Abstract
Most cases of early-onset torsion dystonia (EOTD) are caused by a deletion of one glutamic acid in the carboxyl terminus of a protein named torsinA. The mutation causes the protein to aggregate in perinuclear inclusions as opposed to the endoplasmic reticulum localization of the wild-type protein. Although there is increasing evidence that dysfunction of the dopamine system is implicated in the development of EOTD, the biological function of torsinA and its relation to dopaminergic neurotransmission has remained unexplored. Here, we show that torsinA can regulate the cellular trafficking of the dopamine transporter, as well as other polytopic membrane-bound proteins, including G protein-coupled receptors, transporters, and ion channels. This effect was prevented by mutating the ATP-binding site in torsinA. The dystonia-associated torsinA deletion mutant (ΔE-torsinA) did not have any effect on the cell surface distribution of polytopic membrane-associated proteins, suggesting that the mutation linked with EOTD results in a loss of function. However, a mutation in the ATP-binding site in ΔE-torsinA reversed the aggregate phenotype associated with the mutant. Moreover, the deletion mutant acts as a dominant-negative of wild-type torsinA through a mechanism presumably involving association of wild-type and mutant torsinA. Taken together, our results provide evidence for a functional role for torsinA and a loss of function and a dominant-negative phenotype of the ΔE-torsinA mutation. These properties may contribute to the autosomal dominant nature of the condition.
Keywords: deletion mutant, cell surface, endoplasmic reticulum, transmembrane, polytopic
Early-onset torsion dystonia (EOTD) is the most common and severe form of a group of diseases known as dystonias (1). EOTD is an autosomal-dominant movement disorder characterized by involuntary and sustained muscle contractions affecting several parts of the body and resulting in twisting, abnormal postures, and, in many cases, severe disability (2–4). The disease has been linked to a 3-bp deletion in the DYT1 gene that results in a single glutamate deletion near the carboxyl terminus of the product of the DYT1 gene, a protein termed torsinA (5).
There is increasing evidence indicating that dysfunction of the dopamine system in the basal ganglia underlie the clinical features of EOTD, as well as some other less common forms of dystonia (6, 7). For instance, mutations in two important genes in the biosynthesis of dopamine; the GTP cyclohydrolase (8–10) and tyrosine hydroxylase genes (11–12), were found in patients with dopa-responsive dystonia. Similarly, a mutation in the dopamine D2 receptor (D2DAR) was associated with myoclonus dystonia (13). Consistent with a potential role for the dopamine system in dystonia, the torsinA gene is expressed at high levels in dopamine neurons of the substantia nigra (14–16). However, a link between torsinA or its deletion mutant (ΔE-torsinA) and the dopamine system has not been established.
Based on homology to known proteins, it has been suggested that torsinA might function as a molecular chaperone in cells (3, 5). The primary sequence of torsinA contains a Walker ATP/Mg2+-binding site, including a typical A motif involved in the phosphate binding of ATP. TorsinA is a bona fide member of the AAA+ family of ATPases. Members of this family of proteins participate in a variety of cellular functions, including assembly and disassembly of protein complexes (17–19). However, evidence supporting a definitive role for torsinA as a molecular chaperone is still lacking. Here, we report that torsinA overexpression in several mammalian cell lines reduces the cell-surface distribution of the dopamine transporter (DAT) and of other polytopic membrane-bound proteins. This action of torsinA was abolished when the Walker A ATP-binding consensus site was mutated by replacing a lysine residue at position 108 with a threonine (K108T). As opposed to the wild-type protein, ΔE-torsinA is found in perinuclear aggregates in cells. However, the distribution pattern of the ΔE-torsinA K108T mutant resembles that of the wild-type torsinA protein, suggesting a role for the ATP-binding site in the mislocalization of ΔE-torsinA. Furthermore, we show that ΔE-torsinA is a loss-of-function protein and acts as a dominant-negative of the wild-type torsinA. Taken together, our findings delineate a previously undescribed cellular mechanism for torsinA and ΔE-torsinA.
Materials and Methods
Materials. MEM, FBS, trypsin, and penicillin/streptomycin were purchased from Life Technologies (Grand Island, NY). The [3H]dopamine (31.6 Ci/mmol; 1 Ci = 37 GBq) and 125I labeledcyanopindolol were supplied by PerkinElmer Life Sciences (Boston). Taq polymerase were from Fisher Scientific, restriction enzymes were from Takara Biomedicals (Otsu, Japan), and DNA purification kits were from Qiagen (Valencia, CA). Protease inhibitor mixture and the anti-hemagglutinin (HA) monoclonal antibody were purchased from Roche Diagnostics (Indianapolis). The monoclonal antibody against protein disulfide isomerase was from Stressgen Biotechnologies (Victoria, Canada), the monoclonal Flag antibody was from Sigma, the monoclonal anti-norepinephrine transporter (NET) antibody was from Mab Technologies (Stone Mountain, GA), and the polyclonal anti-torsinA antibody has been described (15). Secondary antibodies conjugated with horseradish peroxidase, FITC, or Texas red were from Jackson ImmunoResearch (West Grove, PA). Sulfo-N-hydroxysuccinimide-SS-biotin, dithiobis(succinimidylpropionate) (DSP), ultralink avidin beads, and the chemiluminescence system were from Pierce. All other chemicals used were from Sigma.
DNA Constructs and Mutagenesis. PCR-based mutagenesis (36 cycles at 94°C for 230 s, 55°C for 30 s, and 72°C for 3 min) was used to introduce mutations into the coding sequence of the human torsinA. After PCR, restriction fragments containing mutated sequences were digested with appropriate restriction enzymes and subcloned into pcDNA3.1. Oligonucleotide primers (Genosys, The Woodlands, TX) were designed to introduce the FLAG epitope (DYKDDDDK) to the carboxyl terminus of wild-type torsinA or ΔE-torsinA immediately upstream of the stop codon. The coding sequence of torsinA or ΔE-torsinA was inserted into the GFP-containing vector pEGFPN3 to create fusion proteins in which GFP is inserted immediately before the stop codon of torsinA or ΔE-torsinA. All constructs were verified by DNA sequencing. The amino-terminal GFP-tagged DAT (GFP-DAT) was generously provided by S. G. Amara (University of Pittsburgh). The GFP-tagged α1B (α1BAR-GFP) and β2 adrenergic receptors (β2AR-GFP) and D2DAR-GFP have been described (20, 21). The human norepinephrine transporter was subcloned into the mammalian expression vector pcDNA3.1. The rat HA-tagged ATP-sensitive potassium channel subunit Kir6.2 and the sulfonylurea receptor subunit SUR1 were provided by E. Cartier and S.-L. Shyng (Oregon Health and Science University, Portland). The GFP-tagged SNAP25 construct was a gift from J. B Sørensen, (Max Planck Institute, Göttingen, Germany). The GFP-tagged EGF receptor was provided by A. Sorkin (University of Colorado Health Sciences Center, Denver), and the GFP-tagged insulin-like growth factor I receptor (IGF-IR) was provided by R. J. Lefkowitz (Duke University Medical Center, Durham, NC). PCR-based mutagenesis and subcloning are described above.
Cell Culture and Transfections. Human embryonic kidney (HEK)293 cells were grown to 60–80% confluency in 100-mm tissue culture dishes and transiently transfected by using the Ca2PO4 precipitation method with 5 μg of total DNA. Cells were incubated with the Ca2PO4-DNA mixture at 37°C for 16 h, followed by a 48-h recovery in MEM supplemented with 10% FBS and 50 units/ml penicillin/streptomycin. Under these conditions, ≈90% of cells are transfected. Subsequent experiments were performed 48–72 h after transfections. COS-7 and PC12-cells were maintained in DMEM and transfected under similar conditions.
Transport Measurements. Forty-eight to 72 h after transfections, medium was removed, and uptake was measured after incubation of cells for 5 min with 250 μl of uptake buffer (5 mM Tris·HCl/7.5 mM Hepes/120 mM NaCl/5.4 mM KCl/1.2 mM CaCl2/1.2 mM MgSO4/1 mM ascorbic acid/5 mM glucose, pH 7.4) containing 20 nM [3H]dopamine (31.6 Ci/mmol) and increasing concentrations of cold dopamine ranging from 100 nM to 30 μM. After rinsing with 1 ml of NaCl-free uptake buffer, cells were solubilized in 0.5 ml of 1% SDS, and the radioactivity incorporated into the cells was measured by liquid scintillation counting. Nonspecific uptake was determined in the presence of 2 μM mazindol. Data are presented as the mean ± SEM. The conditions for dopamine uptake in HEK293 and PC12 cells have been adapted from Giros et al. (22).
Ligand Binding. HEK293 cells were transfected with plasmids containing β2AR and empty vector or β2AR with torsinA cDNAs. Total binding was determined in the presence of 200 pM 125I-labeled cyanopindolol alone. Intracellular receptor binding was determined in the presence of 200 pM 125I-labeled cyanopindolol plus 100 nM CGP12177, whereas nonspecific binding was determined using 1 μM propranolol. Data are presented as the mean ± SEM. The conditions for β2AR binding have been adapted from Barak et al. (23).
Immunocytochemistry and Confocal Microscopy. For immunostaining experiments, transiently transfected cells grown on glass coverslips were placed in six-well dishes at a density of 5 × 105 cells per well, followed by fixation in 4% paraformaldehyde. After three washes with PBS, cells were permeabilized in PBS containing 0.1% Triton X-100 for 10 min and incubated in blocking solution (1% BSA and 5% goat serum in PBS) for 1 h. Cells were incubated with primary antibodies for 1 h at room temperature, followed by incubation with secondary antibodies for another 1 h at room temperature. Cells were then washed three times in PBS, and the coverslips were mounted on glass slides by using Vectashield (Vector Laboratories). Immunofluorescent images were generated by using a Zeiss laser scanning confocal microscope at wavelengths of 585 nm for Texas red and 488 nm for FITC.
Cell-Surface Biotinylation. Transiently transfected monolayers of HEK293 cells were washed three times with PBS and then incubated with gentle agitation for 30 min at 4°C with 1 ml of 1 mg/ml sulfo-N-hydroxysuccinimide-SS-biotin prepared in 150 mM NaCl/2 mM CaCl2/10 mM triethanolamine, pH 7.8. The reaction was quenched by incubating the cells for an additional 10 min with 50 mM glycine in PBS. Cells were then washed three times in PBS and incubated in radioimmune precipitation assay buffer (10 mM Tris·HCl/150 mM NaCl/1 mM EDTA/0.1% SDS/1% Triton X-100/1% sodium deoxycholate, pH 7.4) at 4°C for 1 h. Each sample was divided into two aliquots. One aliquot was used for isolation of biotinylated proteins with ultralink-immobilized neutravidin beads. The second aliquot was used to determine total protein levels. Samples were analyzed by Western blotting with the rat anti-DAT antibody and a horseradish peroxidase-conjugated secondary antibody.
Cross-Linking. HEK293 cells transfected with wild-type or mutant torsinA were incubated with increasing concentrations of DSP, ranging from 1 μM to 10 mM (prepared in dimethyl sulfoxide). After incubation for 30 min at room temperature, the cross-linker was quenched by the addition of 1 M Tris·HCl, pH 7.5, to a final concentration of 20 mM. After quenching, the samples were lysed and analyzed by Western blotting with the torsinA antibody.
Immunoprecipitation and Western Blotting. HEK293 cells expressing torsinA constructs were lysed with 1% Triton X-100 in PBS and incubated with the primary antibody at 4°C overnight. The immunoprecipitated proteins were recovered with a mixture of protein A and G beads, fractionated on 10% acrylamide gels, and transferred to nitrocellulose membranes. Western blotting was performed using anti-torsinA and a secondary antibody conjugated with horseradish peroxidase, and immunoreactive bands were detected with the Super Signal West Pico system (Pierce).
Results
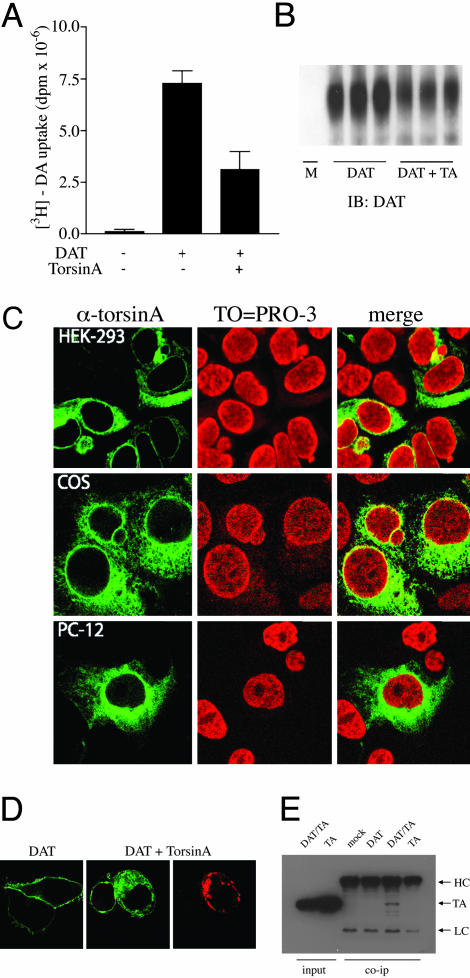
TorsinA Affects the Cell-Surface Expression of the DAT. Because imbalances in the dopamine brain system have been suggested in EOTD, we investigated the potential action of torsinA on dopaminergic neurotransmission by examining whether torsinA could affect the function of the dopamine reuptake system. The rapid DAT-mediated uptake of dopamine from the extracellular space to presynaptic nerve terminals has been shown to be critical in the regulation of dopaminergic transmission (24). Thus, we compared dopamine uptake activity in HEK293 cells transfected with the human DAT alone or in combination with torsinA. Coexpression of torsinA and DAT significantly decreased total DA uptake as compared with cells expressing similar levels of DAT in the absence of torsinA (Vmax = 7.3 ± 0.6 dpm × 10-6 in cells expressing DAT vs. 3.1 ± 0.9 dpm × 10-6 in cells expressing DAT and torsinA; Fig. 1A). Mock-transfected cells did not exhibit detectable levels of dopamine uptake (Fig. 1A). TorsinA overexpression did not induce significant changes in the affinity of DA for the transporter (Km = 2.5 μM in cells expressing DAT vs. 2.8 μM in cells expressing DAT and torsinA), suggesting that torsinA affects dopamine uptake, possibly by interfering with the delivery of DAT to the plasma membrane. We then performed cell-surface biotinylation experiments to test this possibility. Mock-transfected cells or cells transfected with either DAT alone or DAT and torsinA were incubated with sulfo-N-hydroxysuccinimide-SS-biotin, followed by isolation of labeled proteins with avidin beads and analysis by Western blotting using the anti-DAT antibody. Mock-transfected cells did not show any detectable DAT protein (Fig. 1B). The levels of DAT at the cell membrane were significantly reduced in the presence of torsinA, under conditions in which the total levels of transporter protein remained unchanged (data not shown).
Fig. 1.
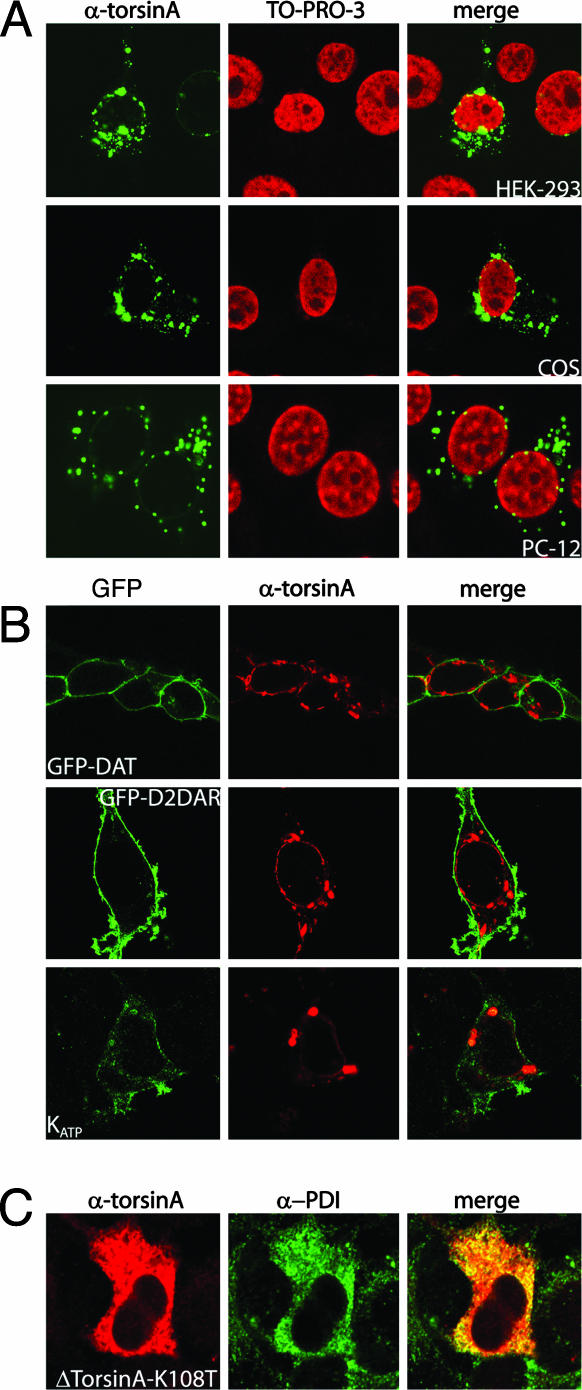
TorsinA overexpression reduces DAT function by decreasing the levels of DAT at the plasma membrane. (A) The [3H]dopamine uptake activity in cells transfected with DAT alone or in combination with torsinA as indicated. Bars correspond to the mean ± SEM of three independent experiments. (B) Cell-surface biotinylation experiments in HEK293 cells mock-transfected or transfected with DAT alone (DAT) or in combination with torsinA (DAT + TA). The immunoblot (IB) was carried out with a rat monoclonal DAT antibody. (C) Distribution of torsinA in different mammalian cell lines. HEK293 (Top), COS (Middle), and PC12 cells (Bottom) were transfected with torsinA and costained with an anti-torsinA antibody (Left) and the DNA-binding dye TO-PRO-3 (Center). (D) Distribution pattern of DAT when expressed alone or in combination with torsinA in HEK293 cells. Images were taken from cells transfected with the GFP-tagged DAT (green) and torsinA immunostained with a rabbit anti-torsinA antibody, followed by a Texas red-conjugated anti-rabbit antibody (red). (E) Coimmunoprecipitation between DAT and torsinA. HEK293 cells were mock-transfected or transfected with DAT, DAT and torsinA, or torsinA alone. Immunoprecipitation was performed in PBS buffer containing 1% Triton X-100 with the DAT antibody, whereas the immunoblot was carried out with the torsinA antibody. TA, torsinA; HC, heavy chain; LC, light chain.
We then examined whether torsinA may affect the cellular trafficking of DAT in cells. Consistent with previous reports (25, 26) in HEK293 cells, torsinA was found in the tubule-vesicular network seen throughout the entire cell (Fig. 1C Left), excluded from the cell nucleus (Fig. 1C Center). As shown (25, 26), torsinA immunoreactivity colocalizes with the endoplasmic reticulum (ER) marker protein disulfide isomerase (data not shown). Similar patterns of distribution for torsinA were observed in COS-7 and PC-12 cells (Fig. 1C). Analysis of DAT subcellular localization by immunofluorescence and confocal microscopy revealed that the transfected transporter is predominantly expressed at the cell surface of HEK293 cells (Fig. 1D). However, when coexpressed with torsinA, DAT is primarily observed in intracellular compartments where it partially colocalized with torsinA (Fig. 1D). Thus, in the presence of torsinA, there was a shift in the localization of DAT from the cell surface to intracellular compartments. To confirm that this observation was not cell-type-specific, we observed similar changes in DAT subcellular localization when coexpressed with torsinA in COS-7 and PC-12 cells (data not shown). In addition, in HEK293 cells cotransfected with torsinA and DAT, immunoprecipitation of DAT resulted in the coprecipitation of torsinA (Fig. 1E), indicating that torsinA forms a protein complex with DAT.
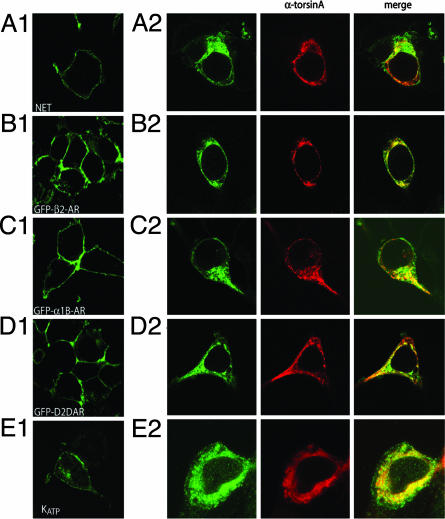
TorsinA Is a General Modulator of Polytopic Membrane-Bound Protein Trafficking. We next explored whether the effect of torsinA was restricted to DAT or was also observed with other membrane-bound proteins. First, we tested the effect of torsinA overexpression on the subcellular localization of the closely related NET transfected in HEK293 cells. As seen with DAT, NET proteins were predominantly observed in intracellular compartments in the presence of torsinA (Fig. 2A2), contrasting with the cell surface distribution observed when the transporter is transfected alone (Fig. 2A1). Because NET is expressed endogenously in PC12 cells, we examined the effect of torsinA overexpression on the function of the native transporter. Overexpression of torsinA caused a significant reduction in norepinephrine uptake compared with control cells (125 ± 14 fmol per mg per min in control cells vs. 67 ± 9 in cells overexpressing torsinA). We next examined the effect of torsinA overexpression on the distribution of several G protein-coupled receptors, including the β2AR, α1AR, and the D2DAR. To facilitate visualization, GFP versions of these receptor proteins were transfected in HEK293 cells. As seen in Fig. 2 B–D, all three GFP-tagged receptors were primarily found at the cell surface when expressed alone in HEK293 cells (Fig. 2 B1–D1). In contrast, in cells cotransfected with any of the different GFP-tagged receptors and torsinA, there was a significant decrease of receptor cell surface expression with a concomitant increase in intracellular retention (Fig. 2 B2–D2). To quantify the effect of torsinA overexpression, we performed binding experiments in cells transfected with the β2AR alone or in combination with torsinA. Whereas torsinA overexpression did not alter total levels of β2AR protein (4.2 ± 0.3 pmol/mg in cells expressing β2AR alone vs. 4.5 ± 0.6 pmol/mg in cells expressing β2AR and torsinA), the intracellular levels of the receptor were significantly increased in the presence of torsinA (0.84 ± 0.1 pmol/mg in cells expressing β2AR alone vs. 2.8 ± 0.3 pmol/mg in cells expressing β2AR and torsinA). Finally, we cotransfected the HA-tagged ATP-sensitive potassium (KATP) channel subunit Kir6.2 with the sulfonylurea receptor subunit SUR1 in HEK293 cells. When coexpressed with SUR1, the Kir6.2 channel subunit is predominantly localized to the cell surface (Fig. 2E1). However, when torsinA is transfected along with HA-Kir6.2 and SUR1, the HA signal of the Kir6.2 channel subunit predominantly appears intracellularly in HEK293 cells (Fig. 2E2).
Fig. 2.
Overexpression of torsinA impairs the cell-membrane expression of several polytopic membrane-bound proteins. HEK293 cells were transfected with NET (A), the GFP-tagged β2AR (B), the GFP-tagged α1BAR (C), the GFP-tagged D2DAR (D), and the HA-tagged Kir6.2 channel (cotransfected with SUR1) (E) individually (A1–E1) or cotransfected with torsinA (A2–E2). In cells transfected with NET and HA-Kir6.2, anti-NET and anti-HA antibodies were used, respectively, followed by a FITC-conjugated secondary antibody (green). TorsinA was stained with a rabbit anti-torsinA antibody, followed by a Texas red-conjugated anti-rabbit antibody (red).
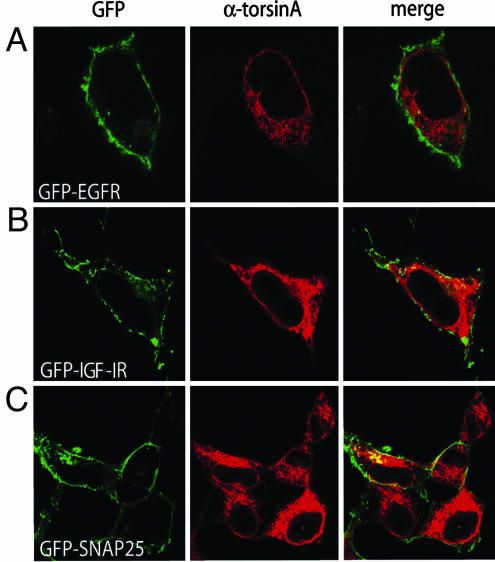
A commonly observed feature of the membrane-associated protein tested thus far is the presence of multiple transmembrane domains. Thus, we tested the effect of torsinA on the cell-surface distribution of membrane-bound proteins containing a single transmembrane domain. Both, the epidermal growth factor receptor (EGFR) and the IGF-IR posses one transmembrane domain. Expression of the GFP-tagged EGFR or GFP-IGF-IR proteins in HEK293 cells resulted in predominant cell-surface distribution of the receptor proteins (Fig. 3 A and B). In contrast to its effects on polytopic membrane proteins, torsinA did not affect the distribution of either the GFP-EGFR or the GFP-IGF-IR in coexpression experiments. Similarly, the cell-membrane localization of the GFP-tagged SNAP-25, an extrinsic membrane protein normally anchored at the plasma membrane through posttranslational palmitoylation, was not altered by torsinA coexpression either (Fig. 3C). Thus, these results suggest that the effect of torsinA is rather selective for polytopic membrane-bound proteins.
Fig. 3.
TorsinA overexpression does not affect the cell-surface distribution of proteins containing one or no transmembrane domains. HEK293 cells were cotransfected with torsinA and the following constructs as indicated: GFP-EGFR (A), GFP-IGF-IR (B), and GFP-SNAP-25 (C). TorsinA was stained with a rabbit anti-torsinA antibody, followed by a Texas red-conjugated anti-rabbit antibody (red).
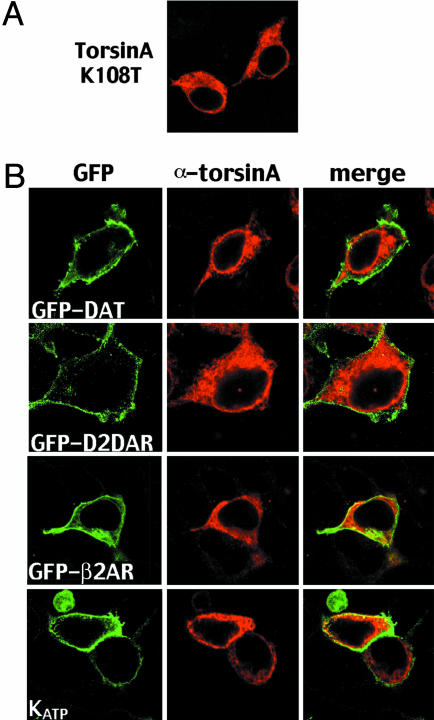
Mutation of a Putative ATP-Binding Motif Abolishes the Effect of TorsinA. Examination of the primary sequence of torsinA reveals the presence of a Walker-A ATP-binding consensus motif that is common to ATPase proteins (5). To test the contribution of this motif in the effect of torsinA, we generated a torsinA mutant in which a highly conserved lysine residue was substituted for a threonine residue. This mutant presumably results in lost of ATP binding, and thus, it might mimic the inactive form of this putative ATPase protein. The distribution pattern of the torsinA K108T mutant was indistinguishable from that of wild-type torsinA when expressed in HEK293 cells (Fig. 4A). The contribution of the ATP-binding domain to the effect of torsinA on polytopic membrane-bound proteins was examined by cotransfecting torsinA K108T with the various cell-surface proteins. As shown in Fig. 4B, overexpression of torsinA K108T did not have any effect on the cell-membrane distribution of GFP-DAT, D2DAR-GFP, β2AR-GFP, or the HA-tagged Kir6.2 (cotransfected with SUR1). No effect on dopamine uptake was observed in cells transfected with DAT alone or in combination with torsinA K108T (Vmax = 6.8 ± 0.4 dpm × 10-6 in cells expressing DAT vs. 7.2 ± 0.7 dpm × 10-6 in cells expressing DAT and torsinA K108T). In addition, immunoprecipitation of DAT failed to coimmunoprecipitate torsinA K108T (Fig. 7, which is published as supporting information on the PNAS web site). These results suggest that the ATP-binding site is required for the effect of torsinA on the cell-membrane expression of polytopic proteins.
Fig. 4.
Mutation of the ATP-binding site in torsinA abolishes the effects of torsinA. (A) HEK293 cells transfected with torsinA K108T and stained with a rabbit anti-torsinA antibody. (B) HEK293 cells were cotransfected with torsinA and the following constructs as indicated: GFP-DAT, GFP-D2DAR, GFP-β2AR, or the HA-tagged Kir6.2. TorsinA was stained with a rabbit anti-torsinA antibody, followed by a Texas red-conjugated anti-rabbit antibody (red), whereas the HA-Kir6.2 channel was stained with an HA monoclonal antibody followed by a FITC-conjugated secondary antibody (green).
Loss of Function and Reversal of the Aggregate Phenotype Linked with the Dystonia-Associated ΔE-TorsinA Mutant. Having established a functional assay for torsinA, we next investigated the effect of ΔE-torsinA on the cell-surface distribution of membrane-bound proteins. As reported (25, 26), transfection of ΔE-torsinA into HEK293, COS-7, and PC-12 cells led to the formation of large torsinA-immunoreactive perinuclear aggregates (Fig. 5A). HEK293 cells were then transfected with DNA constructs encoding GFP-DAT, D2DAR-GFP, and HA-Kir6.2 (cotransfected with SUR1) in the presence or absence of ΔE-torsinA. As seen in Fig. 5B, ΔE-torsinA did not affect the localization of any of the membrane proteins examined. In addition, the cell-membrane distribution of the GFP-EGFR was not altered by ΔE-torsinA overexpression (Fig. 5B). ΔE-torsinA overexpression in HEK293 cells resulted in similar levels of expression when compared with wild-type torsinA (data not shown), indicating that the lack of effect of ΔE-torsinA was not due to reduced levels of expression of the mutant protein.
Fig. 5.
Functional Analysis of the ΔE-torsinA mutant. (A) Subcellular distribution of WT-torsinA in HEK293 (Top), COS (Middle), and PC12 cells (Bottom). Cells were transfected with ΔE-torsinA and costained with an anti-torsinA antibody and an FITC-conjugated anti-rabbit antibody (Left) and the DNA-binding dye TO-PRO-3 (Center). (B) HEK293 cells were cotransfected with ΔE-torsinA and the following constructs as indicated: GFP-DAT (Left), GFP-D2DAR (Center), the HA-tagged Kir6.2 (cotransfected with SUR1) (Right), or the GFP-EGFR. ΔE-TorsinA was stained with a rabbit anti-torsinA antibody, followed by a Texas red-conjugated anti-rabbit antibody (red), whereas the HA-Kir6.2 channel was stained with an HA monoclonal antibody followed by a FITC-conjugated secondary antibody (green). (C) A K108T mutation in ΔE-torsinA reverses the cellular distribution of the mutant into the distribution of the wild-type protein.
To explore the role of the ATP-binding domain in the cellular distribution ΔE-torsinA, we generated the ΔE-torsinA K108T mutant and expressed this construct in HEK293 cells. As seen in Fig. 5C, the cellular aggregate phenotype of the ΔE-torsinA mutant was significantly altered by the K108T mutation and resembled that of the wild-type torsinA. Colocalization experiments with the protein disulfide isomerase marker indicates that the ΔE-torsinA K108T mutant resides primarily in the ER and not in perinuclear inclusions. These results suggest that the ATP-binding site is crucial in the mislocalization and aggregate formation of ΔE-torsinA.
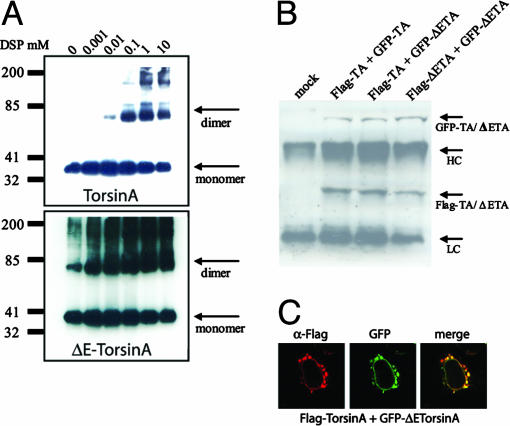
Dominant-Negative Effect of ΔE-torsinA Through a Protein Complex Between Wild-Type TorsinA and ΔE-TorsinA. To explain the loss of function associated with ΔE-torsinA, we searched for mechanistic differences between torsinA and ΔE-torsinA. Self-association is a general and conserved feature of members of the AAA+ family of ATPases and changes in oligomerization have been correlated with changes in function (27). Thus, we examined whether torsinA self-associates and whether this property was altered in ΔE-torsinA. Cells transfected with torsinA or ΔE-torsinA were incubated with the cross-linking membrane-permeant agent DSP for 1 h at room temperature and then analyzed by Western blotting using the anti-torsinA antibody (Fig. 6A). In the absence of DSP, both torsinA and ΔE-torsinA migrated on SDS/PAGE as a monomer of ≈38 kDa (Fig. 6A). In contrast, in cells treated with increasing concentrations of DSP, we observed an additional band of ≈75 kDa, consistent with the formation of dimer complexes. However, in cells transfected with ΔE-torsinA, there was an increased tendency to form dimers. Indeed, the 75-kDa band was observed even in the absence of the cross-linking agent (Fig. 6A Lower). These results suggest that torsinA oligomerizes in cells and that this property is enhanced in ΔE-torsinA. To determine if wild-type torsinA can associate with ΔE-torsinA, we tagged torsinA and ΔE-torsinA with Flag or GFP epitopes. This strategy allows for the discrimination of ΔE-torsinA from wild-type torsinA based on differences in molecular size due to the GFP and Flag tags. In all possible combinations tested, Flag-torsinA and GFP-torsinA (wild-type/wild-type), Flag-torsinA and GFP-ΔE-torsinA (wild-type/mutant), and Flag-ΔE-torsinA and GFP-ΔE-torsinA (mutant/mutant), immunoprecipitation with the Flag antibody resulted in the coprecipitation of the GFP-tagged counterpart (Fig. 6B). Thus, these results demonstrate that torsinA and ΔE-torsinA can form a multimeric protein complex.
Fig. 6.
Interaction of torsinA and ΔE-torsinA. (A). Cross-linking of torsinA (Upper) and ΔE-torsinA (Lower). HEK293 cells expressing either WT-torsinA or ΔE-torsinA were incubated with increasing concentrations of DSP as described in Materials and Methods. Immunoblotting was performed with a polyclonal anti-torsinA antibody. (B) Coimmunoprecipitation between Flag-tagged torsinA and GFP-tagged torsinA, Flag-tagged torsinA and GFP-tagged ΔE-torsinA, and Flag-tagged ΔE-torsinA and GFP-tagged ΔE-torsinA. Immunoprecipitation was performed with the Flag antibody and immunoblotting with the torsinA antibody. TA, torsinA; ΔETA, ΔE-torsinA. (C) Dominant-negative effect of ΔE-torsinA on wild-type torsinA distribution. TorsinA was tagged with the Flag epitope and stained with the Flag antibody, followed by a Texas red secondary antibody (red). ΔE-torsinA was tagged with GFP (green).
An interaction between torsinA and ΔE-torsinA suggests a potential mechanism in which ΔE-torsinA might inhibit the normal function of torsinA through direct or indirect protein–protein interactions. To test this hypothesis, we searched for changes in the subcellular localization of both proteins when cotransfected in HEK293 cells. As seen in Fig. 6C, when coexpressed with ΔE-torsinA, the wild-type torsinA protein was not present in the ER, instead, it was found in large aggregates around the nucleus colocalizing with the mutant ΔE-torsinA. These results suggest that ΔE-torsinA acts as a dominant-negative mutant sequestering wild-type torsinA molecules away from the ER through protein–protein interactions. Although these data are consistent with a direct protein–protein interaction, we cannot exclude the possibility of an intermediate interacting protein.
Discussion
In the present study, we show that torsinA interferes with the expression of polytopic membrane-bound proteins at the cell membrane. This effect appears to be specific for polytopic proteins, because torsinA had no effect on the cellular distribution of membrane-associated proteins containing one or no transmembrane domains. Mutation of the Walker A ATP-binding motif abolishes the effect of torsinA, suggesting that the ATPase activity of torsinA is required for this effect. The finding that ΔE-torsinA behaved as a loss-of-function protein was somewhat expected as the deletion in torsinA causes the protein to mislocalize. More interestingly, we found that ΔE-torsinA interacts directly or indirectly with wild-type torsinA, acting as a dominant-negative by sequestering wild-type torsinA molecules. These results suggest that torsinA might participate in folding, assembly, and/or trafficking of polytopic protein complexes and provide a molecular basis for the effect of the torsinA glutamic acid deletion as a loss of function and a dominant-negative mutant.
Our results also provide insights into the dramatic effect of the deletion in the cellular distribution of the ΔE-torsinA mutant when compared with the wild-type protein. Mutation of a conserved lysine residue in the ATP-binding Walker A motif reverses the cellular phenotype of ΔE-torsinA into the wild-type protein phenotype. It is tempting to speculate that ΔE-torsinA exists in an ATP-bound state that might cause the protein to aggregate. Thus, mutation of the ATP-binding site would release the ATP-bound state and restore the cellular distribution of the mutant. Our findings may have significant clinical implications as the ATP-binding site of ΔE-torsinA could be used to screen for pharmacological compounds that reverse the cellular phenotype of ΔE-torsinA.
A chaperone-like function has been recently suggested for torsinA (28–30). Consistently, our studies suggest that torsinA regulates the assembly/disassembly process of polytopic membrane proteins. Monoamine transporters, channels, and G protein-coupled receptors exist as homomeric, and in some cases, as heteromeric protein complexes (24, 31, 32). There is mounting evidence suggesting that oligomerization of the protein complex precede the plasma membrane trafficking of these proteins. TorsinA might participate directly in the folding, assembly, and/or trafficking of polytopic proteins to the cell membrane, or, alternatively, torsinA might bind a protein that is required for the folding, assembly, and/or trafficking of polytopic proteins to the cell surface. It is also possible that overexpression of the chaperone protein would generate a futile cycle whereby too much protein would inhibit processing. Indeed, there is evidence for the role of molecular chaperones in the disassembly of regulatory protein complexes in cells (33). Alternatively, it is possible that torsinA might interact with another factor required for the assembly of polytopic membrane proteins. The molecular mechanisms by which torsinA affects the early events leading to the assembly/disassembly of these proteins are worthy of further study.
Our results demonstrate a loss of function and a dominant-negative effect associated with the ΔE-torsinA mutant. Thus, these findings also provide a molecular mechanism to explain the genetic dominance observed in patients with EOTD carrying the torsinA mutation. Our findings also explore the molecular mechanisms associated with the dramatic change in cellular distribution associated with ΔE-torsinA. The aggregate-like distribution seen with ΔE-torsinA is reversed to an ER localization when the ATP-binding site is mutated. These results suggest that the aggregation of ΔE-torsinA proteins might be due to an ATP-bound state that is released by the K108T mutation. More importantly, if the aggregation of ΔE-torsinA proteins cause the disease, pharmacological agents that target the ATP-binding site in torsinA might have some therapeutically potential. Taken together, our results are relevant not only to understand the function of torsinA in vivo but also the molecular bases of EOTD associated with ΔE-torsinA.
Note. During the review process for this manuscript, Naismith et al. (34) reported a similar finding and interpretation with respect to the role of the ATP-binding motif in the distribution of torsinA and ΔE-torsinA.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants NS-19576 (to M.G.C.), DA-14150 (to G.E.T.), and NS 43038 (to P.S.). J.-M.B. is supported by a Human Frontiers Science Program Fellowship.
Author contributions: G.E.T. and M.G.C. designed research; G.E.T., A.S., and J.-M.B., performed research; P.S. contributed new reagents/analytic tools; G.E.T. analyzed data; and G.E.T. and M.G.C. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ΔE-torsinA, torsinA deletion mutant; EOTD, early-onset torsion dystonia; DAT, dopamine transporter; HA, hemagglutinin; DSP, dithiobis(succinimidylpropionate); AR, adrenergic receptor; D2DAR, D2 dopamine receptor; HEK, human embryonic kidney; IGF-IR, insulin-like growth factor I receptor; ER, endoplasmic reticulum; NET, norepinephrine transporter; EGFR, epidermal growth factor receptor.
References
- 1.Fahn, S., Bressman, S. B. & Marsden, C. D. (1998) Adv. Neurol. 78, 1-10. [PubMed] [Google Scholar]
- 2.Bragg, D. C., Slater, D. J. & Breakefield, X. O. (2004) Adv. Neurol. 94, 87-93. [PubMed] [Google Scholar]
- 3.Breakefield, X. O., Kamm, C. & Hanson, P. I. (2001) Neuron 31, 9-12. [DOI] [PubMed] [Google Scholar]
- 4.Klein, C., Breakefield, X. O. & Ozelius, L. J. (1999) Semin. Neurol. 19, 271-280. [DOI] [PubMed] [Google Scholar]
- 5.Ozelius, L. J., Hewett, J. W., Page, C. E., Bressman, S. B., Kramer, P. L., Shalish, C., de Leon, D., Brin, M. F., Raymond, D., Corey, D. P., et al. (1997) Nat. Genet. 17, 40-48. [DOI] [PubMed] [Google Scholar]
- 6.Todd, R. D. & Perlmutter, J. S. (1998) Mol. Neurobiol. 16, 135-147. [DOI] [PubMed] [Google Scholar]
- 7.Augood, S. J., Hollingsworth, Z., Albers, D. S., Yang, L., Leung, J., Breakefield, X. O. & Standaert, D. G. (2004) Adv. Neurol. 94, 53-60. [PubMed] [Google Scholar]
- 8.Hirano, M., Tamaru, Y., Nagai, Y., Ito, H., Imai, T. & Ueno, S. (1995) Biochem. Biophys. Res. Commun. 213, 645-651. [DOI] [PubMed] [Google Scholar]
- 9.Bandmann, O., Nygaard, T. G., Surtees, R., Marsden, C. D., Wood, N. W. & Harding, A. E. (1996) Hum. Mol. Genet. 5, 403-406. [DOI] [PubMed] [Google Scholar]
- 10.Furukawa, Y., Shimadzu, M., Rajput, A. H., Shimizu, Y., Tagawa, T., Mori, H., Yokochi, M., Narabayashi, H., Hornykiewicz, O., Mizuno, Y. & Kish, S.J. (1996) Ann. Neurol. 39, 609-617. [DOI] [PubMed] [Google Scholar]
- 11.Knappskog, P. M., Flatmark, T., Mallet, J., Ludecke, B. & Bartholome, K. (1995) Hum. Mol. Genet. 4, 1209-1212. [DOI] [PubMed] [Google Scholar]
- 12.van den Heuvel, L. P., Luiten, B., Smeitink, J. A., de Rijk-van Andel, J. F., Hyland, K., Steenbergen-Spanjers, G. C., Janssen, R. J. & Wevers, R. A. (1998) Hum. Genet. 102, 644-646. [DOI] [PubMed] [Google Scholar]
- 13.Klein, C., Brin, M. F., Kramer, P., Sena-Esteves, M., de Leon, D., Doheny, D., Bressman, S., Fahn, S., Breakefield, X. O. & Ozelius, L. J. (1999) Proc. Natl. Acad. Sci. USA 96, 5173-5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Augood, S. J., Penney, J. B., Jr., Friberg, I. K., Breakefield, X. O., Young, A. B., Ozelius, L. J. & Standaert, D. G. (1998) Ann. Neurol. 43, 669-673. [DOI] [PubMed] [Google Scholar]
- 15.Shashidharan, P., Kramer, B. C., Walker, R. H., Olanow, C. W. & Brin, M. F. (2000) Brain Res. 853, 197-206. [DOI] [PubMed] [Google Scholar]
- 16.Walker, R. H., Brin, M. F., Sandu, D., Gujjari, P., Hof, P. R., Warren Olanow, C. & Shashidharan, P. (2001) Brain Res. 900, 348-354. [DOI] [PubMed] [Google Scholar]
- 17.Ogura, T. & Wilkinson, A. J. (2001) Genes Cells 6, 575-597. [DOI] [PubMed] [Google Scholar]
- 18.Vale, R. D. (2000) J. Cell Biol. 150, F13-FF19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neuwald, A. F., Aravind, L., Spouge, J. L. & Koonin, E. V. (1999) Genome Res. 9, 27-43. [PubMed] [Google Scholar]
- 20.Kim, K. M., Valenzano, K. J., Robinson, S. R., Yao, W. D., Barak, L. S. & Caron, M. G. (2001) J. Biol. Chem. 276, 37409-37414. [DOI] [PubMed] [Google Scholar]
- 21.Wilbanks, A. M., Laporte, S. A., Bohn, L. M., Barak, L. S. & Caron, M. G. (2002) Biochemistry 41, 11981-11989. [DOI] [PubMed] [Google Scholar]
- 22.Giros, B., Wang, Y. M., Suter, S., McLeskey, S. B., Pifl, C. & Caron, M. G. (1994) J. Biol. Chem. 269, 15985-15988. [PubMed] [Google Scholar]
- 23.Barak, L. S., Menard, L., Ferguson, S. S., Colapietro, A. M. & Caron, M. G. (1995) Biochemistry 34, 15407-15414. [DOI] [PubMed] [Google Scholar]
- 24.Torres, G. E., Gainetdinov, R. R. & Caron, M. G. (2003) Nat. Rev. Neurosci. 4, 13-25. [DOI] [PubMed] [Google Scholar]
- 25.Hewett, J., Gonzalez-Agosti, C., Slater, D., Ziefer, P., Li, S., Bergeron, D., Jacoby, D. J., Ozelius, L. J., Ramesh, V. & Breakefield, X. O. (2000) Hum. Mol. Genet. 9, 1403-1413. [DOI] [PubMed] [Google Scholar]
- 26.Kustedjo, K., Bracey, M. H. & Cravatt, B. F. (2000) J. Biol. Chem. 275, 27933-27939. [DOI] [PubMed] [Google Scholar]
- 27.Mogk, A., Schlieker, C., Strub, C., Rist, W., Weibezahn, J. & Bukau, B. (2003) J. Biol. Chem. 278, 17615-17624. [DOI] [PubMed] [Google Scholar]
- 28.Hewett, J., Ziefer, P., Bergeron, D., Naismith, T., Boston, H., Slater, D., Wilbur, J., Schuback, D., Kamm, C., Smith, N., et al. (2003) J. Neurosci. Res. 72, 158-168. [DOI] [PubMed] [Google Scholar]
- 29.Shashidharan, P., Paris, N., Sandu, D., Karthikeyan, L., McNaught, K. S., Walker, R. H. & Olanow, C. W. (2004) J. Neurochem. 88, 1019-1025. [DOI] [PubMed] [Google Scholar]
- 30.Caldwell, G. A., Cao, S., Sexton, E. G., Gelwix, C. C., Bevel, J. P. & Caldwell, K. A. (2003) Hum. Mol. Genet. 12, 307-319. [DOI] [PubMed] [Google Scholar]
- 31.Green, W. N. & Millar, N. S. (1995) Trends Neurosci. 18, 280-287. [PubMed] [Google Scholar]
- 32.Gazi, L., Lopez-Gimenez, J. F. & Strange, P. G. (2002) Curr. Opin. Drug Discov. Devel. 5, 756-763. [PubMed] [Google Scholar]
- 33.Freeman, B. C. & Yamamoto, K. R. (2002) Science 296, 2232-2235. [DOI] [PubMed] [Google Scholar]
- 34.Naismith, T. V., Heuser, J. E., Breakefield, X. O. & Hanson, P. I. (2004) Proc. Natl. Acad. Sci. USA 101, 7612-7617. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.