Abstract
Decorin is a ubiquitous extracellular matrix proteoglycan with a variety of important biological functions that are mediated by its interactions with extracellular matrix proteins, cytokines, and cell surface receptors. Decorin is the prototype of the family of small leucine-rich repeat proteoglycans and proteins (SLRPs), characterized by a protein core composed of leucine-rich repeats (LRRs), flanked by two cysteine-rich regions. We report here the crystal structure of the dimeric protein core of decorin, the best characterized member of the SLRP family. Each monomer adopts the curved solenoid fold characteristic of LRR domains, with a parallel β-sheet on the inside interwoven with loops containing short segments of β-strands, 310 helices, and polyproline II helices on the outside. Two main features are unique to this structure. First, decorin dimerizes through the concave surfaces of the LRR domains, which have been implicated previously in protein-ligand interactions. The amount of surface buried in this dimer rivals the buried surfaces of some of the highest-affinity macromolecular complexes reported to date. Second, the C-terminal region adopts an unusual capping motif that involves a laterally extended LRR and a disulfide bond. This motif seems to be unique to SLRPs and has not been observed in any other LRR protein structure to date. Possible implications of these features for decorin ligand binding and SLRP function are discussed.
Decorin is a small extracellular matrix proteoglycan present in a variety of connective tissues, typically in association with or “decorating” collagen fibrils (1, 2, 3). It is involved in several fundamental biological functions, including the formation and/or organization of collagen fibrils (4, 5) and the modulation of cell adhesion mediated by fibronectin and thrombospondin (6). Decorin also modulates the activity of growth factors, such as transforming growth factor-β (7), and has other, transforming growth factor-β-independent effects on cell proliferation and behavior (8, 9).
Mammalian decorin contains a protein core and a single chondroitin/dermatan sulfate glycosaminoglycan (GAG) chain, attached to a serine residue near the N terminus (10). Decorin is the best characterized member of the growing family of small leucine-rich repeat proteoglycans and proteins (SLRPs) (3, 11), all having a domain of tandem leucine-rich repeats (LRRs), flanked on either side by clusters of conserved Cys residues. Most SLRPs have been grouped into three different classes on the basis of gene organization, amino acid sequence similarity, number of LRRs, and the spacing of Cys residues in the N-terminal segment. Thus, class I includes decorin, biglycan, and asporin; class II includes fibromodulin, osteoadherin, lumican, proline arginine-rich end LRR protein (PRELP), and keratocan; and class III includes opticin, osteoglycin/mimecan, and epiphycan/PGLb (3, 12). Three further proteins, extracellular matrix 2 (ECM2), chondroadherin, and nyctalopin, have LRR domains with significant homology to the SLRP family (12). The structural and functional similarities between different SLRPs suggest that they share biological functions. For instance, several SLRPs are known to regulate collagen fibrillogenesis, and there is evidence that they are able to compensate for each other in studies on knockout mice (11). Conversely, the wide variation in their expression patterns would indicate that their functions are regulated in a cell- or tissue-specific manner.
The LRR motif is very widely distributed and has been found in >100 intracellular, cell surface, and extracellular proteins (the LRR superfamily) (13, 14). Several crystal structures of LRR domains have been determined (15, 16, 17, 18, 19). All of them adopt a curved solenoid fold, with a parallel β-sheet forming the inner concave face and a variety of secondary structure topologies forming the outer convex face. To date, crystal structures of complexes of LRR domains with their protein ligands have shown that the concave surface contains the ligand-binding sites (16, 20, 21). It has been assumed that decorin and SLRPs interact with their ligands in the same way (3, 22). However, biophysical analyses have demonstrated that decorin is dimeric in solution, and low-angle x-ray scattering data have suggested that the concave surfaces are involved in dimerization, potentially making them unavailable for ligand-binding (23). A recent article suggested that decorin is in fact a monomer and that dimerization is artifactual (24). However, the crystal structure of the decorin dimer presented here confirms that the concave surfaces mediate dimerization in a highly specific and conserved manner, almost certainly precluding artifact. The highly specific self-recognition by an LRR domain suggests that current models of decorin-ligand interactions need to be reevaluated.
Materials and Methods
Sample Preparation, Characterization, and Crystallization. Two different decorin samples have been used in this study: a recombinant decorin (DcnR), expressed in HEK 293A cells and purified without chaotropic agents, and a tissue-derived decorin (DcnT), extracted from calfskin and refolded from solutions containing urea. Both forms have been shown to be biologically active as they interact with collagen, inhibit collagen fibrillogenesis, and inhibit fibroblast proliferation (9, 25) (Fig. 5, which is published as supporting information on the PNAS web site). The core proteins were prepared by removal of the GAG chain as described in ref. 23. Further details of the biochemical characterization of both samples are given in Supporting Methods, which is published as supporting information on the PNAS web site. Both light-scattering experiments (23) and sedimentation equilibrium (Fig. 6, which is published as supporting information on the PNAS web site) indicate that decorin is dimeric in solution.
Crystals of both DcnT and DcnR were grown at 20°C by vapor-diffusion methods. Hanging drops (10 μl) were prepared by mixing equal volumes of protein solution (2-3 mg/ml) and 25% (vol/vol) polyethylene glycol 400, both in 0.06 M Tris (pH 7.75)/0.01% β-octyl d-glucoside/0.02% sodium azide. The drops were allowed to equilibrate against 1 ml of the same polyethylene glycol solution. Orthorhombic plate-like crystals appeared within 2 or 3 days and grew to 0.2-0.3 mm in the longest dimension in ≈2 weeks. Crystals suitable for x-ray diffraction were mounted in cryoloops (Hampton Research, Aliso Viejo, CA), flash-cooled, and stored in liquid nitrogen until used for data collection. Derivatives were prepared by soaking crystals overnight in 0.5 mM mercury(II) acetate dissolved in precipitant and then soaking in precipitant solution for several minutes immediately before flash-cooling and x-ray diffraction.
Structure Determination and Refinement. Both decorin forms gave crystals in two different space groups, C2221 and P212121, which were indistinguishable by visual inspection. Several complete sets of data from native and derivatized DcnR and DcnT crystals were collected at different in-house and synchrotron sources, and structures were determined for DcnR and DcnT in each space group. Tables 1 and 2 show the statistics for the two most representative forms, and additional data are summarized in Table 3, which is published as supporting information on the PNAS web site. See Supporting Methods for details of data collection and processing for all crystal forms. The native data for C2221 DcnR were collected with very high redundancy (Table 1), and anomalous data were measured.
Table 1. Data collection statistics and phasing statistics for DcnR in the C2221 form and DcnT in the P212121 form.
| DcnR (recombinant)
|
DcnT (tissue-extracted)
|
||
|---|---|---|---|
| Decorin source | Native C | Hg derivative C | Native P |
| Data collection | |||
| Detector type | R-AXIS IV++* | R-AXIS IV++* | ADSC CCD† |
| Wavelength, Å | 1.5418 | 1.5418 | 0.9780 |
| Resolution, Å | 33.2-2.15 | 44.7-2.30 | 19.7-2.30 |
| Space group | C2221 | C2221 | P212121 |
| a, Å | 55.78 | 55.83 | 52.70 |
| b, Å | 124.15 | 124.10 | 120.95 |
| c, Å | 129.61 | 129.46 | 129.71 |
| Measured reflections‡ | 263,703 (27,165) | 137,083 (13,255) | 208,796 (14,199) |
| Unique reflections‡ | 24,593 (3,266) | 20,244 (2,763) | 34,837 (3024) |
| Completeness,‡ % | 98.6 (93.7) | 99.6 (99.6) | 96.7 (83.5) |
| Multiplicity‡ | 10.7 (8.2) | 6.6 (4.7) | 5.2 (4.7) |
| Rsym,‡ % | 4.8 (36.7) | 5.1 (30.4) | 9.0 (19.9) |
| I/σ‡ | 8.7 (2.1) | 11.5 (2.5) | 6.3 (2.9) |
| Phasing | |||
| Riso, % | 11.5 | ||
| Maximum resolution, Å | 2.70 | ||
| Number of sites | 6 | ||
| Phasing power§ | 0.96/1.16/0.65 | ||
| Figure of merit | 0.599 | ||
Institute for Biomolecular Design, University of Alberta
Synchrotron Radiation Source, beamline 14.2, Daresbury Laboratory (Cheshire, U.K.)
Numbers in parentheses represent values in the highest-resolution shell
Values for centric/acentric/anomalous data
Table 2. Final refinement statistics.
| DcnR C2221 | DcnT P212121 | |
|---|---|---|
| Refinement | ||
| Resolution range, Å | 33.2-2.15 | 19.7-2.30 |
| R factor, % | 20.7 (25.3) | 23.0 (30.9) |
| Rfree, % | 23.5 (28.6) | 27.3 (35.5) |
| Final model | ||
| Protein atoms | 2,370 | 4,732 |
| Sugar atoms | 42 | 70 |
| Water molecules | 165 | 177 |
| rms deviation | ||
| Bond lengths, Å | 0.009 | 0.006 |
| Bond angles, ° | 1.4 | 1.3 |
| Improper torsions, ° | 0.92 | 0.81 |
Phases for the C2221 DcnR form were determined by using single isomorphous replacement and anomalous scattering (SIRAS) methods. A first mercury site was located manually in an isomorphous difference Patterson map. A second major mercury site was located by using xhercules from xtalview (26), and four more (minor) sites were located in difference Fourier maps calculated by using sharp (27). Initial SIRAS phases calculated by using sharp were improved by density modification by using resolve (28) with a nominal solvent content of 50% (optimized by trial and error). The resulting electron density maps showed a solvent-protein boundary and an internal structure in the protein electron density consistent with an LRR domain. Essentially identical maps were obtained for the C2221 DcnT form by using a single mercury derivative (Table 3). All model building was carried out on the DcnR form, using the programs xfit (26) and chain (29). The orientation of the peptide chain and the positions of the N- and C-terminal disulfides were unambiguously assigned with the help of an anomalous Fourier map, calculated by using anomalous differences measured in the DcnR native data (Table 1) and the density-modified SIRAS phases. This anomalous difference map showed clearly 9 of 10 sulfur atoms present in the asymmetric unit. No sulfur peak was observed for Met-21, probably because of N-terminal conformational disorder.
The map obtained from density-modified SIRAS phases was sufficiently connected to build a partial model of the concave side of the central 10 LRRs. To help in model building on the convex side, density-modified SIRAS phases were further improved by repeat-to-repeat map averaging, using the rave suite (30). Model fitting was routinely checked by simulated-annealing omit maps calculated by using cns (31). Rebuilding and extension of the model eventually led to the fitting of residues 22-326 of mature decorin (which correspond to amino acids 52-356 of the unprocessed decorin gene product). Water molecules were added to difference maps at a late stage in the refinement (R factor < 0.25) and kept only when showing both clear density in 2mFo - DFc maps and good hydrogen-bonding connectivity. Final refinement (using cns) was carried out by simulated annealing dynamics and energy minimization, using a maximum likelihood target function. The final model contains residues 22-326 of the mature protein and the first N-acetylglucosamine sugar residue on each of three N-linked oligosaccharides (Asn residues 182, 233, and 274). No ordered density was observed for either the 14-aa propeptide (see Supporting Methods) or residues 1-21 of the mature protein, which contain the GAG attachment site. Western blotting of protein recovered from crystals, using a mAb whose epitope comprises residues 6-10 of mature decorin (32), confirmed that this sequence had not been lost by proteolysis and suggested that the lack of interpretable density was due to conformational disorder. Ribbon and molecular diagrams were prepared with setor (33). Surface representations were prepared with pymol (34).
Structures of DcnT and DcnR in the P212121 crystal form were determined by molecular replacement using amore (35) and molrep (36), with a probe generated from the refined 2.15-Å DcnR C2221. Refinement proceeded as with the C2221 forms, and no significant differences were observed between the DcnT and DcnR types. Final statistics for the P212121 crystal forms are included in Tables 2 (DcnT) and 3 (DcnR).
Results and Discussion
Overall Structure. The refined structures for DcnR and DcnT, although varying in quality, were not significantly different. All figures and the analysis presented hereafter are therefore based on the highest-quality crystal structure, DcnR refined to 2.15 Å in the C2221 space group (Table 2). In all crystal forms, DcnR and DcnT dimerize in the antiparallel arrangement shown in Fig. 1a. In the C2221 crystal forms, the dimer axis is aligned with a twofold crystallographic axis. In the P212121 crystal forms, the dimer axis is tilted ≈10° with respect to the same crystallographic axis (data not shown). Otherwise, the two dimers in the different crystal forms are indistinguishable (dimer-to-dimer Cα rms deviations 0.32-0.36 Å).
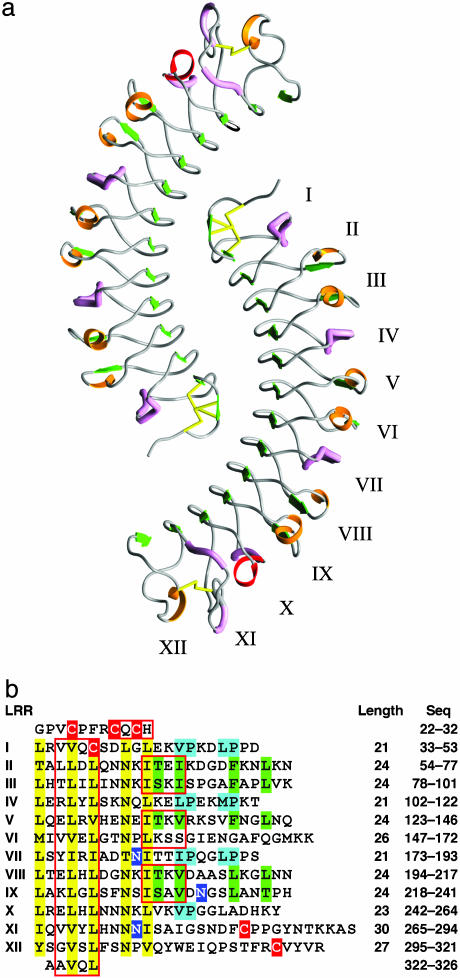
Fig. 1.
Structure of the LRR domain of bovine decorin. (a) Ribbon diagram of the dimeric structure of decorin LRR domain. Green arrows, β-strands; red ribbons, α-helical turns; pink tubes, segments of polyproline II helix; orange ribbons, short segments of 310 helices and β-turns; yellow sticks, disulfide bonds. (b) Internal organization of bovine decorin LRRs (residues 22-326). Yellow highlight, LRR consensus residues; red highlight, Cys residues; green highlight, consensus residues for the 24-aa repeat; cyan highlight, consensus residues for the 21-aa repeat; blue highlight, Asn residues with oligosaccharide substituents; red boxes, amino acids that contribute to β-sheets.
Each monomer is a single-domain structure with the right-handed, curved solenoid fold characteristic of LRR proteins (Fig. 1a). Each domain contains 12 LRRs (numbered I to XII), flanked by a β-hairpin at the N terminus and an additional β-strand at the end of LRR-XII (Fig. 1b). The long β-sheet that forms the inner, concave face comprises 14 β-strands. Each LRR contributes one β-strand (β1-β12) before coiling its way toward the next repeat. The hairpin at the N terminus provides an additional β-strand (β0) that is the only one running antiparallel to the rest, and the final strand β13 knits the C terminus of the solenoid closed. The outer, or convex, face is defined by the less-conserved parts of the LRRs, which adopt a variety of secondary structure motifs (see below).
The 12 LRRs vary in length from 21 to 30 aa, following a short-long-long regular pattern throughout the molecule (Fig. 1b). The first nine LRRs show an almost perfect 21-24-24 pattern, with LRR-VI, at the center of the molecule, having two additional amino acids to give a length of 26. The last three LRRs also show a short-long-long pattern, this time 23-30-27. The structural elements on the convex face vary according to the repeat length. The shortest LRRs (21 residues) show two short segments in a polyproline II conformation. The most common LRRs (24 residues) show one short β-strand followed by a variable region with β-turns and short segments of 310 helix. The rest of the LRRs appear to be intermediate between these two types. Wherever two 24-residue LRRs are adjacent, a short parallel β-sheet occurs at the convex side (Fig. 1b), a feature that also happens between repeats V and VI, which are 24 and 26 residues long, respectively. Based on hydrogen-bonding connectivity, repeat X contains a single turn of α-helix.
The overall curvature of the decorin LRR domain is consistent with that seen in other crystal structures of LRR proteins with repeats of similar length (16, 17, 18, 19). These “banana”-shaped molecules (18) contrast with the more closed “horseshoe”-like structure of ribonuclease inhibitor (RNI), which previously had been used to generate a homology model for decorin (22). RNI differs from these other LRR proteins in that it contains longer repeats (28-29 aa) with α-helices on the convex side imposing a tight curvature.
The N-terminal capping motif contains a cluster of Cys residues conserved in the SLRP family and buries the hydrophobic core of the first LRR. Four Cys residues form a disulfide knot (Cys-25-Cys-31 and Cys-29-Cys-38) between the β-hairpin and LRR-I. This N-terminal capping motif, essentially equivalent to the one seen in the Nogo receptor (18, 19), does not form a separate domain but integrates seamlessly in the LRR architecture.
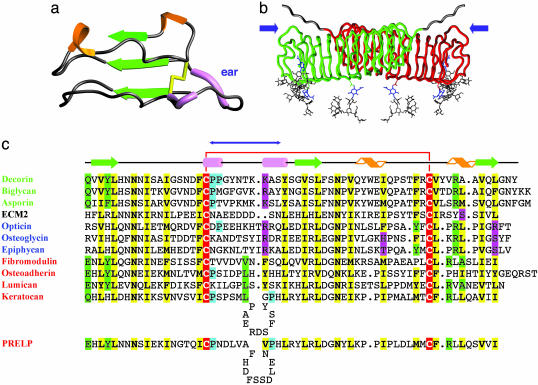
“Ear” Repeats and the C-Terminal Disulfide. LRR-XI is the longest repeat in decorin (Fig. 1b). This repeat, which we will refer to as the “ear” repeat, extends laterally from the main body of the dimer (Fig. 2) and contains a conserved Cys residue that forms a disulfide bond with another Cys residue in the final LRR-XII (Cys-284-Cys-317) (Fig. 2 a and c). The ear repeat, which is always second to last, seems to be a distinctive feature of the SLRP family (Fig. 2c), with the ear itself spanning from the first conserved C-terminal Cys to the beginning of the last LRR. This feature has not been observed in other LRR structures reported to date. Ear repeats have different lengths (Fig. 2c), typically 30 (classes I and III) or 31 (class II) aa. Class II SLRPs keratocan and PRELP have especially long ear repeats, with 38 and 39 aa respectively. ECM2 has an ear repeat of 29 aa. Two other SLRPs, chondroadherin and nyctalopin, seem to use a different type of C-terminal disulfide capping.
Fig. 2.
Topology and secondary structure of the C-terminal capping motif. (a) Ribbon diagram of the C-terminal capping motif of decorin. This motif includes LRR-XI, LRR-XII, and the additional C-terminal strand β13. Secondary structural elements are depicted as in Fig. 1. (b) Rope diagram of a glycosylated, extended model of the decorin dimer, showing the position of the two ear repeats (blue arrows). Extended oligosaccharide chains have been modeled (gray) onto the three N-acetylglucosamine residues (blue) to illustrate their approximate size and general positioning relative to the dimer surface. Both N termini also have been extended a few residues (gray) to illustrate the general sense of directionality. The GAG chains are not included in the model. (c) Structure-based alignment for the C-terminal motifs. Green text, class I SLRPs; red text, class II SLRPs; blue text, class III SLRPs. Yellow, conserved residues; red, conserved Cys residues; green, partially conserved residues; cyan, partially conserved Pro residues indicative of polyproline II conformation; magenta, polar residues occupying hallmark LRR hydrophobic sites. The blue double-headed arrow indicates the extent of the ear. Ear repeats in keratocan and PRELP are significantly longer than those for all other SLRPs and, therefore, are shown as containing insertion loops. The decorin sequence is that of the bovine protein; all others are of human proteins.
The first 18-19 residues in each ear repeat follow a conserved pattern similar to that in other LRRs, with hydrophobic residues pointing in and polar residues pointing out. Residues within the ear per se are not highly conserved (Fig. 2c), suggesting the possibility of functional specialization, such as ligand binding, in different SLRPs. The decorin ear contains two short segments of polyproline II conformation, but the structure is probably different in other SLRPs. The last four residues return to a more conventional LRR architecture.
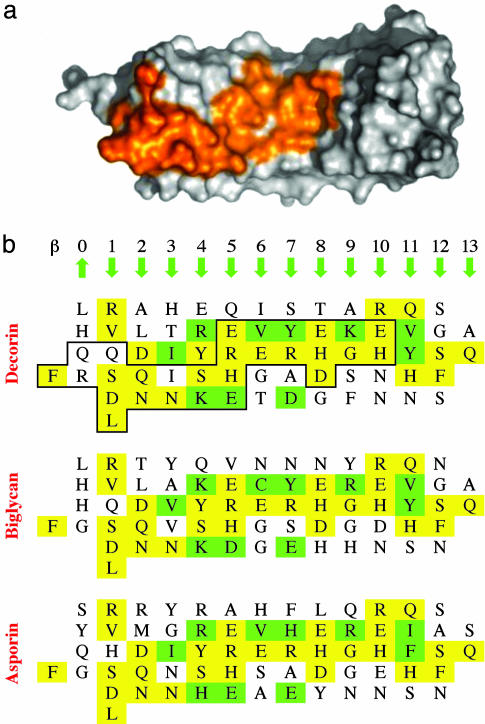
The Dimer Interface. The two monomers in the dimer interact through their concave faces (Fig. 1). The dimer interface is contiguous and extends from the N-terminal capping to more than three-quarters of the length of the concave face β-sheet (Fig. 3). The total buried surface in the dimer is ≈2,300 Å2, comparable to that in complexes of RNI with ribonuclease or angiogenin (2,600 Å2) or glycoprotein Ibα with von Willebrand factor A1 domain (2,100 Å2) (16, 20, 21). Such large contact surfaces are associated with very high binding affinities, such as in the RNI-angiogenin interaction (Ki < 1 fM) (21). The large contact area in the decorin dimer interface thus seems consistent with the high affinity (subnanomolar) estimated from light-scattering experiments (23). Conservation of surface residues at the concave face of class I SLRPs follows closely the footprint of the dimer interface (Fig. 3b), suggesting that both biglycan and asporin may dimerize in the same way (see below).
Fig. 3.
Extent and sequence conservation of the dimer interface. (a) View of the concave side of a decorin monomer. Residues that are buried from solvent in the dimer are shown in orange. (b) Two-dimensional representation of the surface residues at the concave side of class I SLRPs. Yellow, residues fully conserved in all three SLRPs; green, partially conserved residues; black outline, the footprint of the decorin dimerization interface. The relative positions and directions of the 14 β-strands that form the concave side β-sheet are indicated.
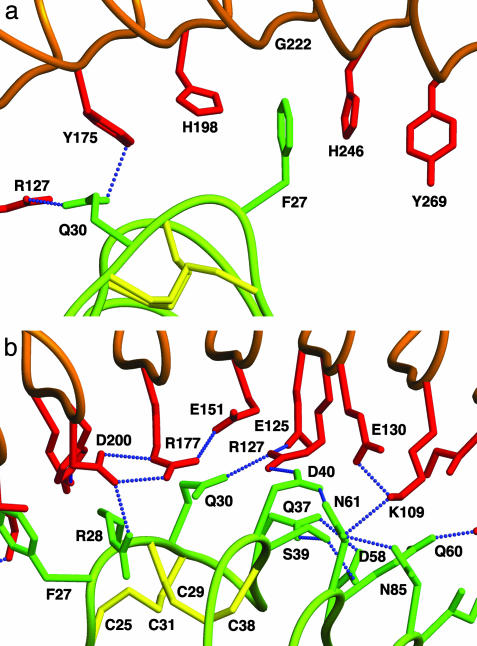
Amino acids involved in direct contacts at the dimer interface belong to the first β-turn in the N-terminal capping and to strands from β0 to β10. The extent and nature of these interactions is consistent with a high-affinity complex: several aromatic and hydrophobic residues are buried, and there are several salt bridges and extensive hydrogen-bonding between the two monomers (Fig. 4). Moreover, many structural water molecules participate in the dimer interaction by bridging additional groups from the two monomers (Fig. 7a, which is published as supporting information on the PNAS web site). The aromatic ring of Phe-27, in the N-terminal capping of one monomer, is sandwiched between two His aromatic rings from β8 and β10 of the other monomer (Fig. 4a). Rings from His-246 and Phe-27 are stacked against each other, whereas the interaction between His-198 and Phe-27 is a CH···π hydrogen bond. Strand β10 carries residue Gly-222 in the position that corresponds to the two His residues above, leaving exactly the space required for Phe-27 to intercalate. This sandwich of rings is part of a six-layered group of residues forming a hydrophobic array (Fig. 4a). This noteworthy structural feature is contiguous to an extensive network of hydrogen bonds, salt bridges, and additional hydrophobic interactions between the monomers (Fig. 4b).
Fig. 4.
Molecular interactions at the dimer interface. (a) The aromatic ring of Phe-27 in one monomer (green) becomes intercalated between the aromatic rings of two His residues in the other monomer (red). This hydrophobic sandwich is part of an extended hydrophobic array (see text). (b) Extensive hydrogen-bonding networks (blue dotted lines) occur between the two monomers.
It is of interest that His residues are critically involved in the dimer interface, because their ionization state could potentially modulate dimer formation (pI 7.47). We did, however, obtain crystals with identical morphology over a wide range of pH values, between 6 and 9 (data not shown). Furthermore, the light and x-ray scattering experiments were performed at pH 7.0 (23). These observations would argue against the charge of His residues having a critical effect on dimer stability.
The arrangement of monomers in the crystal structure is consistent with the x-ray scattering profile of decorin core protein in solution (23). This finding strongly suggests that the core protein of decorin crystallizes from preexisting dimers in solution identical to those in the crystalline lattice. This dimer arrangement is perfectly compatible with the GAG-containing intact proteoglycan, because both N-terminal ends point away from the dimer interface (Fig. 2b).
Biglycan has been reported to form dimers reversibly in solution (37), and recent evidence from our laboratories indicates that opticin, a class III SLRP, also exists as a stable dimer in solution (38). An analysis of the conservation of residues involved in decorin dimerization across the SLRP family strongly suggests that the three class I SLRPs dimerize in the same way (Fig. 3b). In particular, most of the residues depicted in Fig. 4 are conserved in biglycan and asporin, including the intercalation pair Phe-27-Gly-222. An interesting exception is Arg-28 (Gly in both biglycan and asporin). This residue adopts a strained αL conformation in decorin (data not shown), which could be released by a change to Gly in the same position.
A recent paper (24) suggests that biologically active decorin is monomeric and that dimerization is an artifact of lyophilization. We used the same decorin forms for biophysical solution studies (23) (Fig. 6) and crystallization and have shown them to be biologically active (Fig. 5). In addition, we have prepared DcnR without freezing or lyophilization at any stage and verified that it is entirely dimeric (P.G.S., unpublished data). The crystal structure now confirms the quaternary arrangement proposed on the basis of x-ray scattering (23) and shows a very specific dimerization interface, which is incompatible with nonspecific multimerization.
Implications for Decorin-Ligand Interactions. It has been widely assumed that horseshoe-shaped SLRPs interact with collagen molecules through their concave surfaces and that the inner space in the horseshoe molecule can accommodate only a single collagen triple helix (11, 22). Our findings challenge this view. The decorin LRR domains are banana-shaped rather than horseshoe-shaped, which is likely to be the case for all SLRPs. This more open structure simply results from the shorter LRR length in all SLRPs compared with the LRR length in the RNI structure, and it seems incompatible with a tight interaction with a single collagen triple helix. Furthermore, at least in the case of decorin, the concave surface is involved in a high-affinity dimer interaction and, therefore, is unlikely to be available for ligand binding, although available data cannot rule out the possibility of dimer-to-monomer transitions in decorin-ligand interactions. Analysis of electrostatic charge distribution does not indicate any obvious clustering of charged residues that could suggest a ligand-binding site (Fig. 7b). Assuming that decorin binds ligands as a dimer, the disposition of N-oligosaccharides across one side of the dimer (Fig. 2b) suggests that this surface is not involved in protein-protein interactions. Conservation analysis across class I SLRPs does reveal a clustering of partially conserved residues on the sugar-free surface of LRRs IV-VI (Fig. 7c), a region that has been implicated in collagen binding (39). Further biochemical studies are needed to explore the molecular basis of the interactions between decorin and its many ligands. This crystal structure provides a valuable foundation for such studies.
Supplementary Material
Acknowledgments
This work was supported by the Canadian Institutes of Health Research (P.G.S.), The Wellcome Trust (P.G.S., P.N.B., and J.B), and the United Kingdom Biotechnology and Biological Sciences Research Council (P.A.M. and J.B.). Access to synchrotron beam time in Canada (Alberta Synchrotron Institute), the United States (Advanced Light Source, Lawrence Berkeley National Laboratory, Berkeley, CA), and the United Kingdom (Daresbury Laboratory, Cheshire, U.K.) was supported by the Alberta Heritage Foundation for Medical Research, the Alberta Science and Research Authority, the U.S. National Science Foundation, and the United Kingdom Biotechnology and Biological Sciences Research Council.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: DcnR, recombinant decorin; DcnT, tissue-derived decorin; GAG, chondroitin/dermatan sulfate glycosaminoglycan; LRR, leucine-rich repeat; RNI, ribonuclease inhibitor; SIRAS, single isomorphous replacement and anomalous scattering; SLRP, small leucine-rich repeat proteoglycans and protein.
Data deposition: The atomic coordinates and structure factors have been deposited in the KU/CD Protein Data Bank, www.pdb.org (PDB ID codes 1XKU, 1XEC, and 1XCD).
References
- 1.Krusius, T. & Ruoslahti, E. (1986) Proc. Natl. Acad. Sci. USA 83, 7683-7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bianco, P., Fisher, L. W., Young, M. F., Termine, J. D. & Robey, P. G. (1990) J. Histochem. Cytochem. 38, 1549-1563. [DOI] [PubMed] [Google Scholar]
- 3.Hocking, A. M., Shinomura, T. & McQuillan, D. J. (1998) Matrix Biol. 17, 1-19. [DOI] [PubMed] [Google Scholar]
- 4.Scott, J. E. (1992) FASEB J. 6, 2639-2645. [PubMed] [Google Scholar]
- 5.Danielson, K. G., Baribault, H., Holmes, D. F., Graham, H., Kadler, K. E. & Iozzo, R. V. (1997) J. Cell Biol. 136, 729-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Winnemoller, M., Schon, P., Vischer, P. & Kresse, H. (1992) Eur. J. Cell Biol. 59, 47-55. [PubMed] [Google Scholar]
- 7.Yamaguchi, Y., Mann, D. M. & Ruoslahti, E. (1990) Nature 346, 281-284. [DOI] [PubMed] [Google Scholar]
- 8.Iozzo, R. V., Moscatello, D. K., McQuillan, D. J. & Eichstetter, I. (1999) J. Biol. Chem. 274, 4489-4492. [DOI] [PubMed] [Google Scholar]
- 9.Hakkinen, L., Strassburger, S., Kahari, V. M., Scott, P. G., Eichstetter, I., Iozzo, R. V. & Larjava, H. (2000) Lab. Invest. 80, 1869-1880. [DOI] [PubMed] [Google Scholar]
- 10.Chopra, R. K., Pearson, C. H., Pringle, G. A., Fackre, D. S. & Scott, P. G. (1985) Biochem. J. 232, 277-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ameye, L. & Young, M. F. (2002) Glycobiology 12, 107R-116R [DOI] [PubMed] [Google Scholar]
- 12.Henry, S. P., Takanosu, M., Boyd, T. C., Mayne, P. M., Eberspaecher, H., Zhou, W., de Crombrugghe, B., Hook, M. & Mayne, R. (2001) J. Biol. Chem. 276, 12212-12221. [DOI] [PubMed] [Google Scholar]
- 13.Kobe, B. & Deisenhofer, J. (1994) Trends Biochem. Sci. 19, 415-421. [DOI] [PubMed] [Google Scholar]
- 14.Buchanan, S. G. S. C. & Gay, N. J. (1996) Prog. Biophys. Mol. Biol. 65, 1-44. [DOI] [PubMed] [Google Scholar]
- 15.Kobe, B. & Kajava, A. V. (2001) Curr. Opin. Struct. Biol. 11, 725-732. [DOI] [PubMed] [Google Scholar]
- 16.Huizinga, E. G., Tsuji, S., Romijn, R. A., Schiphorst, M. E., de Groot, P. G., Sixma, J. J. & Gros, P. (2002) Science 297, 1176-1179. [DOI] [PubMed] [Google Scholar]
- 17.Uff, S., Clemetson, J. M., Harrison, T., Clemetson, K. J. & Emsley, J. (2002) J. Biol. Chem. 277, 35657-35663. [DOI] [PubMed] [Google Scholar]
- 18.Barton, W. A., Liu, B. P., Tzvetkova, D., Jeffrey, P. D., Fournier, A. E., Sah, D., Cate, R., Strittmatter, S. M. & Nikolov, D. B. (2003) EMBO J. 22, 3291-3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xe, X. L., Bazan, J. F., McDermott, G., Park, J. B., Wang, K., Tessier-Lavigne, M., He, Z. & Garcia, K. C. (2003) Neuron 38, 177-185. [DOI] [PubMed] [Google Scholar]
- 20.Kobe, B. & Deisenhofer, J. (1995) Nature 374, 183-186. [DOI] [PubMed] [Google Scholar]
- 21.Papageorgiou, A. C., Shapiro, R. & Acharya, K. R. (1997) EMBO J. 16, 5162-5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weber, I. T., Harrison, R. W. & Iozzo, R. V. (1996) J. Biol. Chem. 271, 31767-31770. [DOI] [PubMed] [Google Scholar]
- 23.Scott, P. G., Grossmann, J. G., Dodd, C. M., Sheehan, J. K. & Bishop, P. N. (2003) J. Biol. Chem. 278, 18353-18359. [DOI] [PubMed] [Google Scholar]
- 24.Goldoni, S., Owens, R. T., McQuillan, D. J., Shriver, Z., Sasisekharan, R., Birk, D. E., Campbell, S. & Iozzo, R. V. (2004) J. Biol. Chem. 279, 6606-6612. [DOI] [PubMed] [Google Scholar]
- 25.Scott, P. G., Winterbottom, N., Dodd, C. M., Edwards, E. & Pearson, C. H. (1986) Biochem. Biophys. Res. Commun. 138, 1348-1354. [DOI] [PubMed] [Google Scholar]
- 26.McRee, D. E. (1999) J. Struct. Biol. 125, 155-165. [DOI] [PubMed] [Google Scholar]
- 27.de La Fortelle, E. & Bricogne, G. (1997) Methods Enzymol. 276, 472-494. [DOI] [PubMed] [Google Scholar]
- 28.Terwilliger, T. C. (2001) Acta Crystallogr. D 57, 1763-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sack, J. S. (1988) J. Mol. Graphics 6, 224-225. [Google Scholar]
- 30.Kleywegt, G. J. & Jones, T. A. (1994) in From First Map to Final Model, eds. Bailey, S., Hubbard, R. & Waller, D. A. (Science and Engineering Research Council Daresbury Lab., Daresbury, U.K.), pp. 59-66.
- 31.Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kuntsleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 32.Scott, P. G., Dodd, C. M. & Pringle, G. A. (1993) J. Biol. Chem. 268, 11558-11564. [PubMed] [Google Scholar]
- 33.Evans, S. V. (1993) J. Mol. Graphics 11, 134-138. [DOI] [PubMed] [Google Scholar]
- 34.DeLano, W. L. (2002) The pymol Molecular Graphics System (DeLano Scientific, San Carlos, CA).
- 35.Navaza, J. (1994) Acta Crystallogr. A 50, 157-163. [Google Scholar]
- 36.Vagin, A. & Teplyakov, A. (1997) J. Appl. Crystallogr. 30, 1022-1025. [Google Scholar]
- 37.Liu, J., Laue, T. M., Choi, H. U., Tang, L. H. & Rosenberg, L. (1994) J. Biol. Chem. 269, 28366-28373. [PubMed] [Google Scholar]
- 38.Le Goff, M. M., Hindson, V. J., Jowitt, T. A., Scott, P. G. & Bishop, P. N. (2003) J. Biol. Chem. 278, 45280-45287. [DOI] [PubMed] [Google Scholar]
- 39.Svensson, L., Heinegard, D. & Oldberg, A. (1995) J. Biol. Chem. 270, 20712-20716. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.