabled us to examine whether the C. elegans reorganizes in the absence of spindle microtubules. Subsequent staining of these embryos with antibodies against HCP-3/CeCENP-A (green) and HCP-1/CeCENP-F (red) (e and f) revealed that the kinetochore reorganizes under these conditions, indicating that this process occurs in a microtubule-independent manner. We also examined the effects of anoxia (g and h) or shorter exposure to 40 μg/ml nocodazole (10 min) (i and j) on the mitotic spindle. After these treatments, embryos were stained with DAPI (blue), and α-tubulin (green) and α-HCP-3 (red) antibodies. In both cases, the array of astral microtubules was severely disrupted, but the mitotic looked relatively normal. Bar, 1 μm.
Reorganization of the C. elegans Kinetochore in Response to Checkpoint Activation
Activation of the spindle checkpoint induces alterations in the mammalian kinetochore that are distinct from those that occur after microtubule attachment (Thrower et al., 1996; Martinez-Exposito et al., 1999; Hoffman et al., 2001). To determine whether the C. elegans kinetochore exhibits a similar distinction, we examined the distribution of kinetochore proteins after activating the spindle checkpoint. Two independent methods were used to induce the checkpoint in C. elegans embryos. First, similar to the previous experiment, we incubated early embryos in nocodazole, which has been shown to activate a checkpoint response in C. elegans (Nystul et al., 2003). After a short exposure to nocodazole (10 min at 40 μg/ml), we observed mitotic blastomeres that contained no astral microtubules, indicating that nocodazole had entered the cell and promoted microtubule depolymerization. However, these cells contained a robust array of spindle microtubules and the chromosomes had organized into a metaphase plate that looked relatively normal (Figure 5, i and j). Second, we exposed embryos to an anoxic environment for 24 h, which has recently been described as an alternate mechanism for activating the checkpoint pathway (Nystul et al., 2003). In this case as well, we observed mitotic cells containing no astral microtubules, but with a well-formed mitotic spindle and chromosomes that had aligned into a metaphase plate (Figure 5, g and h). Both of these conditions are sufficient to activate the spindle checkpoint (Nystul et al., 2003), so we examined the metaphase plates in these cells to determine whether there were alterations in kinetochore structure.
In these embryos, HCP-3/CeCENP-A was detectable as tight lines of centromere staining associated with each chromosome, identical to what is normally seen at metaphase (Figure 6a). In contrast, HCP-1/CeCENP-F exhibited a dramatic reorganization under these conditions. Although the majority of the HCP-1/CeCENP-F protein localized to the poleward face of the metaphase plate, as described above, a significant fraction formed spike-like protrusions that projected out laterally from the metaphase plate (Figure 6, b and c, nocodazole-treated embryo). These bodies, which we refer to as “flares,” constitute a major rearrangement of the C. elegans kinetochore in response to checkpoint activation. Although flares were never detected under normal conditions (metaphases in four cell embryos, n = 77), they were present at a very high frequency in both nocodazole-(65%, n = 25) and anoxia (92%, n = 37)-treated embryos. This pattern of kinetochore staining is also evident when metaphase plates are viewed from the perpendicular perspective (Figure 6, j-l, nocodazole-treated embryo). Finally, it should be noted that embryos exhibited complete survival after treatment with either nocodazole or anoxia, indicating that the observed kinetochore reorganizations are not detrimental to embryonic viability.
Figure 6.
The C. elegans kinetochore exhibits a distinct reorganization in response to checkpoint stimuli. After exposure to either anoxia (a-f, j-l) or nocodazole (g-i), embryos were stained with antibodies against HCP-3/CeCENP-A (a-c), HIM-10/CeNUF2 (d-f), and BUB-1 (g-l) (green), as well as HCP-1/CeCENP-F (a-c, g-l) and SAN-1/CeMAD3 (d-f) (red). Metaphase plates in four cell embryos were examined to determine the effect that checkpoint activation would have on kinetochore organization. The centromere did not change in response to this treatment as seen by HCP-3/CeCENP-A staining (a). However, the kinetochore underwent significant rearrangements after exposure to either nocodazole or anoxia. This was characterized by the localization of all of the kinetochore proteins in large flares projecting out from the metaphase plate (b-i). The flares also could be detected when metaphase plates were observed from the perpendicular perspective (j-l), appearing as aggregates of staining lying above or below the metaphase plate. White arrows in the merged panels highlight the location of the flares. Bar, 1 μm.
Because HIM-10/CeNUF2, SAN-1/CeMAD3, and BUB-1 all reorganize in a manner analogous to HCP-1/CeCENP-F at prometaphase, we predicted that these proteins would display a similar rearrangement in checkpoint activated embryos. Like HCP-1/CeCENP-F, in checkpoint-activated embryos, these three kinetochore components localized to flares projecting out from the metaphase plate (Figure 6, d-f, nocodazole-treated embryo; g-i, anoxic embryo). The frequency of these flares was very similar to that observed for HCP-1/CeCENP-F, both in anoxia and nocodazole (our unpublished data). Furthermore, by staining with combinations of antibodies, we demonstrated that multiple kinetochore proteins localize to each flare. That HCP-1/CeCENP-F, HIM-10/CeNUF2, SAN-1/CeMAD3, and BUB-1 exhibit the same patterns of reorganization in response to checkpoint activation highlights the dramatic nature of kinetochore reorganization in response to checkpoint stimuli. Furthermore, the fact that both san-1(RNAi) and him-10(e1511ts) embryos exhibit increased sensitivity to anoxia-induced checkpoint activation (Nystul et al., 2003; our unpublished data), suggest that these reorganizations may play a role in mediating the checkpoint response.
hcp-1/hcp-2(RNAi) and him-10(RNAi) Embryos Exhibit Defects at Metaphase
Several of the kinetochore proteins that we observed reorganizing during prometaphase and in response to checkpoint activation play an important role in ensuring accurate chromosome segregation during mitosis. Both hcp-1/hcp-2(RNAi) and him-10(RNAi) embryos exhibit severe missegregation phenotypes (Moore et al., 1999; Howe et al., 2001), and the lethality observed in bub-1(RNAi) embryos seems to correlate with subtle defects in chromosome segregation (our unpublished data). If the mitotic defects observed in these backgrounds exhibited a common feature, it might suggest a shared function for these proteins and provide insight into the significance of their reorganization at prometaphase. To investigate this question, we examined the phenotypes of hcp-1/hcp-2(RNAi) and him-10(RNAi) embryos, focusing in particular on chromosome-related defects.
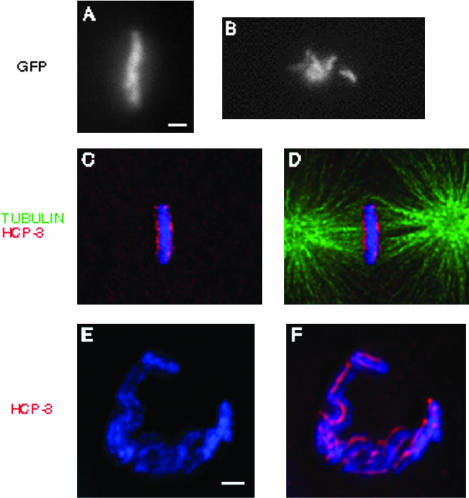
To better characterize the chromosome segregation phenotype in hcp-1/hcp-2(RNAi) embryos, we took four-dimensional movies of chromosome dynamics during the first cleavage division by using a transgenic strain containing a GFP::histone H2B fusion protein. In wild-type embryos, the chromosomes congress to the center of the cell and organize into a metaphase plate (Supplemental Figure 2 and Figure 7a). However, in hcp-1/hcp-2(RNAi) embryos, the chromosomes moved about the cell, indicating that they were interacting with the spindle, but they never align to form a metaphase plate (Figure 7b). As a result, they never reached anaphase and eventually segregate into the daughter cells in an extremely disorganized manner (Supplemental Figure 2; Moore et al., 1999). Severe congression defects also were observed when hcp-1/hcp-2(RNAi) embryos were examined by immunofluorescence microscopy. Although 11% of wild-type one-cell embryos were in metaphase (n = 382), <2% of the RNAi embryos achieved this stage of mitosis (n = 93). The chromosomes became mono-oriented with respect to the mitotic spindle and ended up amassed around the poles. These experiments also revealed that additional kinetochore proteins, including HIM-10, MCAK, and BUB-1 were targeted normally to hcp-1/hcp-2(RNAi) chromosomes (Supplemental Figure 2; our unpublished data), indicating that these phenotypes cannot be attributed to a global defect in kinetochore assembly. We found it interesting that the primary defect seen in hcp-1/hcp-2(RNAi) embryos occurs at the prometaphase/metaphase transition, when the HCP-1/CeCENP-F protein normally dissociates from the centromere.
Figure 7.
hcp1/hcp2(RNAi) and him-10(RNAi) embryos exhibit defects at metaphase. In the histone H2B::GFP background, wild-type (a) and hcp1/hcp2(RNAi) (b) chromosomes were visualized in one cell embryos shortly after pronuclear fusion. We filmed the progression of these embryos from prometaphase to anaphase (see Supplemental Figure 2), and the frames shown here represent the maximum level of congression observed in these backgrounds. Whereas wild-type chromosomes congress to the center of the cell and align properly at metaphase (a), hcp1/hcp2(RNAi) chromosomes fail to form a metaphase plate. him-10(RNAi) (c-f) embryos were stained with DAPI (blue), α-tubulin (green), and α-HCP-3 (red). Whereas him-10(RNAi) chromosomes are able to congress to the center of the cell, as seen from the perpendicular perspective (c and d), they do not organize into a proper metaphase plate, as seen from the centrosomal perspective (e and f). Instead of the solid plate observed in wild-type (see Figure 2), the chromosomes assemble into an open, horseshoe-shaped structure that is indicative of a failure to organize metaphase chromosomes. Bar, 1 μm.
Although him-10(RNAi) chromosomes migrate to the center of the cell at metaphase, they exhibit distinct defects in their ability to organize properly. As seen from the perpendicular perspective, the chromosomes seemed to assemble into a relatively normal metaphase plate. Markers for the centromere and kinetochore, including HCP-3/CeCENP-A, HCP-4, HCP-6, MCAK, and BUB-1 often localized to the poleward faces of the metaphase chromosomes, and the mitotic spindle seemed robust (Figure 7, c and d; our unpublished data). However, as seen from the centrosomal perspective, him-10(RNAi) chromosomes assemble into an open, horseshoe-shaped structure, instead of the circular pattern observed in wild type. The chromosomes are concentrated together around the rim of the horseshoe and seem to overlap and often lie atop of one another (Figure 7, e and f). Eighty-six percent of the him-10(RNAi) metaphase plates (n = 21) examined from this perspective exhibited this defect, which represents a significant failure to organize chromosomes at metaphase. Closer examination of these metaphase structures also revealed a dramatic decrease in the levels of HCP-1 staining (Supplemental Figure 3), indicating that him-10(RNAi) chromosomes are unable to recruit the full complement of kinetochore proteins. These embryos also display a variety of anaphase defects, presumably related to the inability of him-10(RNAi) chromosomes to assemble into a proper metaphase plate (our unpublished data; Howe et al., 2001). The fact that both hcp-1/hcp-2(RNAi) and him-10(RNAi) embryos exhibit a defect in properly aligning their chromosomes before metaphase raises the possibility that the reorganization of these proteins at prometaphase facilitates the formation of a metaphase plate.
DISCUSSION
In this work, we show that several C. elegans kinetochore proteins dissociate from the centromere during prometaphase. Thus, the C. elegans kinetochore, like the mammalian kinetochore, undergoes a significant reorganization during the transition from prophase to metaphase. The fact that such a rearrangement is conserved throughout evolution suggests that this process may represent an important functional aspect of metazoan kinetochores. The gene products described here, as well as their homologues in other organisms, have been shown to regulate multiple facets of chromosome biology, including activation of the spindle checkpoint and organization of the metaphase plate. We hypothesize that the reorganization of these proteins plays a role in mediating these functions.
Flares and the Spindle Checkpoint
In addition to the kinetochore reorganization at prometaphase, we also have described a more extreme rearrangement in response to checkpoint stimuli. After exposure to either nocodazole or anoxia, HCP-1/CeCENP-F, HIM-10/CeNUF2, BUB-1, and SAN-1/CeMAD3 all localize to flares that project out from the metaphase plate. The changes in shape and increased size of the kinetochore under these conditions are reminiscent of the exaggerated ring kinetochores observed in mammalian cells after prolonged periods of nocodazole treatment (Thrower et al., 1996; Martinez-Exposito et al., 1999; Hoffman et al., 2001). It is possible that this reorganization is not a direct result of checkpoint activation, but rather arises in response to the mitotic arrest after checkpoint activation. However, we can observe flares after very short (<5-min) exposures to nocodazole (our unpublished data). Thus, the rapid formation of flares leads us to favor the possibility that this reorganization is a direct consequence of checkpoint activation.
If the rearrangement of these kinetochore proteins does contribute to checkpoint activation, we would expect that disrupting these gene products would result in an increased sensitivity to checkpoint stimuli. Consistent with this idea, it has previously been demonstrated that san-1(RNAi) embryos exhibit reduced viability compared with wild type when exposed to anoxic environments (Nystul et al., 2003). Similarly, we observe that a temperature-sensitive allele of him-10, him-10(e1511), also shows increased embryo lethality after anoxic treatment (our unpublished data). Disruption of either hcp-1 or hcp-2 does not result in a similar phenotype, although this may be due to the fact that these gene products play functionally redundant roles within the cell (Moore et al., 1999), and the checkpoint pathway, like the chromosome segregation machinery, may not require the functions of both HCP-1/CeCENP-F and HCP-2. Thus, we show a number of proteins that are important for mediating the spindle checkpoint pathway also undergo a dramatic reorganization after checkpoint activation. This correlation suggests a possible role for these reorganizations in modulating the spindle checkpoint.
Function of HCP-1/CeCENP-F, HCP-2, and HIM-10/CeNUF2 during Mitosis
Previous reports demonstrated that both hcp-1/hcp-2(RNAi) (Moore et al., 1999) and him-10(RNAi) (Howe et al., 2001; Desai et al., 2003) embryos exhibit defects in chromosome segregation. Our observation that HCP-1/CeCENP-F and HIM-10/CeNUF2 exhibited a similar pattern of reorganization at prometaphase prompted us to ask whether these proteins might play similar roles in regulating chromosome dynamics. We undertook a more detailed analysis of these defects and showed that in both cases the phenotypes center around a failure in organizing the chromosomes at metaphase. These results are consistent with previous work from other systems that describe congression defects after disruption of Nuf2 (DeLuca et al., 2002; McCleland et al., 2003). More recently, studies in C. elegans have reported that disrupting HIM-10/CeNUF2 produces disorganized metaphase plates (Desai et al., 2003). Our results, in conjunction with these data, indicate that HIM-10/CeNUF2 and HCP-1/CeCENP-F help establish the proper alignment of chromosomes at metaphase. We suggest that the observed reorganization of these proteins during prometaphase may play a role in this process.
For example, alterations in kinetochore structure could modulate the establishment of proper microtubule/chromosome attachment and promote the appropriate capture of chromosomes by the mitotic spindle. Supporting this model are previous studies that suggest HIM-10/CeNUF2 and its homologues mediate chromosome/microtubule attachment at the kinetochore (Howe et al., 2001; DeLuca et al., 2002; McCleland et al., 2003). Although both hcp-1/hcp-2(RNAi) and him-10(RNAi) embryos exhibited chromosomal behavior that is indicative of functional microtubule attachment (Supplemental Figures 2 and 3; our unpublished data), it is possible that these chromosomes have a decreased capacity for mediating kinetochore/microtubule attachment, leading to the defects in chromosome segregation that we observed. Similar phenotypes have been observed after disruption of CENP-E activity in mammalian cells (Schaar et al., 1997). If the inability of hcp-1/hcp-2(RNAi) and him-10(RNAi) embryos to assemble metaphase plates is caused by a defect in microtubule attachment, this phenotype cannot be attributed to a global defect in kinetochore assembly. Both hcp-1/hcp-2(RNAi) and him-10(RNAi) chromosomes are able to recruit other kinetochore proteins, such as HCP-4, MCAK, BUB-1, and in the case of hcp-1/hcp-2(RNAi), HIM-10/CeNUF2 (Supplemental Figures 2 and 3; our unpublished data). Although him-10(RNAi) chromosomes exhibit a defect in HCP-1/CeCENP-F assembly (Supplemental Figure 3), because disruption of hcp-1 function generates no phenotype, this fact alone cannot account for the metaphase defect we observe. Furthermore, as mentioned above, many aspects of chromosomal behavior are normal in hcp-1/hcp-2(RNAi) and him-10(RNAi) embryos, which would not be the case if these chromosomes were unable to assemble a functional kinetochore.
Another model to explain how the kinetochore reorganization could affect the alignment of chromosomes at metaphase centers around the idea that physical associations are established between neighboring chromosomes during mitosis. In many systems, preparations of prometaphase chromosomes are “sticky,” due to interconnections between the chromosomes (Braselton, 1971; Bokhari, 1980; Rieder et al., 1990). Manipulations of chromosomes in human cells have suggested that the entire complement of chromosomes is physically connected to one another at metaphase (Maniotis et al., 1997). Finally, studies of metaphase plates in several holocentric organisms have revealed physical connections between adjacent chromosomes on the metaphase plate (Braselton, 1971; Bokhari, 1980; Rieder et al., 1990; Gonzalez-Garcia et al., 1996). These connections have been proposed to assist in the organization of the chromosomes into the metaphase plate, as well as coordinating the movement of the chromosomes at anaphase (Rieder et al., 1990; Nagele et al., 1995; Maniotis et al., 1997). It is thus possible that the observed reorganization of the C. elegans kinetochore at prometaphase represents the establishment of physical connections between adjacent chromosomes and that these associations are required for the proper organization of chromosomes at metaphase.
Supplementary Material
Acknowledgments
We thank Sue Biggins, Brian Buchwitz, Dana Miller, and especially Jesse Goldmark for critical reading of this manuscript. We thank Tony Hyman for the generous gift of BUB-1 and MCAK antibodies as well as the use of laboratory space and equipment, and we acknowledge Barbara Meyer for providing the HIM-10 antibody. Some of the strains used here were provided by the Caenorhabditis Genetics Center. This work was supported by National Institutes of Health grant R01GM-48435 to M.B.R.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04-06-0486. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-06-0486.
The online version of this article contains supplementary material accessible through http://www.molbiolcell.org.
References
- Albertson, D.G., and Thomson, J.N. (1982). The kinetochores of Caenorhabditis elegans. Chromosoma 86, 409-428. [DOI] [PubMed] [Google Scholar]
- Bokhari, M.B.E.G.F.S. (1980). The ultrastructure of the diffuse kinetochore in Luzula nivea. Chromosoma 79, 125-136. [Google Scholar]
- Braselton, J. (1971). The ultrastructure of the non-localized kinetochore of Luzula and Cyperus. Chromosoma 36, 89-99. [Google Scholar]
- Buchwitz, B.J., Ahmad, K., Moore, L.L., Roth, M.B., and Henikoff, S. (1999). A histone-H3-like protein in C. elegans. Nature 401, 547-548. [DOI] [PubMed] [Google Scholar]
- Cassimeris, L., Rieder, C.L., Rupp, G., and Salmon, E.D. (1990). Stability of microtubule attachment to metaphase kinetochores in PtK1 cells. J. Cell Sci. 96, 9-15. [DOI] [PubMed] [Google Scholar]
- Chen, R.H., Waters, J.C., Salmon, E.D., and Murray, A.W. (1996). Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science 274, 242-246. [DOI] [PubMed] [Google Scholar]
- Cleveland, D.W., Mao, Y., and Sullivan, K.F. (2003). Centromeres and kinetochores. From epigenetics to mitotic checkpoint signaling. Cell 112, 407-421. [DOI] [PubMed] [Google Scholar]
- DeLuca, J.G., Moree, B., Hickey, J.M., Kilmartin, J.V., and Salmon, E.D. (2002). hNuf2 inhibition blocks stable kinetochore-microtubule attachment and induces mitotic cell death in HeLa cells. J. Cell Biol. 159, 549-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg, A.F. (2001). Here, there, and everywhere: kinetochore function on holocentric chromosomes. J. Cell Biol. 153, F33-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai, A., Rybina, S., Muller-Reichert, T., Shevchenko, A., Hyman, A., and Oegema, K. (2003). KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev. 17, 2421-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire, A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E., and Mello, C.C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806-811. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Garcia, J.M., Benavente, R., and Rufas, J.S. (1996). Cytochemical and immunocytochemical characterization of kinetochores in the holocentric chromosomes of Graphosoma italicum. Eur. J. Cell Biol. 70, 352-360. [PubMed] [Google Scholar]
- Hoffman, D.B., Pearson, C.G., Yen, T.J., Howell, B.J., and Salmon, E.D. (2001). Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol. Biol. Cell 12, 1995-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe, M., McDonald, K.L., Albertson, D.G., and Meyer, B.J. (2001). HIM-10 is required for kinetochore structure and function on Caenorhabditis elegans holocentric chromosomes. J. Cell Biol. 153, 1227-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonski, S.A., Chan, G.K., Cooke, C.A., Earnshaw, W.C., and Yen, T.J. (1998). The hBUB1 and hBUBR1 kinases sequentially assemble onto kinetochores during prophase with hBUBR1 concentrating at the kinetochore plates in mitosis. Chromosoma 107, 386-396. [DOI] [PubMed] [Google Scholar]
- King, J.M., Hays, T.S., and Nicklas, R.B. (2000). Dynein is a transient kinetochore component whose binding is regulated by microtubule attachment, not tension. J. Cell Biol. 151, 739-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa, R., and Rose, A.M. (1999). Components of the spindle-assembly checkpoint are essential in Caenorhabditis elegans. Nat. Cell Biol. 1, 514-521. [DOI] [PubMed] [Google Scholar]
- Li, Y., and Benezra, R. (1996). Identification of a human mitotic checkpoint gene: hsMAD2. Science 274, 246-248. [DOI] [PubMed] [Google Scholar]
- Maney, T., Hunter, A.W., Wagenbach, M., and Wordeman, L. (1998). Mitotic centromere-associated kinesin is important for anaphase chromosome segregation. J. Cell Biol. 142, 787-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniotis, A.J., Bojanowski, K., and Ingber, D.E. (1997). Mechanical continuity and reversible chromosome disassembly within intact genomes removed from living cells. J. Cell Biochem. 65, 114-130. [PubMed] [Google Scholar]
- Martinez-Exposito, M.J., Kaplan, K.B., Copeland, J., and Sorger, P.K. (1999). Retention of the BUB3 checkpoint protein on lagging chromosomes. Proc. Natl. Acad. Sci. USA 96, 8493-8498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleland, M.L., Gardner, R.D., Kallio, M.J., Daum, J.R., Gorbsky, G.J., Burke, D.J., and Stukenberg, P.T. (2003). The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev. 17, 101-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, L.L., Morrison, M., and Roth, M.B. (1999). HCP-1, a protein involved in chromosome segregation, is localized to the centromere of mitotic chromosomes in Caenorhabditis elegans. J. Cell Biol. 147, 471-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, L.L., and Roth, M.B. (2001). HCP-4, a CENP-C-like protein in Caenorhabditis elegans, is required for resolution of sister centromeres. J. Cell Biol. 153, 1199-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio, A., and Hardwick, K.G. (2002). The spindle checkpoint: structural insights into dynamic signalling. Nat. Rev. Mol. Cell. Biol. 3, 731-741. [DOI] [PubMed] [Google Scholar]
- Nagele, R., Freeman, T., McMorrow, L., and Lee, H.Y. (1995). Precise spatial positioning of chromosomes during prometaphase: evidence for chromosomal order. Science 270, 1831-1835. [DOI] [PubMed] [Google Scholar]
- Nystul, T.G., Goldmark, J.P., Padilla, P.A., and Roth, M.B. (2003). Suspended animation in C. elegans requires the spindle checkpoint. Science 302, 1038-1041. [DOI] [PubMed] [Google Scholar]
- Oegema, K., Desai, A., Rybina, S., Kirkham, M., and Hyman, A.A. (2001). Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 153, 1209-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla, P.A., Nystul, T.G., Zager, R.A., Johnson, A.C., and Roth, M.B. (2002). Dephosphorylation of cell cycle-regulated proteins correlates with anoxia-induced suspended animation in Caenorhabditis elegans. Mol. Biol. Cell 13, 1473-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson, C.G., Maddox, P.S., Zarzar, T.R., Salmon, E.D., and Bloom, K. (2003). Yeast kinetochores do not stabilize Stu2p-dependent spindle microtubule dynamics. Mol. Biol. Cell 14, 4181-4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder, C.L., Bowser, S.S., Cole, R., Rupp, G., Peterson, A., and Alexander, S.P. (1990). Diffuse kinetochores and holokinetic anaphase chromatin movement during mitosis in the hemipteran Agallia constricta (leafhopper) cell line AC-20. Cell Motil. Cytoskeleton 15, 245-259. [DOI] [PubMed] [Google Scholar]
- Rieder, C.L., and Salmon, E.D. (1998). The vertebrate cell kinetochore and its roles during mitosis. Trends Cell Biol. 8, 310-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaar, B.T., Chan, G.K., Maddox, P., Salmon, E.D., and Yen, T.J. (1997). CENP-E function at kinetochores is essential for chromosome alignment. J. Cell Biol. 139, 1373-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp-Baker, H., and Chen, R.H. (2001). Spindle checkpoint protein Bub1 is required for kinetochore localization of Mad1, Mad2, Bub3, and CENP-E, independently of its kinase activity. J. Cell Biol. 153, 1239-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stear, J.H., and Roth, M.B. (2002). Characterization of HCP-6, a C. elegans protein required to prevent chromosome twisting and merotelic attachment. Genes Dev. 16, 1498-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, S.S., Ha, E., and McKeon, F. (1998). The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J. Cell Biol. 142, 1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrower, D.A., Jordan, M.A., and Wilson, L. (1996). Modulation of CENP-E organization at kinetochores by spindle microtubule attachment. Cell Motil. Cytoskeleton. 35, 121-133. [DOI] [PubMed] [Google Scholar]
- Waters, J.C., Chen, R.H., Murray, A.W., and Salmon, E.D. (1998). Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J. Cell Biol. 141, 1181-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, X., Anderson, K.L., and Cleveland, D.W. (1997). The microtubule-dependent motor centromere-associated protein E (CENP-E) is an integral component of kinetochore corona fibers that link centromeres to spindle microtubules. J. Cell Biol. 139, 435-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkowski, R.P., Meyne, J., and Brinkley, B.R. (1991). The centromere-kinetochore complex: a repeat subunit model. J. Cell Biol. 113, 1091-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.