Abstract
Lipoxygenases are a class of dioxygenases that form hydroperoxy fatty acids with distinct positional and stereo configurations. Several amino acid residues influencing regiospecificity have been identified, whereas the basis of stereocontrol is not understood. We have now identified a single residue in the lipoxygenase catalytic domain that is important for stereocontrol; it is conserved as an Ala in S lipoxygenases and a Gly in R lipoxygenases. Our results with mutation of the conserved Ala to Gly in two S lipoxygenases (mouse 8S-LOX and human 15-LOX-2) and the corresponding Gly–Ala substitution in two R lipoxygenases (human 12R-LOX and coral 8R-LOX) reveal that the basis for R or S stereo-control also involves a switch in the position of oxygenation on the substrate. After the initial hydrogen abstraction, antarafacial oxygenation at one end or the other of the activated pair of double bonds (pentadiene) gives, for example, 8S or 12R product. The Ala residue promotes oxygenation on the reactive pentadiene at the end deep in the substrate binding pocket and S stereochemistry of the product hydroperoxide, and a Gly residue promotes oxygenation at the proximal end of the reactive pentadiene resulting in R stereochemistry. A model of lipoxygenase reaction specificity is proposed in which product regiochemistry and stereochemistry are determined by fixed relationships between substrate orientation, hydrogen abstraction, and the Gly or Ala residue we have identified.
Lipoxygenases are a class of nonheme iron oxygenases that catalyze the conversion of arachidonic acid (AA) and other polyunsaturated fatty acids to their hydroperoxy derivatives (1, 2). The products are involved in a series of biological events such as inflammation (3, 4), cell development, and differentiation (5, 6). Lipoxygenase isoforms of differing regiospecificity and stereospecificity are widespread in both the animal (7, 8) and plant kingdoms (9–11).
Lipoxygenase regiospecificity has been studied extensively, and there is now experimental evidence to support the control of substrate binding as critical to positional specificity (12). Both the depth of substrate entry into the catalytic domain of the protein and the head-to-tail orientation of the substrate are known to be important. The difference in positional specificity of 12S-LOX and 15S-LOX, for example, is due to a “frame shift” of the substrate, where the position of oxygenation is determined by how deeply the substrate enters into the active site (13–15). Positional specificity is also determined by the substrate orientation, whereby different oxygenation products are formed if the substrate enters the active site with the carboxylic or the methyl end first (e.g., 8S-LOX, 15S-LOX) (16–19). All these changes have been demonstrated with S lipoxygenases, and in these experiments the oxygenation remained S-specific with the change in positional specificity. The mechanism controlling the R or S stereospecificity in lipoxygenases is less understood, and although a model has been proposed to explain how structurally related enzymes can form products with different stereoconfiguration (20), no experimental data are available to support the proposed mechanisms (21).
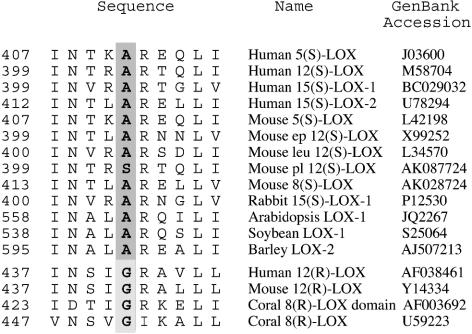
R and S lipoxygenases share a high degree of sequence identity in their primary structure (≤50%), and the percent identity is even higher in the active site. In addition, the iron ligands are well conserved in lipoxygenases, suggesting that there is no major difference in the overall architecture of the active site of R and S lipoxygenases. In investigation of the basis of R and S specificity, our initial experiments, where helices in the region of the active site of the human 15-LOX-2 were swapped with those from the human 12R-LOX, resulted in either a loss of catalytic activity or no change in stereospecificity (unpublished data). As an alternative approach, we searched for conserved differences between R and S lipoxygenases in residues in and around the active site. This search resulted in identification of a candidate residue conserved as an Ala in S lipoxygenases and Gly in R lipoxygenases (Fig. 1). The aim of this study was to test the effect of mutations of this residue to bulkier or smaller residues on a number of R and S lipoxygenases and observe the effect of these changes on the stereospecificity and regiospecificity of the lipoxygenase enzymes and determine whether the Ala/Gly residue is a stereodeterminant in lipoxygenase catalysis.
Fig. 1.
Alignment of R and S lipoxygenases: identification of a potential stereodeterminant. A representative selection of S lipoxygenase enzymes are aligned with all of the R lipoxygenases of known primary structure. The highlighted residue is conserved as an Ala in all of the S lipoxygenases except for the mouse platelet 12S-LOX, where it is replaced by Ser. The equivalent residue in R lipoxygenase is conserved as Gly. Ep 12S-LOX, epidermal-type 12-LOX; leu 12S-LOX, leukocyte-type 12-LOX; pl 12S-LOX, platelet-type 12-LOX; coral 8R-LOX domain, the lipoxygenase domain of the peroxidase-lipoxygenase fusion protein of Plexaura homomalla (26).
Experimental Procedures
Site-Directed Mutagenesis. Site-directed mutagenesis of lipoxygenase enzymes was performed by using the QuikChange site-directed mutagenesis kit (Stratagene) with overlapping mismatching oligonucleotides as primers designed according to the manufacturer's instructions. The His-tagged human 12R-LOX (in the pCW expression vector), coral 8R-LOX [in the pET3a (Novagen) expression vector], human 15-LOX-2 [in the pET3a (Novagen) expression vector], and the non-His-tagged mouse 8S in either the pcDNA3.1 (Invitrogen) or pET28 (Novagen) expression vectors were used as templates. PCRs were performed according to the manufacturer's instructions, and the product was analyzed on a 1.2% agarose gel stained with ethidium bromide. PCRs (1–5 μl) were used to transform Escherichia coli XL1-Blue cells (Stratagene). Correctly mutated clones were identified by sequencing.
Expression of Recombinant Lipoxygenases in E. coli. WT 8S-LOX, WT 15-LOX-2, WT 12R-LOX, WT 8R-LOX, and mutants were expressed in E. coli Novagen BL21 (DE3) as His6-tagged proteins and purified on nickel-nitrilotriacetic acid agarose (Qiagen, Valencia, CA) according to the manufacturer's instructions. Fractions collected from the affinity column were assayed by SDS/PAGE, and fractions containing recombinant lipoxygenases were dialyzed against 50 mM sodium phosphate (pH 7.0)/150 mM NaCl buffer containing 20% glycerol.
Expression and Assay System of Mouse 8S-LOX and Mutants in HeLa Cells. The cDNAs of the WT mouse 8S-LOX enzyme and the mutants in the expression vector pcDNA3.1 were expressed in HeLa cells as described in ref. 22. Harvested cells were washed with PBS and sonicated on ice for 5 s in 100 μl of 50 mM Tris buffer (pH 7.5)/150 mM NaCl, followed by the addition of [1-14C]AA (100 μM final concentration) and incubation at 37°C on a shaker for 45 min. Incubations were stopped by the addition of methanol, and metabolites were extracted by using the Bligh and Dyer method (23). Hydroperoxy derivatives were resuspended in methanol and reduced to hydroxy fatty acids by the addition of 10 μg of triphenylphosphine (TPP) before HPLC analysis.
Assay System for Human 15-LOX-2 Mutants. His-tagged purified recombinant 15-LOX-2 WT or mutants were incubated with [1-14C]AA (100 μM final concentration) in 100 μl of 50 mM Tris buffer (pH 7.5)/150 mM NaCl on a shaker at 37°C for 45 min. For some experiments 15-LOX-2 and the Ala-416 → Gly (Ala416Gly) mutant were incubated with 1-palmitoyl-2-arachidonoyl phosphatidylcholine (C16/AA-PC; 100 μM final concentration) in 100 μl of 150 mM NaCl/Tris buffer (pH 7.5)/0.5 mM deoxycholate, on a shaker at 37°C for 45 min. Incubations were stopped by the addition of methanol, and the products were extracted by using the Bligh and Dyer method (23). Hydroperoxy derivatives were resuspended in methanol and reduced to hydroxy fatty acids by the addition of 1 μl of TPP (10 mg/ml) before HPLC analysis. Samples incubated with C16/AA-PC were transmethylated with NaOMe before HPLC analysis (24).
Assay System for Human 12R-LOX Mutants. His-tagged purified recombinant 12R-LOX WT or mutants were incubated with [1-14C]AA (106 cpm) in 100 μl of 100 mM Mes buffer (pH 6)/200 mM NaCl on a shaker at 37°C for 45 min. Incubations were stopped by the addition of methanol, and the products were extracted by using the Bligh and Dyer method (23). Hydroperoxy derivatives were resuspended in methanol and reduced to hydroxy fatty acids by the addition of 1 μl of TPP (10 mg/ml) before HPLC analysis. To estimate the enzymatic activity of the human 12R-LOX and the Gly441Ala mutant, purified proteins were incubated at 37°C for 45 min with 10 μg of AA in 100 μl of 100 mM Mes buffer (pH 6)/200 mM NaCl. Incubations were stopped by the addition of methanol, and products were extracted by using the Bligh and Dyer method (23). Reaction products were quantified by using a PerkinElmer Lambda 35 UV-visible spectrophotometer.
Assay System for Coral 8R-LOX Mutants. Recombinant WT 8R-LOX and mutants were purified as His-tagged proteins and incubated in 100 μl of 50 mM Tris buffer at pH 8 containing 500 mM NaCl, 2 mM CaCl2, and 0.01% Emulphogen (Sigma) with [1-14C]AA for 5 min at room temperature. Incubations were stopped by the addition of methanol, and samples were reduced by addition of 1 μl of TTP (10 mg/ml) before HPLC analysis. For some experiments, 8R-LOX and the Gly416Ala mutant were incubated with C16/AA-PC (200 μM final concentration) in 100 μl of 50 mM Tris buffer (pH 8)/0.5 mM deoxycholate on a shaker at room temperature for 10 min. Incubations were stopped by the addition of methanol, and products were extracted by using the Bligh and Dyer method (23).
HPLC Analysis. Products of the reaction of lipoxygenase enzymes and mutants with [1-14C]AA were analyzed on an Agilent (Palo Alto, CA) 1100 HPLC equipped with a diode array detector connected online to a Flo-One A-100 radioactive detector (Radiomatic Instruments and Chemical, Meridian, CT). Metabolites were analyzed with a Whatman Partisil 5 silica column (0.46 × 25 cm) eluted at a flow rate of 1 ml/min with hexane/isopropyl alcohol/acetic acid (100:2:0.1, by volume) with UV detection at 235 nm. Metabolites from incubation with C16/AA-PC were transesterified by using NaOMe, and the methyl esters were analyzed on a Waters Symmetry C18 5-μm column by using a solvent of methanol/water/acetic acid (85:15:0.01, by volume) eluted at 1 ml/min with UV detection at 235 nm. In some cases, metabolites were analyzed by using a Waters Symmetry C18 5-μm column (0.46 × 25 cm) eluted at a flow rate of 1 ml/min with methanol/water/acetic acid (80:20:0.01, by volume) for 22 min and finally with methanol to elute unreacted AA with UV detection at 235 nm. Chiral analysis was performed on hydroxyeicosatetraenoic acid (HETE) methyl esters by using a Chiralpak AD (Daicel Chemical Industries, Tokyo) (0.46 × 25 cm) eluted at a flow rate of 1 ml/min with hexane/methanol (100:2, by volume), with UV detection at 235 nm (25).
Kinetic Analysis. Enzymatic activity of purified enzymes was determined by monitoring the increase of the signal at 235 nm in an UV-visible spectrophotometer with AA concentrations ranging from 0.5 to 100 μM. Rates of reaction were calculated from the initial linear part of the curve. The enzymatic assay for the human 15-LOX-2 and the Ala416Gly mutant was performed in 0.5 ml of 50 mM Tris (pH 7.5)/150 mM NaCl buffer. Assay of the mouse 8S-LOX and the Ala417Gly mutant used 0.5 ml of 50 mM Tris at pH 7.0 containing 150 mM NaCl and 0.03% Tween 20 buffer. The assay for the coral 8R-LOX and the Gly427Ala mutant was performed in 0.5 ml of 50 mM Tris (pH 8)/500 mM NaCl, with 0.2 μg of 8R-hydroperoxyeicosatetraenoic acid as hydroperoxide activator. Km and kcat values were calculated with the Michaelis–Menten equation.
Results
A review of conserved differences in the active site of lipoxygenases revealed one candidate amino acid conserved as an Ala in S lipoxygenases and as a Gly in R lipoxygenases (Fig. 1). We selected four enzymes to examine the importance of this residue in lipoxygenase stereospecificity. The two selected S lipoxygenases were the mouse 8S-LOX and the human 15-LOX-2; the two R lipoxygenases were an 8R-LOX from the coral Plexaura homomalla (26) and the human 12R-LOX. In each enzyme, we examined the Ala/Gly substitution and also the effects of replacement with Val, Ser, and Thr. The enzymes were incubated with AA, the products were analyzed by HPLC, and in most cases the rates of reaction were measured by UV spectroscopy. Individual HETE products were identified by their characteristic UV spectrum and cochromatography with authentic standards, and their stereochemistry was determined by chiral phase HPLC analysis.
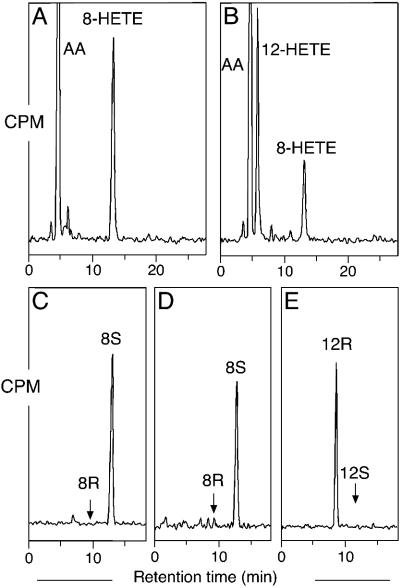
Mouse 8S-LOX Mutants. Lysates of HeLa cells expressing WT 8S-LOX converted [1-14C]AA to 8-hydroperoxyeicosatetraenoic acid (which was reduced to HETE for analysis) as the sole HETE metabolite, as shown by straight-phase (SP)-HPLC (Fig. 2A). Chiral analysis of the 8-HETE methyl ester showed, as expected, that the metabolite is exclusively the S enantiomer (Fig. 2C). The Ala417Gly mutant converted [1-14C]AA mainly to 12-HETE with lower amounts of 8-HETE (1.8:1 ratio) (Fig. 2B). Chiral HPLC analysis of the products showed that the 12-HETE had the R stereoconfiguration (>99%), whereas the 8-HETE retained S stereochemistry (>95%) (Fig. 2 D and E). Comparison of the rates of reaction of the WT and the Ala417Gly mutant by UV analysis of the increase in absorbance at 235 nm showed kcat of 18 min-1 for the WT enzyme and 12 min-1 for the mutant and Km values of 8 μM and 14 μM, respectively, together indicating that the mutant retained 38% of the catalytic efficiency (kcat/Km) of the WT 8S-LOX.
Fig. 2.
SP-HPLC and chiral HPLC analysis of products formed by WT and mutant mouse 8S-LOX. (A) SP-HPLC analysis for WT. (B) SP-HPLC analysis for Ala417Gly mutant. (C) Chiral HPLC analysis of 8-HETE-Me (8-HETE methyl ester) from WT. (D) Chiral HPLC analysis of 8-HETE-Me from Ala417Gly mutant. (E) Chiral HPLC analysis for 12-HETE-Me from Ala417Gly mutant. [1-14C]AA (100 μM) was incubated with equal amounts of HeLa cell homogenates expressing WT and mutant mouse 8S-LOX as described under Experimental Procedures. Products were analyzed by using a Whatman Partisil 5 silica column (0.46 × 25 cm) eluted at a flow rate of 1 ml/min with hexane/isopropyl alcohol/acetic acid (100:2:0.1, by volume). Individual metabolites purified by both RP-HPLC and SP-HPLC were analyzed on a Daicel Chiralpak AD (0.46 × 25 cm) eluted at a flow rate of 1 ml/min with hexane/methanol (100:2, by volume) (25).
When Ala-417 was mutated to Ser, the enzyme retained the same oxygenase specificity and similar catalytic activity to WT mouse 8S-LOX. Mutation of Ala-417 to the more space-filling Val or Thr residues resulted in complete loss of enzymatic activity (data not shown).
Human 15-LOX-2 Mutants. WT 15-LOX-2 converted [1-14C]AA exclusively to 15-HETE (Fig. 8A, which is published as supporting information on the PNAS web site), and chiral HPLC analysis of the product showed that there was 99% S stereoconfiguration (Fig. 8C). The Ala416Gly mutant (equivalent in position to Ala-417 of mouse 8S-LOX), converted [1-14C]AA to 11-HETE and 15-HETE in a 1.5:1 ratio (Fig. 8B). Chiral HPLC analysis of the products after methylation with diazomethane showed that 11-HETE had the R stereoconfiguration (>99%), and the 15-HETE had the S stereoconfiguration (94%) (Fig. 8 D and E). Rates of reactions for the WT and Ala416Gly mutants in two independent experiments gave kcat values of 14 min-1 and 3 min-1, respectively, with no significant change in Km (≈1 μM) and a 5-fold reduction in overall catalytic efficiency (kcat/Km) for the mutant enzyme.
As found for mouse 8S-LOX, the Ala-416/Ser mutation of human 15-LOX-2 gave the same product profile as the WT, with 15S-HETE as the only product (data not shown). Again, replacement of Ala-416 with the more space-filling Val or Thr resulted in complete loss of activity (data not shown).
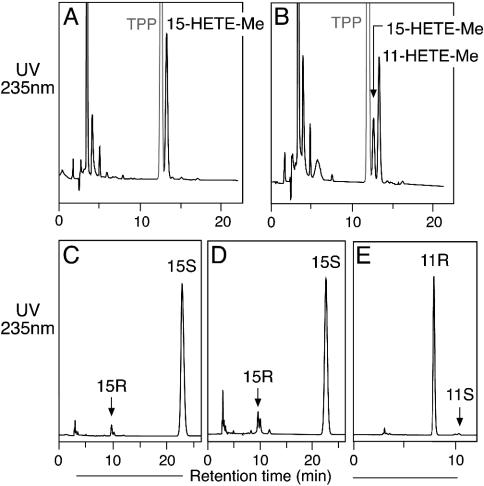
To investigate the orientation of substrate binding with 15-LOX-2, we examined the product profile obtained by using C16/AA-PC as a substrate. Because of the bulkiness of the phosphatidylcholine (PC) head group, the carbon chain of the esterified arachidonate can enter the active site only by a tail-first entry. The products of the incubation of WT 15-LOX-2 and the Ala416Gly mutant with C16/AA-PC were analyzed by RP-HPLC after reduction with TPP and transesterification to form the methyl ester derivatives. Analysis of the incubation with WT enzyme indicated that C16/AA-PC was metabolized exclusively to 15-HETE-PC (Fig. 3A), and, as expected, chiral analysis showed that the stereoconfiguration for 15-HETE was S (Fig. 3C). The Ala416Gly mutant formed both 11-HETE-PC and 15-HETE-PC (Fig. 3B) in the same 1.5:1 ratio as in the incubation with free AA, and chiral HPLC analysis established the stereoconfigurations as 11R and 15S (Fig. 3 D and E). This result establishes that both the 11R-HETE and 15S-HETE products are formed with the substrate entering the active site in the same tail-first orientation as in the WT 15-LOX-2.
Fig. 3.
Metabolism of C16/AA-PC substrate by WT and mutant 15-LOX-2. (A) RP-HPLC analysis of WT. (B) RP-HPLC analysis of Ala416Gly mutant. (C) Chiral HPLC of 15-HETE-Me from WT. (D) Chiral HPLC of 15-HETE-Me from Ala416Gly mutant. (E) Chiral HPLC of 11-HETE-Me from Ala416Gly mutant. Products from the incubation of C16/AA-PC (100 μM) with WT and Ala416Gly 15-LOX-2 were reduced with TPP, transesterified to the methyl esters, and analyzed on a Waters Symmetry C18 5-μm column by using a solvent of methanol/water/acetic acid (85:15:0.01, by volume) and eluting at 1 ml/min with UV detection at 235 nm. Chiral HPLC analysis was carried out as described in the legend to Fig. 2.
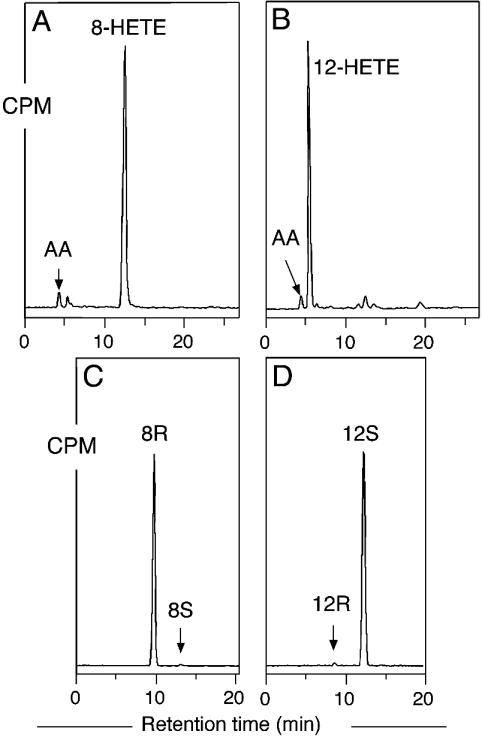
Coral 8R-LOX Mutants. On the order of 100 S-specific lipoxygenase sequences are reported to date, whereas there are only a few reported R lipoxygenases. One of the R lipoxygenases we selected for mutation is the 8R-LOX domain of the peroxidase-lipoxygenase fusion protein from the coral Plexaura homomalla (26, 27). SP-HPLC analysis of incubations of WT coral 8R-LOX with [1-14C]AA showed the formation of 8-HETE as the only metabolite (Fig. 4A), and, as expected, the stereoconfiguration was 8R (99%) (Fig. 4C). Mutation of Gly-427 to Ala resulted in an enzyme that converted [1-14C]AA almost exclusively to 12-HETE as shown by SP-HPLC analysis (Fig. 4B), and the 12-HETE was shown to have S stereochemistry (98%) (Fig. 4D). The kcat of the Gly427Ala mutant was reduced 5-fold compared with WT (30 vs. 140 s-1), and the calculated Km increased from 23 to 73 μM, together giving a reduction in kcat/Km of 15-fold for the mutant enzyme.
Fig. 4.
SP-HPLC and chiral HPLC analysis of products formed by WT and mutant coral 8R-LOX. (A) SP-HPLC analysis of WT. (B) SP-HPLC analysis of Gly427Ala mutant. (C) Chiral HPLC analysis of 8-HETE-Me from WT. (D) Chiral HPLC analysis of 12-HETE-Me from Gly427Ala mutant. [1-14C]AA (100 μM) was incubated with aliquots of WT and mutant coral 8R-LOX as described in Experimental Procedures. SP-HPLC and chiral HPLC analyses were carried out as described in the legend to Fig. 2.
In contrast to the other enzymes studied here, replacement of Gly-427 with Val resulted in the formation of a catalytically active enzyme. The Val mutant gave the same product profile as the Gly427Ala enzyme (data not shown). Substitution with the bulkier Thr gave an enzyme with weak but measurable catalytic activity; it converted AA mainly to 12-HETE with a small fraction of 8-HETE (7.4:1 ratio), as shown by both SP-HPLC and RP-HPLC analysis (data not shown). Chiral analysis of the methyl esters of the metabolites showed that the 12-HETE was mainly the S enantiomer (92%), and the minor 8-HETE was predominantly 8R (70%) (data not shown).
The coral 8R-LOX is the second of the two enzymes studied here that is predicted to react with AA esterified in PC (a topic covered further in Discussion). We confirmed that WT 8R-LOX converted C16/AA-PC to the 8R-hydroperoxyeicosatetraenoic acid PC ester, and the Gly427Ala mutant formed the 12S-hydroperoxyeicosatetraenoic acid derivative (for details see Supporting Text, which is published as supporting information on the PNAS web site).
Human 12R-LOX Mutants. The second R lipoxygenase we selected for mutation was the human 12R-LOX, where the equivalent Gly-441 was mutated to Ala, Val, Ser, and Thr. SP-HPLC analysis of the incubation of the purified His-tagged WT human 12R-LOX with [1-14C]AA showed the formation of 12R-HETE as the only HETE metabolite (Fig. 9 A and C, which is published as supporting information on the PNAS web site). The Gly441Ala mutant converted [1-14C]AA to both 8-HETE and 12-HETE (1.4:1 ratio) as shown by SP-HPLC analysis (Fig. 9B). Chiral HPLC analysis of the HETE methyl esters showed that the 8-HETE was formed mainly as the S enantiomer (91%), whereas the 12-HETE was mainly the R enantiomer (92%) (Fig. 9 D and E). The rates of reaction could not be reliably measured by direct monitoring of absorbance at 235 nm and, therefore, the percent of conversions to product was determined for the WT and mutant enzymes. By this criterion, the Gly441Ala mutant gave 26 ± 3% (SD, n = 3) conversion, indistinguishable from WT (25 ± 1%).
Mutation of Gly-441 to Ser had the same effect as the Ala substitution (data not shown), producing a mixture of 8S-HETE (major) and 12R-HETE (minor). Replacement of Gly-441 with either a Val or a Thr resulted in complete loss of enzymatic activity.
Discussion
Here we report the role of a single residue in controlling the stereochemistry of the reaction catalyzed by lipoxygenases. The residue we have identified is highly conserved as an Ala in S lipoxygenases and a Gly in all R lipoxygenases. The high conservation of this residue and the consistent results obtained by its mutation on four different lipoxygenases strongly suggests its role as a universal determinant of lipoxygenase stereospecificity. Among S-LOX enzymes, the one known exception to the conservation of Ala in the critical position is the natural occurrence of a Ser in mouse platelet-type 12S-LOX. The cDNA sequence of this mouse 12S-LOX was reported originally to have a Thr in the equivalent position (28), but this amino acid has been corrected to Ser in a more recent database entry (GenBank accession number AK087724). In our mutagenesis experiments, we found that changing WT Ala to Ser in mouse 8S-LOX or 15S-LOX had no effect on the product profile or the S stereospecificity. Thus, either Ala or Ser can confer S specificity. By contrast, mutation to Val or Thr resulted in loss of catalytic activity.
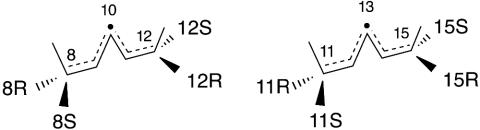
Our results reveal that the basis for R and S stereocontrol also involves a switch in the position of oxygenation on the substrate. The basis for this switch is readily understood from an appreciation of the relationship between R and S on a typical pentadiene (Fig. 5). Oxygenation at one face of the pentadiene produces, for example, 8R product at one end and 12S at the other. In the same way, there is a relationship between 8S and 12R or 11R and 15S. All of the changes in stereochemistry we found are based on this switch in the position of oxygenation to opposite ends of one face of the activated substrate.
Fig. 5.
Relationship between R and S stereospecificity in lipoxygenases. Reaction is illustrated at the 8,11 (Left) and 11,14 (Right) double bonds of a polyunsaturated fatty acid substrate such as AA. In a lipoxygenase-catalyzed reaction, after the initial hydrogen abstraction, oxygen can react on the opposite face of the substrate at one or the other end of the pentadiene radical. Thus, there is a relationship between 8S/12R and 8R/12S (Left) and, in the same way, between 11R/15S and 11S/15R (Right).
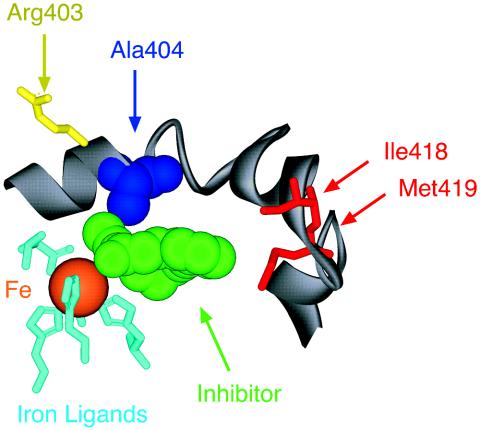
The location of the critical residue we have identified can be illustrated by using the x-ray structure of the rabbit reticulocyte 15-LOX-1 (29). The equivalent amino acid in this 15S-LOX enzyme is Ala-404; it resides in the active site between the nonheme iron and a likely entrance and exit channel for substrate (Fig. 6). In this view, all of the amino acids in the foreground have been stripped away to reveal the nonheme iron that resides in the center of the catalytic domain. The iron ligands (light blue) are shown without the polypeptide chains that hold these amino acids in place. Beyond the iron lies the key residue, Ala-404 (deep blue). A lipoxygenase inhibitor (green) cocrystallized with the enzyme (29) should occupy some of the space normally occupied by the fatty acid substrate, and part of its structure sits between the iron and Ala-404.
Fig. 6.
Location of the conserved Ala-404 residue in the active site of rabbit reticulocyte 15-LOX-1. In this detailed view from the crystal structure (29), iron (orange) in the center of the catalytic domain is held in place by conserved ligands (light blue). The sixth coordination position faces an open cavity, occupied here by a lipoxygenase inhibitor (green) and is presumably the location of fatty acid during catalysis. The conserved Ala-404 (blue) lies beyond the iron in a suitable position for influencing oxygenation of the activated substrate. On the far side of the protein, there is visible access into the active site over the polypeptide chain of Ala-404. Residues on the lower-right side include Ile-418 and Met-419 (red), shown earlier to influence binding of the ω chain of substrate AA (14). On the top left is Arg-403 (yellow), a residue exposed on the far side of the protein surface and suggested to interact with the carboxylic end of AA in this enzyme (32).
Although there is no x-ray structure of a lipoxygenase enzyme with a bound fatty acid substrate, some aspects of the fatty acid binding can be deduced from other lines of evidence. From the fact that arachidonoyl PC is an acceptable substrate for 15S-LOX enzymes (30), we can deduce that the fatty acid enters the 15S-LOX active site tail-first and the carboxyl end remains outside the active site. Here we establish that enzymes with the ability to metabolize fatty acid esterified in PC retain this ability after their positional and stereospecificity is switched by the Ala/Gly mutagenesis. This result shows that the Ala–Gly substitution does not change the tail-first orientation of the substrate in the active site of the 8R-LOX and 15S-LOX enzymes. We tested the other enzymes used here for the ability to metabolize PC substrates and found no 12R or 8S products, in line with the predicted carboxyl-end orientation in the lipoxygenase active site (C. Schneider, G.C., and A.R.B., unpublished observations). Thus, the PC experiments provide important experimental support for modes of substrate binding. In the 15-LOX-1 structure (Fig. 6), it follows that oxygenation occurs deep into the binding pocket at C-15, the site of hydrogen abstraction at C-13 is positioned close to the iron, whereas the proximal end of the reacting pentadiene on the AA substrate (C-11) is positioned close to Ala-404. Visible access from the exterior of the enzyme into the rabbit 15-LOX-1 active site lies adjacent to Ala-404, and, therefore, the carboxyl end of the substrate carbon chain will extend beyond Ala-404 and onto the exterior surface of the enzyme (15).
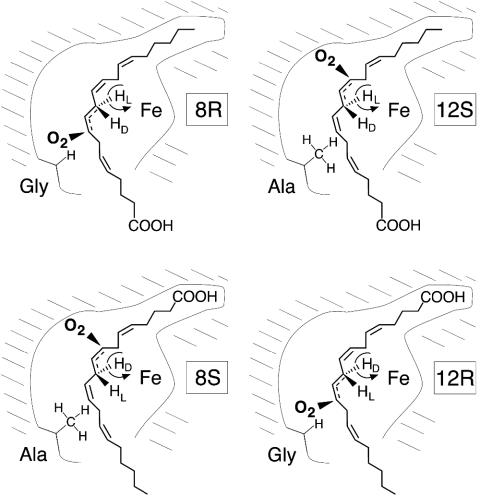
Our results provide experimental support for a model proposed earlier to explain the control of positional and stereospecificity among lipoxygenases (20). The key elements are two modes of substrate orientation (tail-first entry into the active site or carboxyl end first), which in turn dictate a specific hydrogen abstraction (l- or d-hydrogen, respectively), and two positions of antarafacial oxygenation (for S lipoxygenase deep in the substrate binding pocket and R lipoxygenase more shallow in the pocket). Each of the four enzymes we selected for the current experiments represents one of these possibilities. For any given pair of double bonds, there will be four enzymatic specificities; Fig. 7 illustrates the concept for the four possible combinations of 8-LOX and 12-LOX, namely 8S, 12R, 8R, and 12S. Part of the idea is that S and R specificity is conferred by oxygenation at opposite ends of one face of the reactive pentadiene on the substrate (compare Fig. 5); in fact, it is reported from a close examination of mammalian 12S-LOX isozymes that some form traces of 8R-HETE as a minor by-product (31). Fig. 7 illustrates the concepts that S specificity is associated with oxygenation deep in the active site, and that R specificity entails oxygenation at the proximal end of the pentadiene, as well as the recognition of a fixed relationship between substrate orientation and hydrogen abstraction. Previously we had no information to support this basis for stereocontrol. The significant findings are that 8S specificity can be changed to 12R, 8R to 12S, and so on, by the critical Ala/Gly substitution and, that, as indicated by the experiments with PC substrate, binding orientation is preserved in the process. The model proposed can be applied to any activated pentadiene on the substrate. In this study we showed the relationship between 15S-LOX and 11R. In the same way it can be predicted that a 5S-LOX is related to 9R and similarly for other lipoxygenases.
Fig. 7.
A basis for R or S stereospecificity in the lipoxygenase active site. Formation of four different products is represented in lipoxygenase active sites of related structure. In 8R-LOX (Upper Left) and 12S-LOX (Upper Right), AA has the same tail-first orientation in the active site, and reaction is initiated with the same l-hydrogen abstraction from one face of the substrate molecule. A Gly residue in the critical position in the proximal area of the active site allows antarafacial oxygenation at the proximal end of the reactive pentadiene in the 8R configuration (Upper Left). Substitution with the larger Ala residue prevents oxygen insertion at C-8 and, instead, the antarafacial oxygenation occurs deep in the binding pocket in the 12S configuration (Upper Right). In 8S-LOX (Lower Left) and 12R-LOX (Lower Right), the substrate binds in the reverse orientation with the carboxyl end in the active site, allowing removal of the d-hydrogen from C-10. A Gly residue allows oxygenation at the proximal end of the reacting substrate in the 12R configuration (Lower Right), whereas the larger Ala residue prevents this, and the antarafacial oxygenation occurs deeper in the active site in the 8S configuration (Lower Left). The CH2 hydrogens are labeled as d or l, because unlike the corresponding pro-R or pro-S designations, the Fischer nomenclature has a fixed right or left connotation all along the carbon chain (33).
The consequences of mutation of this conserved Ala/Gly residue are dramatic and point to effects beyond a simple opening or closing of space near the proximal end of the reactive pentadiene. Certainly, the presence of the larger Ala residue in S lipoxygenases may shield the proximal oxygenation position and, thus, prevent R oxygenation, but its mutation to the smaller Gly is associated with reduced S oxygenation at the deeper end of the pentadiene, as well as in the case of 15-LOX-2, with a significant reduction in overall catalytic efficiency. This result implies that the mutation may induce further structural changes around the active site and/or affect the optimal alignment of the substrate and exposure of the deeper end to molecular oxygen. Finally, we note that of the four enzymes we tested, the effects of the point mutation on overall catalytic efficiency were mixed, ranging from little effect to an ≈15-fold drop in activity. It can be anticipated that there is a secondary role for additional amino acids in optimizing performance of a specific R or S lipoxygenase with a natural Gly or Ala residue.
In summary, in this study we have established a structural basis for the difference between R- and S-specific lipoxygenases. Our results reveal both a key element of the primary structure characteristic of R or S oxygenation (Gly in R lipoxygenase and Ala in S lipoxygenase) and the fundamental concepts of modes of substrate binding together with the occurrence of S oxygenation deep in the catalytic pocket, and R oxygenation more superficially, closer to the entrance/exit and the surface of the active site of the enzyme. These insights have potential application in the design of selective lipoxygenase inhibitors. In addition, the results elucidate the simple evolutionary concept that the fundamental difference between R and S lipoxygenases is but a single nucleotide switch, the GGN codon of glycine to GCN of alanine.
Supplementary Material
Acknowledgments
We thank Dr. Richard Kim and Brenda Leake for help with the HeLa cell transfection experiments and Dr. Mitsuo Jisaka for providing the appropriate plasmid and optimized expression system for mouse 8S-LOX. This work was supported by National Institutes of Health Grant GM-53638.
Author contributions: G.C. and A.R.B. designed research, performed research, analyzed data, and wrote the paper.
Abbreviations: AA, arachidonic acid; HETE, hydroxyeicosatetraenoic acid; PC, phosphatidylcholine; RP, reversed-phase; SP, straight-phase; TPP, triphenylphosphine; C16/AA-PC, 1-palmitoyl-2-arachidonoyl phosphatidylcholine.
References
- 1.Brash, A. R. (1999) J. Biol. Chem. 274, 23679-23682. [DOI] [PubMed] [Google Scholar]
- 2.Kühn, H. & Thiele, B. J. (1999) FEBS Lett. 449, 7-11. [DOI] [PubMed] [Google Scholar]
- 3.Funk, C. D. (2001) Science 294, 1871-1875. [DOI] [PubMed] [Google Scholar]
- 4.Levy, B. D., De Sanctis, G. T., Devchand, P. R., Kim, E., Ackerman, K., Schmidt, B. A., Szczeklik, W., Drazen, J. M. & Serhan, C. N. (2002) Nat. Med. 8, 1018-1023. [DOI] [PubMed] [Google Scholar]
- 5.Shureiqi, I. & Lippman, S. M. (2001) Cancer Res. 61, 6307-6312. [PubMed] [Google Scholar]
- 6.Marks, F., Muller-Decker, K. & Furstenberger, G. (2000) Toxicology 153, 11-26. [DOI] [PubMed] [Google Scholar]
- 7.De Petrocellis, L. & Di Marzo, V. (1994) Prostaglandins Leukotrienes Essent. Fatty Acids 51, 215-229. [DOI] [PubMed] [Google Scholar]
- 8.Funk, C. D. (1996) Biochim. Biophys. Acta 1304, 65-84. [DOI] [PubMed] [Google Scholar]
- 9.Gerwick, W. H. (1994) Biochim. Biophys. Acta 1211, 243-255. [DOI] [PubMed] [Google Scholar]
- 10.Grechkin, A. (1998) Prog. Lipid Res. 37, 317-352. [DOI] [PubMed] [Google Scholar]
- 11.Oliw, E. H. (2002) Prostaglandins Other Lipid Mediat. 68–69, 313-323. [DOI] [PubMed] [Google Scholar]
- 12.Kühn, H. (2000) Prostaglandins Other Lipid Mediat. 62, 255-270. [DOI] [PubMed] [Google Scholar]
- 13.Kühn, H., Sprecher, H. & Brash, A. R. (1990) J. Biol. Chem. 265, 16300-16305. [PubMed] [Google Scholar]
- 14.Sloane, D. L., Leung, R., Craik, C. S. & Sigal, E. (1991) Nature 354, 149-152. [DOI] [PubMed] [Google Scholar]
- 15.Borngräber, S., Browner, M., Gillmor, S., Gerth, C., Anton, M., Fletterick, R. & Kühn, H. (1999) J. Biol. Chem. 274, 37345-37350. [DOI] [PubMed] [Google Scholar]
- 16.Egmond, M. R., Vliegenthart, J. F. G. & Boldingh, J. (1972) Biochem. Biophys. Res. Commun. 48, 1055-1060. [DOI] [PubMed] [Google Scholar]
- 17.Prigge, S. T., Gaffney, B. J. & Amzel, L. M. (1998) Nat. Struct. Biol. 5, 178-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hornung, E., Walther, M., Kühn, H. & Feussner, I. (1999) Proc. Natl. Acad. Sci. USA 96, 4192-4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jisaka, M., Kim, R. B., Boeglin, W. E. & Brash, A. R. (2000) J. Biol. Chem. 275, 1287-1293. [DOI] [PubMed] [Google Scholar]
- 20.Brash, A. R., Boeglin, W. E., Chang, M. S. & Shieh, B.-H. (1996) J. Biol. Chem. 271, 20949-20957. [DOI] [PubMed] [Google Scholar]
- 21.Schneider, C. & Brash, A. R. (2002) Prostaglandins Other Lipid Mediat. 68–69, 291-301. [DOI] [PubMed] [Google Scholar]
- 22.Blakely, R. D., Clark, J. A., Rudnick, G. & Amara, S. G. (1991) Anal. Biochem. 194, 302-308. [DOI] [PubMed] [Google Scholar]
- 23.Bligh, E. G. & Dyer, W. J. (1959) Can. J. Biochem. Physiol. 37, 911-917. [DOI] [PubMed] [Google Scholar]
- 24.Takahashi, Y., Glasgow, W. C., Suzuki, H., Taketani, Y., Yamamoto, S., Anton, M., Kuhn, H. & Brash, A. R. (1993) Eur. J. Biochem. 218, 165-171. [DOI] [PubMed] [Google Scholar]
- 25.Schneider, C., Boeglin, W. E. & Brash, A. R. (2000) Anal. Biochem. 287, 186-189. [DOI] [PubMed] [Google Scholar]
- 26.Koljak, R., Boutaud, O., Shieh, B.-H., Samel, N. & Brash, A. R. (1997) Science 277, 1994-1996. [DOI] [PubMed] [Google Scholar]
- 27.Boutaud, O. & Brash, A. R. (1999) J. Biol. Chem. 274, 33764-33770. [DOI] [PubMed] [Google Scholar]
- 28.Chen, X.-S., Kurre, U., Jenkins, N. A., Copeland, N. G. & Funk, C. D. (1994) J. Biol. Chem. 269, 13979-13987. [PubMed] [Google Scholar]
- 29.Gillmor, S. A., Villaseñor, A., Fletterick, R., Sigal, E. & Browner, M. F. (1997) Nat. Struct. Biol. 4, 1003-1009. [DOI] [PubMed] [Google Scholar]
- 30.Takahashi, Y., Glasgow, W. C., Suzuki, H., Taketani, Y., Yamamoto, S., Anton, M., Kühn, H. & Brash, A. R. (1993) Eur. J. Biochem. 218, 165-171. [DOI] [PubMed] [Google Scholar]
- 31.Burger, F., Krieg, P., Marks, F. & Fürstenberger, G. (2000) Biochem. J. 348, 329-335. [PMC free article] [PubMed] [Google Scholar]
- 32.Gan, Q.-F., Browner, M. F., Sloane, D. L. & Sigal, E. (1996) J. Biol. Chem. 271, 25412-25418. [DOI] [PubMed] [Google Scholar]
- 33.Brash, A. R. & Hawkins, D. J. (1990) Methods Enzymol. 187, 187-195. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.