Abstract
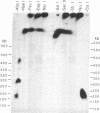
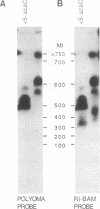
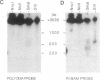
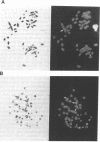
The organization of amplified DNA in mammalian cells in the form of inverted repeats rather than tandem repeats was first observed and studied in the 3B rat cell line. The structure and chromosomal location of the amplified inverted duplications in this cell line have been further analyzed by cloning, long-range mapping, and fluorescence in situ hybridization. The amplification unit is at least 450 kilobases in size and all of the amplicons are located in a single chromosomal location of approximately 10 or 11 megabases. No heterogeneity in either size or molecular structure is detected between the 3B amplicons, indicating that the 20- to 40-fold amplification occurred in a single event and not through a series of events, which would result in heterogeneity among the amplicons. Thus the amplification in 3B cells may reflect more closely the situation seen in tumors containing amplified oncogenes/protooncogenes than the amplifications present in cell lines after multiple selections with cytotoxic drugs. The progenitor Rat-2 cell line contains three alleles of the region of DNA that is amplified in 3B cells; two are located on the two normal homologues of rat chromosome 2 and the third is at the equivalent position on a marker chromosome, der(3)t(2;3). 3B cells contain only one of the two normal homologues of chromosome 2 in addition to chromosome der(3)t(2;3). All of the amplified DNA is located on a new marker chromosome, M2, whose amplified DNA region does not resemble chromosome 2. These results are consistent with the amplification model proposed by Passananti et al. [Passananti, C., Davies, B., Ford, M. & Fried, M. (1987) EMBO J. 6, 1697-1703], in which the excision from a chromosome of the DNA to be amplified results in the loss of rearrangement of that chromosome. In this model the excised DNA can be amplified extrachromosomally during a single S phase before becoming stabilized by integration into a chromosome, probably at a different location to that of its unamplified allele.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alitalo K., Schwab M. Oncogene amplification in tumor cells. Adv Cancer Res. 1986;47:235–281. doi: 10.1016/s0065-230x(08)60201-8. [DOI] [PubMed] [Google Scholar]
- Chu G., Vollrath D., Davis R. W. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science. 1986 Dec 19;234(4783):1582–1585. doi: 10.1126/science.3538420. [DOI] [PubMed] [Google Scholar]
- Ford M., Davies B., Griffiths M., Wilson J., Fried M. Isolation of a gene enhancer within an amplified inverted duplication after "expression selection". Proc Natl Acad Sci U S A. 1985 May;82(10):3370–3374. doi: 10.1073/pnas.82.10.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford M., Fried M. Large inverted duplications are associated with gene amplification. Cell. 1986 May 9;45(3):425–430. doi: 10.1016/0092-8674(86)90328-4. [DOI] [PubMed] [Google Scholar]
- Frischauf A. M., Lehrach H., Poustka A., Murray N. Lambda replacement vectors carrying polylinker sequences. J Mol Biol. 1983 Nov 15;170(4):827–842. doi: 10.1016/s0022-2836(83)80190-9. [DOI] [PubMed] [Google Scholar]
- Heard E., Fried M. The use of 5-azacytidine to increase cleavage of methylation sensitive rare cutting restriction enzymes sites in amplified DNA. Nucleic Acids Res. 1990 Oct 25;18(20):6147–6148. doi: 10.1093/nar/18.20.6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heartlein M. W., Latt S. A. Amplified inverted duplications within and adjacent to heterologous selectable DNA. Nucleic Acids Res. 1989 Feb 25;17(4):1697–1716. doi: 10.1093/nar/17.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt J. D., Valentine M., Tereba A. Excision of N-myc from chromosome 2 in human neuroblastoma cells containing amplified N-myc sequences. Mol Cell Biol. 1990 Feb;10(2):823–829. doi: 10.1128/mcb.10.2.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien O., Debatisse M., Buttin G., de Saint Vincent B. R. The multicopy appearance of a large inverted duplication and the sequence at the inversion joint suggest a new model for gene amplification. EMBO J. 1988 Feb;7(2):407–417. doi: 10.1002/j.1460-2075.1988.tb02828.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legouy E., Fossar N., Lhomond G., Brison O. Structure of four amplified DNA novel joints. Somat Cell Mol Genet. 1989 Jul;15(4):309–320. doi: 10.1007/BF01534970. [DOI] [PubMed] [Google Scholar]
- Levan G., Ahlström U., Mitelman F. The specificity of chromosome A2 involvement in DMBA-induced rat sarcomas. Hereditas. 1974;77(2):263–280. doi: 10.1111/j.1601-5223.1974.tb00939.x. [DOI] [PubMed] [Google Scholar]
- Levan G. Nomenclature for G-bands in rat chromosomes. Hereditas. 1974;77(1):37–52. doi: 10.1111/j.1601-5223.1974.tb01352.x. [DOI] [PubMed] [Google Scholar]
- Looney J. E., Hamlin J. L. Isolation of the amplified dihydrofolate reductase domain from methotrexate-resistant Chinese hamster ovary cells. Mol Cell Biol. 1987 Feb;7(2):569–577. doi: 10.1128/mcb.7.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalbantoglu J., Meuth M. DNA amplification--deletion in a spontaneous mutation of the hamster aprt locus: structure and sequence of the novel joint. Nucleic Acids Res. 1986 Nov 11;14(21):8361–8371. doi: 10.1093/nar/14.21.8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passananti C., Davies B., Ford M., Fried M. Structure of an inverted duplication formed as a first step in a gene amplification event: implications for a model of gene amplification. EMBO J. 1987 Jun;6(6):1697–1703. doi: 10.1002/j.1460-2075.1987.tb02420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkel D., Straume T., Gray J. W. Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc Natl Acad Sci U S A. 1986 May;83(9):2934–2938. doi: 10.1073/pnas.83.9.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz J. C., Wahl G. M. Formation of an inverted duplication can be an initial step in gene amplification. Mol Cell Biol. 1988 Oct;8(10):4302–4313. doi: 10.1128/mcb.8.10.4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito I., Stark G. R. Charomids: cosmid vectors for efficient cloning and mapping of large or small restriction fragments. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8664–8668. doi: 10.1073/pnas.83.22.8664. [DOI] [PMC free article] [PubMed] [Google Scholar]