Abstract
Angiotensin-converting enzyme 2 (ACE2) is a receptor for SARS-CoV, the novel coronavirus that causes severe acute respiratory syndrome [Li, W. Moore, M. J., Vasilieva, N., Sui, J., Wong, S. K., Berne, M. A., Somasundaran, M., Sullivan, J. L., Luzuriaga, K., Greenough, T. C., et al. (2003) Nature 426, 450–454]. We have identified a different human cellular glycoprotein that can serve as an alternative receptor for SARS-CoV. A human lung cDNA library in vesicular stomatitis virus G pseudotyped retrovirus was transduced into Chinese hamster ovary cells, and the cells were sorted for binding of soluble SARS-CoV spike (S) glycoproteins, S590 and S1180. Clones of transduced cells that bound SARS-CoV S glycoprotein were inoculated with SARS-CoV, and increases in subgenomic viral RNA from 1–16 h or more were detected by multiplex RT-PCR in four cloned cell lines. Sequencing of the human lung cDNA inserts showed that each of the cloned cell lines contained cDNA that encoded human CD209L, a C-type lectin (also called L-SIGN). When the cDNA encoding CD209L from clone 2.27 was cloned and transfected into Chinese hamster ovary cells, the cells expressed human CD209L glycoprotein and became susceptible to infection with SARS-CoV. Immunohistochemistry showed that CD209L is expressed in human lung in type II alveolar cells and endothelial cells, both potential targets for SARS-CoV. Several other enveloped viruses including Ebola and Sindbis also use CD209L as a portal of entry, and HIV and hepatitis C virus can bind to CD209L on cell membranes but do not use it to mediate virus entry. Our data suggest that the large S glycoprotein of SARS-CoV may use both ACE2 and CD209L in virus infection and pathogenesis.
Severe acute respiratory syndrome (SARS) is caused by a novel coronavirus called SARS-CoV (1–3). The virus causes atypical pneumonia with diffuse alveolar damage with an overall mortality of ≈10% that ranges from 0% in children and 50% in persons over 65 (2). Coronaviruses bind to their glycoprotein receptors by the ≈200-kDa spike glycoprotein, S, on the viral envelope. Identification of virus receptors can provide insight into mechanisms of virus entry, tissue tropism, pathogenesis, and host range.
Several types of receptors were previously identified for coronavirus S glycoproteins. The receptors for the murine coronavirus mouse hepatitis virus are murine carcinoembryonic antigen cell adhesion molecule 1 (CEACAM1) and related murine glycoproteins in the carcinoembryonic antigen family in the Ig superfamily (4). The receptors for human coronavirus 229E (HCoV-229E), transmissible gastroenteritis virus of swine, and feline coronavirus in genetic group 1 are aminopeptidase N (APN) glycoproteins (5–8).
Angiotensin-converting enzyme 2 (ACE2) was found to be an efficient receptor for the S glycoprotein of SARS-CoV (9, 10). Because the Vero line of rhesus monkey kidney cells is highly susceptible to infection with SARS-CoV, Li et al. (9) used a codon-optimized soluble SARS-CoV spike glycoprotein (amino acids 12–672) fused to the Fc domain of human IgG1 to immunoprecipitate a putative receptor glycoprotein from Vero cell membranes and identified the simian ACE2 protein by mass spectrometry. They showed that expression of recombinant human ACE2 greatly enhanced the susceptibility of human 293T cells to infection by SARS-CoV. Wang et al. (10) independently demonstrated that human ACE2 is a receptor for SARS-CoV by transducing HeLa cells with a retrovirus library of cDNAs from Vero E6 cells and by using flow cytometry to select transduced cells that bound to purified soluble SARS-CoV S glycoprotein (amino acids 14–502) with a 6-histidine tag. They found that the simian cDNA in these cells, which encoded triosephosphate isomerase, enhanced expression of human ACE2 by inserting into the HeLa cell genome immediately upstream of the ACE2 ORF. Murine NIH 3T3 cells expressing recombinant human ACE2, but not those expressing recombinant triosephosphate isomerase, were susceptible to infection by HIV pseudovirus expressing SARS-CoV S protein.
In this report, we describe the discovery of an additional receptor for SARS-CoV, CD209L (also called L-SIGN, DC-SIGNR, and DC-SIGN2) (11). Our strategy for identifying a SARS-CoV receptor was to transduce a human lung cDNA library carried by a retroviral vector into Chinese hamster ovary (CHO) cells and use flow cytometry to select transduced CHO cells that bound soluble, codon-optimized, c-myc-tagged SARS-CoV S590 glycoprotein (amino acids 1–590) expressed in 293T cells. The SARS-CoV S590-binding cells were then challenged with infectious SARS-CoV, and infection was demonstrated by detection of subgenomic viral RNA synthesis and immunofluorescence with antiviral antibody. The finding that CD209L is expressed in human type II alveolar cells and lung endothelial cells that may be targets for SARS-CoV (12, 13) suggests that ACE2 and CD209L may both participate in SARS-CoV entry.
Materials and Methods
Virus. The Urbani strain of SARS-CoV was kindly provided by W. Bellini and T. Ksiazek (Centers for Disease Control and Prevention, Atlanta) (1) and propagated in Vero E6 cells (from American Type Culture Collection) in a Biosafety Level 3 laboratory. The inocula used for these experiments were the 24- to 28-h supernatant media from the second passage of virus in our laboratories, centrifuged at 1,000 rpm in a Beckman Coulter Allegra 64R refrigerated centrifuge with a Beckman Coulter GH3.8 rotor in Aerosolve aerosol-resistant canisters, aliquoted, flash frozen, and stored at -80°C. Cells were inoculated with virus at a multiplicity of infection (moi) of 0.01. Virus was adsorbed for 1 h at 37°C in serum-free DMEM, and the inoculum was removed and replaced with DMEM, 10% FBS, and 2% penicillin, streptomycin, and fungisone (PSF) (Invitrogen).
Antibodies. Rabbit-polyclonal anti-N Ig to detect SARS-CoV N protein was purchased from Imgenex (San Diego). Monoclonal IgGs to DC-SIGN (MAB161), CD209L (MAB162), or DC-SIGN plus CD209L (MAB1621), ACE2 (MAB933), and a goat IgG against ACE2 (AF933) were purchased from R & D Systems. Control monoclonal IgG specific for cholera toxin was kindly provided by R. K. Holmes (University of Colorado Health Sciences Center). Polyclonal-goat IgG (N-17 or C-17), specific for the N and C termini of CD209L, respectively, were purchased from Santa Cruz Biotechnology. FITC- and phycoerythrin-conjugated goat anti-mouse, goat anti-rabbit, and donkey anti-goat IgGs were purchased from Jackson ImmunoResearch. Mouse monoclonal and rabbit polyclonal anti-c-myc IgGs were purchased from Santa Cruz Biotechnology. Rabbit polyclonal anti-6-his IgG was purchased from Abcam (Cambridge, MA). Normal goat serum IgG was purchased from R & D Systems, and normal rabbit serum IgG was purchased from Bethyl Laboratories (Montgomery, TX). Biotinylated rabbit anti-goat IgG was purchased from Vector Laboratories.
Expression and Purification of SARS-CoV S Glycoproteins. Full-length spike protein contains 1,255 aa. The expression and purification of soluble, codon-optimized, c-myc-tagged S590 glycoprotein of SARS-CoV (amino acids 1–590, containing the receptor binding domain) expressed in 293T cells is described in ref. 14. The cDNA encoding the soluble, His6-tagged S1180 glycoprotein of SARS-CoV (amino acids 1–1180, containing most of the 1,191-aa ectodomain) was subcloned from cDNA of the Urbani strain of SARS-CoV (kindly provided by W. Bellini) (1), expressed in TN5 cells (Invitrogen) from baculovirus in the University of Colorado Health Sciences Center Tissue Culture/Monoclonal Antibody Core, and purified by nickel affinity chromatography.
Preparation of Pseudotypes Containing Human Lung cDNA Library. Pseudotypes were produced in ΦNX cells, a second-generation retroviral packaging cell line with an integrated Moloney murine leukemia virus (MoMuLV) gag-pol that expresses retroviral capsids but not viral envelope glycoproteins (15). These cells were used to produce pseudotyped viruses by cotransfection with the Viraport (Stratagene) human lung cDNA library plasmids and a plasmid that expresses vesicular stomatitis virus G glycoprotein, according to the manufacturer's instructions. Pseudotypes in culture supernatants were centrifuged at 1,000 × g for 10 min to remove cell debris and used immediately to transduce CHO cells or BHK cells at a moi of 10. Pseudotypes were treated with poly(N,N,N′,N′-tetramethyl-N-trimethylenehexamethylenediammonium dibromide) (Polybrene) (Sigma) and adsorbed onto cells for 4 h at 37°C. Medium was replaced by DMEM with 10% FBS/2% PSF.
Flow Cytometry. A Beckman Coulter XL cytofluorometer in the University of Colorado Flow Cytometry and Immunology Core Laboratory was used for flow cytometry. Cells were incubated for 1 h in 100 μl of fluorescence-activated cell sorting (FACS) buffer (PBS containing 2% normal serum from the same species as the corresponding secondary antibody) with 0.4 mg/ml purified SARS-CoV c-myc-tagged S590 or His6-tagged S1180, then washed three times with FACS buffer. S protein binding was detected by using the primary antibodies specific for the tags on the glycoproteins. The primary antibodies were adsorbed to 106 cells in suspension for 1 h at 4°C in FACS buffer. Cells were washed three times in FACS buffer at 4°C and adsorbed with phycoerythrin-conjugated secondary antibody directed against the primary antibody. The cells were fixed with 1.5% formaldehyde, and FACS was performed. Controls, which were all negative, included cells without primary antibody, cells without S protein, cells labeled with control primary mAb to an irrelevant antigen, and normal goat IgG.
Immunofluorescent Labeling and Immunoblotting. To detect viral N protein, cells were fixed with 75% methanol 25% acetic acid at -20°C, rehydrated in PBS, labeled with rabbit anti-SARS N antibody (Imgenex), and washed three times with PBS containing 0.1% normal goat serum. FITC-conjugated goat anti-rabbit IgG antibody was added for 1 h, and the cells were washed and observed with an Olympus BS-2 fluorescence microscope. To detect cells expressing DC-SIGN, CD209L or ACE2, cells were fixed and processed as described above. For immunoblots, cells were treated with lysis buffer (50 mM Tris·HCl, pH 8.0/5 mM EDTA/150 mM NaCl/1% Triton X-100) for 5 min at room temperature, and cell debris was removed by centrifugation at 16,100 × g for 10 min in an Eppendorf 5415D centrifuge. Supernatant in Laemmli buffer was boiled in saturated sodium chloride solution for 10 min, and proteins were separated on 10% SDS/PAGE then blotted onto poly(vinylidene difluoride) membranes with a Bio-Rad minitransblot cell at 50 V for 50 min.
Immunohistochemistry. Immunohistochemistry with horseradish peroxidase- or fluorescein-labeled antibodies was performed as reported in ref. 16. These are standard methods with the exception that the tissue was fixed overnight in 4% paraformaldehyde and the sections were treated with 6 M guanidine HCl for 30 min at room temperature, followed by 0.2 mg/ml trypsin for 30 min at 37°C before antibodies were applied. The goat anti-CD209L IgG N-17 or control goat IgG was incubated overnight at 4°C. The sections were rinsed and then incubated either with biotinylated rabbit anti-goat IgG, followed by streptavidin-biotin-horseradish peroxidase and then diaminobenzidine, or with FITC-labeled donkey anti-goat IgG.
Multiplex RT-PCR. To detect replication of SARS-CoV, multiplex RT-PCR was performed by using primers that amplify nucleotide 279-1069 of GAPDH, nucleotide 21263 in gene 1b to 21593 in S gene of SARS-CoV genomic RNA, and nucleotide 1 in the viral leader RNA to nucleotide 21593 in the viral S gene of the subgenomic viral RNA, as described in ref. 17.
DNA Sequencing. The DNA isolated from cloned cell lines was analyzed on 1% agarose gels in 1× TAE buffer (40 mM Tris-acetate/1 mM EDTA), and bands were visualized with ethidium bromide and excised from the gel. DNAs were sequenced at the University of Colorado Cancer Center DNA Sequencing and Analysis Core Facility on a Prism 3100 DNA fluorescent capillary automated sequencer (Applied Biosystems), using LTR and internal primers provided by the supplier of the ViraPort system (Stratagene).
Plasmids and Expression of CD209L Glycoprotein. The 1,100-bp cDNA encoding CD209L from clone 2.27 was cloned into the pENTR D/TOPO vector (Invitrogen) by PCR with primers specific for the 5′ and 3′ LTRs of MoMuLV provided by the manufacturer. The clone was shuttled into pDEST40 vector (Invitrogen) by using the LR clonase enzyme provided by the manufacturer. The insert was fully sequenced and found to encode a CD209L isotype that was 99% identical to GenBank entry NM_014257. New primers (forward, CACCATGAGTGACTCCAAGGAACC; reverse, CTATTCGTCTCTGAAGCAGGC) were made that were specific for CD209L cDNA, and PCR was performed to clone the cDNA without the MoMuLV 5′ and 3′ LTRs. The PCR product was cloned into pENTR D/TOPO and shuttled into pDEST40. The 3′ primer contained a terminal stop codon to prevent the V5/His6 from being incorporated into the CD209L gene product. The integrity of the clone (pDEST40 CD209L) was confirmed by sequencing. The cloned CD209L cDNA was transfected into CHO cells by using Lipofectamine 2000 (Invitrogen) reagent according to the manufacturer's directions. Transiently transfected cells were used 48 h after transfection. Stably transfected cells were selected by propagation in 500 μg/ml G418 for 10 days and sorted by flow cytometry for expression of CD209L.
Results
The Viraport human lung cDNA library (Stratagene) was used to identify a SARS-CoV receptor. This library contains 2.4 × 106 cDNA copies of mRNA isolated from normal lungs of five male and female white donors, ages 15–40, in a retrovirus vector derived from MoMuLV. The library allows for quick and efficient transduction of human cDNA into cells by retroviral pseudotypes that contain the vesicular stomatitis virus G protein to mediate entry. CHO cells were transduced with the pseudotypes at a moi of 10 to ensure that all cells were transduced with at least one virus particle. The cells were grown for 2 days and sorted by flow cytometry to detect binding of soluble codon optimized SARS-CoV S590 glycoprotein (14) to cells that express putative receptors (data not shown). Control CHO cells that were not transduced with retroviral pseudotypes were negative for S590 binding (data not shown). Sorting for S590 binding was repeated two more times. In the third sort, single positive cells were dispersed into 96-well plates. The 40 expanded clones were stored in liquid nitrogen.
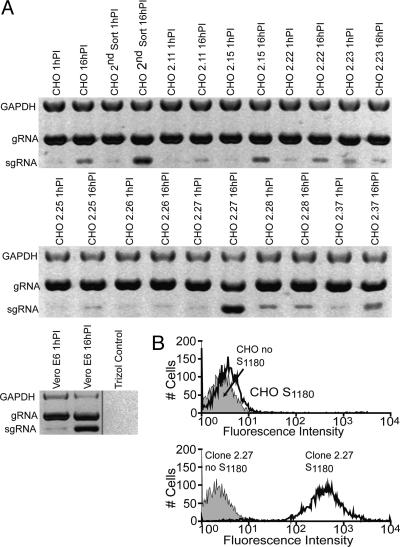
Under Biosafety Level 3 conditions, eight pools of five human lung cDNA-transduced CHO cell clones were challenged with infectious SARS-CoV at a moi of 0.01 plaque-forming units per cell, and RNA was isolated at 1 and 16 h after virus inoculation (data not shown). SARS-CoV subgenomic RNA was detected by using primers in the leader sequence and the body of the ORF encoding the S glycoprotein. These primers differentiate between input virus that contains only genomic RNA and infected cells that contain both genomic and subgenomic RNA. The SARS-CoV inoculum prepared from the 24-h supernatant of infected Vero E6 cells contained a minute amount of subgenomic RNA from lysed cells. The five pools of cloned CHO cells expressing human lung cDNA were challenged with SARS-CoV at a moi of 0.01 plaque-forming units per cell and tested for infection by multiplex RT-PCR at 1 h postinoculation (PI) and 16 h PI (data not shown). Viral subgenomic RNA increased markedly from 1 to 16 h after virus inoculation in two of the five pools (data not shown), indicating that virus replication had occurred. Each of the clones from the two RNA positive pools was then separately challenged with SARS-CoV. Fig. 1A shows that synthesis of viral RNA increased markedly from 1 to 16 h in transduced CHO cells from the second sort, in Vero E6 cells, and in human lung cDNA-transduced CHO clones 2.15, 2.22, 2.27, and 2.37, but not in other S-binding clones. Thus, four clones expressed human lung proteins that facilitated virus entry and the subsequent synthesis of subgenomic viral RNA.
Fig. 1.
Expression of SARS-CoV genomic and messenger RNA in CHO cells transduced with a human lung cDNA library, and binding of SARS-CoV spike glycoprotein to cell membranes. (A) Multiplex RT-PCR at 1 and 16 h PI with SARS-CoV. Multiplex RT-PCR was performed as in ref. 17. Control CHO cells or CHO cells transduced with retrovirus pseudotypes containing a human lung cDNA library were inoculated with SARS-CoV or mock inoculated. The viral genomic and subgenomic RNAs and cellular mRNA encoding GAPDH were reverse transcribed and amplified. Negative images of the amplicons are shown. The bottom gel shows Vero E6 monkey kidney cells, a positive control for SARS-CoV infection, in which subgenomic viral RNA increased at 16 h PI. The minute amount of subgenomic viral RNA in the 1-h sample was probably due to a low level of subgenomic viral RNA contamination of the input virus. In the fourth lane of the upper gel, CHO library cells that were sorted twice for S590 binding also showed an increase in subgenomic viral RNA at 16 h PI. These cells were further sorted for S590 binding, and single-cell clones were produced. Four of these cell lines showed an increase in subgenomic viral RNA at 16 h PI: clones 2.15, 2.22, 2.27, and 2.37. The proviral integrants were amplified by RT-PCR of RNA from the cloned cell lines. Sequencing results in Table 1 showed that all four of the SARS-CoV-susceptible cell lines contained CD209L. Clone 2.27 cells that had the largest increase in subgenomic viral RNA were further studied. (B) Flow cytometric analysis of soluble SARS-CoV S1180 protein binding to CHO (Upper) and clone 2.27 (Lower) cells.
DNA was extracted with TRIzol (Invitrogen) from human lung cDNA-containing CHO clones that bound SARS-CoV S protein, and the integrated human cDNAs in the proviruses were amplified by PCR using oligonucleotide primers derived from the MoMuLV LTRs in the Viraport cDNA library. The amplicons were partially sequenced, and blast searches were performed to identify human lung cDNAs that were transduced by the pseudotyped retroviral library into each of the S590-binding CHO clones (Table 1). Expression of several human proteins facilitated binding of S590 to transduced CHO cell clones, but only four clones (clones 2.15, 2.22, 2.27, and 2.37; Fig. 1 A) showed an increase in SARS-CoV subgenomic RNA, indicating that virus had entered the cells and begun subgenomic RNA synthesis. All four SARS S-binding clones contained cDNAs, of several sizes, that encoded human CD209L, a C-type lectin also called L-SIGN, DC-SIGNR, and DC-SIGN2 (11, 18). Transduction of BHK cells with the human lung cDNA library also yielded cells that bound S590 protein and expressed human CD209L protein (data not shown).
Table 1. Human cDNA expressed in SARS-CoV S glycoprotein binding cloned CHO cells transduced with human lung library.
| Clone | Approximate fragment size, bp | Nucleotides sequenced | Predicted protein product* |
|---|---|---|---|
| 2.11 | 1,100 | 886 | Unknown |
| 2.15 | 2,500 | 874 | CD209L |
| 2.22 | 3,500 | 769 | CD209L |
| 1,700 | 940 | CD209L | |
| 1,500 | 954 | Unknown | |
| 1,100 | 942 | Unknown | |
| 2.23 | 1,800 | 881 | CD209L |
| 1,100 | 878 | SPC† | |
| 2.25 | 2,700 | 596 | FcγRI |
| 1,500 | 928 | FcγRI | |
| 900 | 863 | FcγRI | |
| 2.26 | 2,800 | 531 | FcγRI |
| 1,500 | 866 | FcγRI | |
| 2.27 | 1,600 | 826 | CD209L |
| 1,100 | 531 | CD209L | |
| 2.28 | 1,650 | 652 | Calpain |
| 1,600 | 493 | FcγRI | |
| 2.37 | 1,700 | 806 | CD209L |
BLAST searches were done by using the nucleotide sequence from each fragment as the query. The protein encoded by the cDNA with the E value of 0.0 is shown
Surfactant protein C
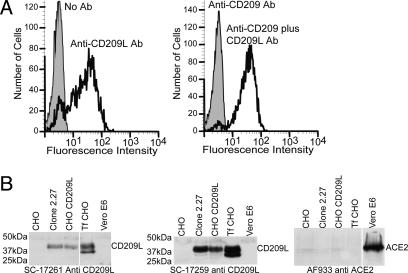
Flow cytometric analysis was performed by using purified His6-tagged SARS-CoV S1180 protein expressed in TN5 cells (19) from a baculovirus vector. More than 90% of clone 2.27 cells, which contain human CD209L cDNA, bound the S1180 glycoprotein (Fig. 1B Lower), whereas control CHO cells (Fig. 1B Upper) did not bind S1180. FACS analysis was performed with three monoclonal antibodies that are specific for either DC-SIGN or CD209L, or both CD209L and DC-SIGN (R & D Systems). Only anti-CD209L and anti-CD209L plus DC-SIGN reacted with clone 2.27 (Fig. 2A), showing that these cells expressed CD209L but not DC-SIGN. In addition, immunoblotting (Fig. 2B) and immunofluorescence (data not shown) showed that CD209L was expressed in clone 2.27 cells but not in Vero E6 cells or CHO cells. Immunoblotting (Fig. 2B) and immunofluorescence (data not shown) showed that ACE2 was expressed only on Vero E6 cells and not on clone 2.27 cells or CHO cells. These observations show that binding of SARS-CoV S590 glycoprotein to clone 2.27 cells was not due to increased expression of human ACE2, as in the experiments of Wang et al. (10).
Fig. 2.
Expression of CD209L protein by CHO cells transduced with a human lung cDNA library. (A) Flow cytometric analysis of clone 2.27 cells with antibodies specific for CD209L and DC-SIGN. Antibodies specific for DC-SIGN, CD209L, and an antibody that recognizes both DC-SIGN and CD209L were used to determine which glycoprotein was expressed on clone 2.27 cells. Only the antibodies that recognized CD209L bound to the clone 2.27 cells. (B) Immunoblots for ACE2 or CD209L of cell lines used in this study. Cell lysates from CHO cells, clone 2.27 cells, CHO cells stably expressing CD209L, CHO cells transiently transfected (Tf) with CD209L, and Vero E6 cells were run on a 10% SDS/PAGE gel and blotted onto poly(vinylidene difluoride). The blots were probed with polyclonal antibodies specific for the N or C termini of CD209L or for ACE2. The antibodies were detected by horseradish peroxidase-conjugated donkey anti-goat IgG antibodies and chemiluminescence.
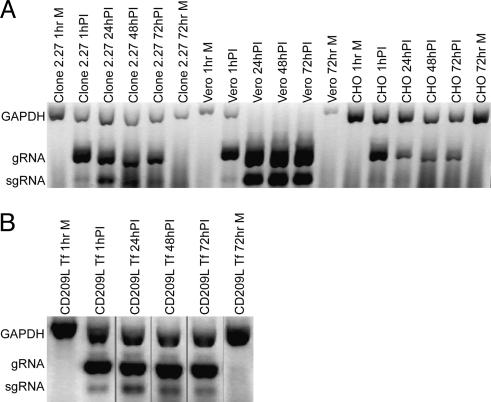
To determine whether CD209L was the only human protein in clone 2.27 cells required for binding of S590 and S1180 and for infection by SARS-CoV, we transiently expressed human CD209L cDNA in CHO cells and detected expression of CD209L protein by immunofluorescence (data not shown) and immunoblotting (Fig. 2B). The double band of CD209L in the transiently transfected CHO lanes probably represents differences in protein glycosylation due to the high level of expression in the transiently transfected cells. Vero E6 cells, control CHO cells, clone 2.27 cells, and CHO cells transiently transfected with human CD209L cDNA were challenged with SARS-CoV at a moi of 0.1, and multiplex RT-PCR was performed from samples at 1, 24, 48, and 72 h after virus inoculation (Fig. 3). This experiment showed that, like Vero E6 and clone 2.27 cells, CHO cells transiently expressing human CD209L were susceptible to virus infection, whereas nontransfected CHO cells were not susceptible.
Fig. 3.
Multiplex RT-PCR analysis of cell lines at various time points after SARS-CoV inoculation or mock inoculation. (A) Multiplex RT-PCR analysis was performed as in Fig. 1 on samples collected at 1, 24, 48, and 72 h PI from clone 2.27, Vero E6, and CHO cells (A) and from CHO cells transiently transfected (Tf) with a plasmid containing CD209L cDNA (B). Negative images are shown.
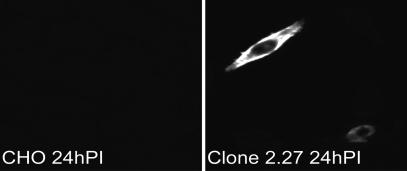
An immunofluorescence assay was performed to detect SARS-CoV N protein in CHO or clone 2.27 cells 24 h after virus inoculation. Fig. 4 shows cytoplasmic expression of viral N protein in <1% of clone 2.27 cells but not in control CHO cells. These data indicated that some but not all of the CD209L-expressing cells are susceptible to SARS-CoV infection. In cell lines that transiently express ACE2 cDNA, >50% can be infected by SARS-CoV (9, 10). Thus, CD209L is a less efficient receptor for SARS-CoV than ACE2.
Fig. 4.
Immunofluorescent labeling of SARS-CoV N protein in CHO and clone 2.27 cells inoculated with SARS-CoV. Scattered infected cells showed cytoplasmic expression of the virus nucleocapsid protein.
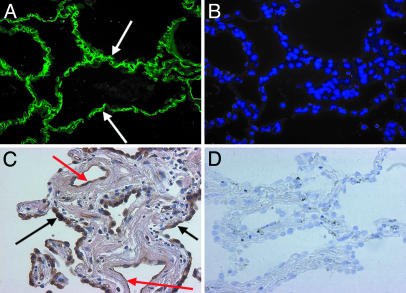
The cells in the lung that express CD209L were identified. Immunohistochemistry with anti-CD209L antibody N-17 showed that in human lung, CD209L is expressed in type II alveolar cells and endothelial cells (Fig. 5).
Fig. 5.
Expression of CD209L in human lung. (A) Localization of CD209L to the capillaries (arrows) in the alveolar wall by immunohistochemistry. (B) DAPI (4′,6-diamidino-2-phenylindole) staining to demonstrate cell nuclei along the alveolar wall. (C) Expression of CD209L in hyperplastic type II alveolar (arrow) and endothelial (red arrow) cells by peroxidase immunohistochemistry in a lung of a cigarette smoker. (D) There was no labeling of these cells with control goat IgG.
Discussion
In this paper, we showed that human CD209L (L-SIGN) binds purified soluble SARS-CoV S glycoproteins expressed in human 293T cells and in insect cells, and that CD209L can serve as a portal of entry for infectious SARS-CoV. Our strategy was based on expression of human lung cDNAs in CHO cells from a retroviral library, selection for cells that bound the SARS-CoV S glycoprotein, and identification of the human lung cDNAs expressed in those cells. Cloned S-binding cells were challenged with infectious SARS-CoV, and virus infection was demonstrated by increased expression of subgenomic viral RNA using multiplex RT-PCR and by immunofluorescence with antiviral antibody.
CD209L is a type II transmembrane glycoprotein in the C-type lectin family (18, 20). The isoform of CD209L identified in this paper as a SARS-CoV receptor contains 376 aa and is composed of a short cytoplasmic tail, a transmembrane domain, an extracellular stalk consisting of seven repeats of a 23-aa sequence (KAAVGELxEKSKxQEIYQELTxL), and a large C-terminal carbohydrate-recognition domain (CRD). The CRDs of the CD209L isoforms bind specifically to high-mannose glycans on glycoproteins (21). We do not yet know whether the SARS-CoV S protein binds to the CRD of CD209L by its carbohydrate moieties or recognizes amino acid residues on CD209L.
Six membrane-bound isoforms and three soluble isoforms of CD209L are variably expressed on sinusoidal endothelial cells in the liver, endothelial cells and macrophage-like cells in lymph nodes, and capillary endothelial cells in the placenta (18, 22, 23), as well as in human Peyer's patches and on capillaries in the villous lamina propria of the terminal ileum (24). CD209L is expressed at low levels on the KG1 myeloid dendritic cell line and endometrium (18, 23). We isolated cDNA encoding CD209L from a human lung cDNA library and showed by immunohistochemistry that CD209L is expressed in human type II alveolar cells and endothelial cells (Fig. 5). The principal targets for SARS-CoV infection in humans are the lung and gastrointestinal tract (25, 26). SARS-CoV or viral antigens have been detected in type II alveolar cells in lung, and in liver, kidney, sweat glands, parathyroid, brain, pancreas, and adrenal glands (26). We postulate that expression of CD209L, perhaps in combination with ACE2, in lymph nodes, Peyer's patches, and lung may contribute to the spread of virus in vivo.
Human CD209L is 77% identical to human DC-SIGN (also called CD209) (20), which is expressed on dendritic cells (DCs), monocytes, alveolar macrophages, placenta, and rectal mucosa (18). Both CD209L and DC-SIGN bind in a calcium-dependent manner to ICAM-3 and to HIV gp120 (22, 27). Although these C-type lectins on cell membranes can bind HIV virions, they do not mediate HIV infection of DCs. DCs expressing DC-SIGN carry infectious HIV virions to lymph nodes where they mediate infection in trans of T lymphocytes that express CD4 and chemokine receptor CXCR4 (27). CD209L expressed on endothelial cells and Peyer's patches in the rectum might mediate transfer of HIV1 virus in trans to dendritic or T cells (24).
Both DC-SIGN and CD209L interact with Ebola virus glycoprotein and mediate infection of susceptible endothelial cells both in cis and in trans (28, 29). Both CD209L and DC-SIGN mediate attachment of Sindbis glycoprotein via their CRDs and mediate productive infection by Sindbis virus, especially by virus produced in insect cells that has high levels of high mannose glycans on the envelope glycoproteins compared with virus produced in mammalian cells (30). DC-SIGN also mediates productive infection of human dendritic cells by dengue virus (31, 32). DC-SIGN and CD209L glycoproteins bind hepatitis C virus glycoproteins E1 and E2 (33). DC-SIGN was recently shown to bind SARS-CoV S glycoprotein but does not initiate SARS-CoV infection. DC-SIGN can, however, mediate SARS-CoV infection in trans of cells that express human ACE2 (34).
Human ACE2 is an efficient receptor for SARS-CoV. It is a zinc-containing carboxypeptidase that converts angiotensin I to angiotensin II in the renin–angiotensin system and regulates heart function, mainly by maintaining blood pressure and fluid and electrolyte homeostasis (35, 36). Northern blotting showed that ACE2 mRNA is expressed abundantly in endothelial cells of the heart, kidneys, and testis (37). In addition, RT-PCR and immunocytochemistry showed that ACE2 mRNA and protein are expressed on oral, nasal, and intestinal mucosa, alveolar epithelial cells in lung, stomach, skin, lymph nodes, thymus, bone marrow, spleen liver, kidney, and brain, and arterial and venous endothelial cells and arterial smooth muscle cells in many organs (38–41). These sites for ACE2 expression include most of the sites for SARS-CoV replication in autopsy specimens (26). Our data show that human CD209L can also mediate infection by SARS-CoV, although it is a much less efficient receptor than human ACE2. Fig. 5 shows that CD209L is expressed in human lung on type II alveolar cells, which are an important target for SARS-CoV infection (13, 25), and on endothelial cells.
Many different strategies can be used to identify virus receptors, coreceptors, and/or ligands. The strategies used by Li et al. (9) and Wang et al. (10) to identify ACE2 as a receptor for SARS-CoV were based on interactions of the virus or spike glycoprotein with Vero monkey kidney cells. Neither of these approaches would have found CD209L or DC-SIGN because Vero E6 cells do not express these glycoproteins. Our strategy focused on SARS-CoV infection of human lung. In the human lung cDNA library, we identified a number of proteins that bound the SARS-CoV S glycoprotein, but all of the S-binding clones of transduced CHO cells that were susceptible to virus entry contained human CD209L cDNA from the library. From the human lung library we repeatedly identified transduced cDNAs of several sizes that encoded CD209L, but we did not identify any human cDNAs that encoded ACE2 or DC-SIGN. In addition, RT-PCR of the human lung library that we used detected neither ACE2 nor DC-SIGN cDNA (data not shown), perhaps because of limited complexity in the cDNAs in the lung library.
Many viruses, including HIV, herpes simplex virus, and measles viruses, use multiple alternative receptors and/or coreceptors for entry into host cells (27, 42–45). Multiple interactions of a viral envelope glycoprotein with different receptors may facilitate infection of different tissues or different hosts (46). Different spike/receptor interactions may occur sequentially on a single cell type or independently on different cell types (44). Because the S glycoproteins of coronaviruses are some of the largest viral spike glycoproteins, it is possible that different domains within a single S protein could recognize multiple alternative receptors. Further experiments are needed to determine whether interactions between ACE2 and CD209L play important roles in SARS-CoV infection and pathogenesis.
Acknowledgments
We thank M. K. Smith, Brian C. Turner, and Megan W. Howard for scientific discussions. Excellent technical assistance was provided by Christian Pontillo. Portions of this work were performed at the University of Colorado Cancer Center DNA Sequencing and Analysis Core Facility, Tissue Culture/Monoclonal Antibody Core, and Flow Cytometry and Immunology Core Laboratory, which are supported by National Institutes of Health (NIH)/National Cancer Institute Cancer Core Support Grant P30 CA046934. This work was supported by a supplement to NIH Grant RO1 AI-25231, NIH Grants RO1 HL-67671 and PO1 AI59576, NIH Contracts N01-AI25489 and N01-AI25490, Department of Health and Human Services and Centers for Disease Control and Prevention (CDC) Grant U901 CCU216988, and National Institute of Allergy and Infectious Diseases (NIAID) Contract N01-AI65315. L.G.-R. was supported by the CDC Emerging Infectious Disease fellowship program. S.A.J. was supported by NIAID Training Grant T32 AI 07537-04.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CHO, Chinese hamster ovary; CoV, coronavirus; FACS, fluorescence-activated cell sorting; moi, multiplicity of infection; MoMuLV, Moloney murine leukemia virus; PI, postinoculation; SARS, severe acute respiratory syndrome.
References
- 1.Rota, P. A., Oberste, M. S., Monroe, S. S., Nix, W. A., Campagnoli, R., Icenogle, J. P., Penaranda, S., Bankamp, B., Maher, K., Chen, M. H., et al. (2003) Science 300, 1394-1399. [DOI] [PubMed] [Google Scholar]
- 2.Peiris, J. S., Yuen, K. Y., Osterhaus, A. D. & Stohr, K. (2003) N. Engl. J. Med. 349, 2431-2441. [DOI] [PubMed] [Google Scholar]
- 3.Marra, M. A., Jones, S. J., Astell, C. R., Holt, R. A., Brooks-Wilson, A., Butterfield, Y. S., Khattra, J., Asano, J. K., Barber, S. A., Chan, S. Y., et al. (2003) Science 300, 1399-1404. [DOI] [PubMed] [Google Scholar]
- 4.Dveksler, G. S., Dieffenbach, C. W., Cardellichio, C. B., McCuaig, K., Pensiero, M. N., Jiang, G. S., Beauchemin, N. & Holmes, K. V. (1993) J. Virol. 67, 1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wentworth, D. E. & Holmes, K. V. (2001) J. Virol. 75, 9741-9752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yeager, C. L., Ashmun, R. A., Williams, R. K., Cardellichio, C. B., Shapiro, L. H., Look, A. T. & Holmes, K. V. (1992) Nature 357, 420-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delmas, B., Gelfi, J., L'Haridon, R., Vogel, L. K., Sjostrom, H., Noren, O. & Laude, H. (1992) Nature 357, 417-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tresnan, D. B., Levis, R. & Holmes, K. V. (1996) J. Virol. 70, 8669-8674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li, W., Moore, M. J., Vasilieva, N., Sui, J., Wong, S. K., Berne, M. A., Somasundaran, M., Sullivan, J. L., Luzuriaga, K., Greenough, T. C., et al. (2003) Nature 426, 450-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang, P., Chen, J., Zheng, A., Nie, Y., Shi, X., Wang, W., Wang, G., Luo, M., Liu, H., Tan, L., et al. (2004) Biochem. Biophys. Res. Commun. 315, 439-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pohlmann, S., Soilleux, E. J., Baribaud, F., Leslie, G. J., Morris, L. S., Trowsdale, J., Lee, B., Coleman, N. & Doms, R. W. (2001) Proc. Natl. Acad. Sci. USA 98, 2670-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hwang, D. M., Chamberlain, D. W., Poutanen, S. M., Low, D. E., Asa, S. L. & Butany, J. (2004) Mod. Pathol., 10.1038/modpathol.3800247.
- 13.To, K. F. & Lo, A. W. (2004) J. Pathol. 203, 740-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Babcock, G. J., Esshaki, D. J., Thomas, W. D., Jr., & Ambrosino, D. M. (2004) J. Virol. 78, 4552-4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grignani, F., Kinsella, T., Mencarelli, A., Valtieri, M., Riganelli, D., Lanfrancone, L., Peschle, C., Nolan, G. P. & Pelicci, P. G. (1998) Cancer Res. 58, 14-19. [PubMed] [Google Scholar]
- 16.Mason, R. J., Kalina, M., Nielsen, L. D., Malkinson, A. M. & Shannon, J. M. (2000) Am. J. Pathol. 156, 175-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gillim-Ross, L., Taylor, J., Scholl, D. R., Ridenour, J., Masters, P. S. & Wentworth, D. E. (2004) J. Clin. Microbiol. 42, 3196-3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bashirova, A. A., Geijtenbeek, T. B., van Duijnhoven, G. C., van Vliet, S. J., Eilering, J. B., Martin, M. P., Wu, L., Martin, T. D., Viebig, N., Knolle, P. A., et al. (2001) J. Exp. Med. 193, 671-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wickham, T. J. & Nemerow, G. R. (1993) Biotechnol. Prog. 9, 25-30. [DOI] [PubMed] [Google Scholar]
- 20.Mummidi, S., Catano, G., Lam, L., Hoefle, A., Telles, V., Begum, K., Jimenez, F., Ahuja, S. S. & Ahuja, S. K. (2001) J. Biol. Chem. 276, 33196-33212. [DOI] [PubMed] [Google Scholar]
- 21.Geijtenbeek, T. B., Engering, A. & Van Kooyk, Y. (2002) J. Leukocyte Biol. 71, 921-931. [PubMed] [Google Scholar]
- 22.Soilleux, E. J., Morris, L. S., Rushbrook, S., Lee, B. & Coleman, N. (2002) Hum. Pathol. 33, 652-659. [DOI] [PubMed] [Google Scholar]
- 23.Soilleux, E. J., Barten, R. & Trowsdale, J. (2000) J. Immunol. 165, 2937-2942. [DOI] [PubMed] [Google Scholar]
- 24.Jameson, B., Baribaud, F., Pohlmann, S., Ghavimi, D., Mortari, F., Doms, R. W. & Iwasaki, A. (2002) J. Virol. 76, 1866-1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chow, K. C., Hsiao, C. H., Lin, T. Y., Chen, C. L. & Chiou, S. H. (2004) Am. J. Clin. Pathol. 121, 574-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding, Y., He, L., Zhang, Q., Huang, Z., Che, X., Hou, J., Wang, H., Shen, H., Qiu, L., Li, Z., et al. (2004) J. Pathol. 203, 622-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clapham, P. R. & McKnight, A. (2001) Br. Med. Bull. 58, 43-59. [DOI] [PubMed] [Google Scholar]
- 28.Simmons, G., Reeves, J. D., Grogan, C. C., Vandenberghe, L. H., Baribaud, F., Whitbeck, J. C., Burke, E., Buchmeier, M. J., Soilleux, E. J., Riley, J. L., et al. (2003) Virology 305, 115-123. [DOI] [PubMed] [Google Scholar]
- 29.Alvarez, C. P., Lasala, F., Carrillo, J., Muniz, O., Corbi, A. L. & Delgado, R. (2002) J. Virol. 76, 6841-6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klimstra, W. B., Nangle, E. M., Smith, M. S., Yurochko, A. D. & Ryman, K. D. (2003) J. Virol. 77, 12022-12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tassaneetrithep, B., Burgess, T. H., Granelli-Piperno, A., Trumpfheller, C., Finke, J., Sun, W., Eller, M. A., Pattanapanyasat, K., Sarasombath, S., Birx, D. L., et al. (2003) J. Exp. Med. 197, 823-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marovich, M., Grouard-Vogel, G., Louder, M., Eller, M., Sun, W., Wu, S. J., Putvatana, R., Murphy, G., Tassaneetrithep, B., Burgess, T., et al. (2001) J. Invest. Dermatol. Symp. Proc. 6, 219-224. [DOI] [PubMed] [Google Scholar]
- 33.Pohlmann, S., Zhang, J., Baribaud, F., Chen, Z., Leslie, G. J., Lin, G., Granelli-Piperno, A., Doms, R. W., Rice, C. M. & McKeating, J. A. (2003) J. Virol. 77, 4070-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang, Z. Y., Huang, Y., Ganesh, L., Leung, K., Kong, W. P., Schwartz, O., Subbarao, K. & Nabel, G. J. (2004) J. Virol. 78, 5642-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crackower, M. A., Sarao, R., Oudit, G. Y., Yagil, C., Kozieradzki, I., Scanga, S. E., Oliveira-dos-Santos, A. J., da Costa, J., Zhang, L., Pei, Y., et al. (2002) Nature 417, 822-828. [DOI] [PubMed] [Google Scholar]
- 36.Eriksson, U., Danilczyk, U. & Penninger, J. M. (2002) Curr. Biol. 12, R745-R752. [DOI] [PubMed] [Google Scholar]
- 37.Danilczyk, U., Eriksson, U., Crackower, M. A. & Penninger, J. M. (2003) J. Mol. Med. 81, 227-234. [DOI] [PubMed] [Google Scholar]
- 38.Harmer, D., Gilbert, M., Borman, R. & Clark, K. L. (2002) FEBS Lett. 532, 107-110. [DOI] [PubMed] [Google Scholar]
- 39.Komatsu, T., Suzuki, Y., Imai, J., Sugano, S., Hida, M., Tanigami, A., Muroi, S., Yamada, Y. & Hanaoka, K. (2002) DNA Seq. 13, 217-220. [DOI] [PubMed] [Google Scholar]
- 40.Hamming, I., Timens, W., Bulthuis, M. L., Lely, A. T., Navis, G. J. & van Goor, H. (2004) J. Pathol. 203, 631-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan, K. H., Poon, L. L., Cheng, V. C., Guan, Y., Hung, I. F., Kong, J., Yam, L. Y., Seto, W. H., Yuen, K. Y. & Peiris, J. S. (2004) Emerg. Infect. Dis. 10, 294-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zaitseva, M., Peden, K. & Golding, H. (2003) Biochim. Biophys. Acta 1614, 51-61. [DOI] [PubMed] [Google Scholar]
- 43.Yanagi, Y., Ono, N., Tatsuo, H., Hashimoto, K. & Minagawa, H. (2002) Virology 299, 155-161. [DOI] [PubMed] [Google Scholar]
- 44.Manchester, M., Naniche, D. & Stehle, T. (2000) Virology 274, 5-10. [DOI] [PubMed] [Google Scholar]
- 45.Yanagi, Y. (2001) Rev. Med. Virol. 11, 149-156. [DOI] [PubMed] [Google Scholar]
- 46.Spear, P. G. (2004) Cell. Microbiol. 6, 401-410. [DOI] [PubMed] [Google Scholar]