Abstract
Approximately 14% of genetic mutations in patients with ataxia-telangiectsia (A-T) are single-nucleotide changes that result in primary premature termination codons (PTCs), either UAA, UAG, or UGA. The purpose of this study was to explore a potential therapeutic approach for this subset of patients by using aminoglycosides to induce PTC read-through, thereby restoring levels of full-length ATM (A-T mutated) protein. In experiments using a modified in vitro cDNA coupled transcription/translation protein truncation test, 13 A-T cell lines carrying PTC mutations in different contexts exhibited read-through expression of ATM fragments, with three of four aminoglycosides tested. In ex vivo experiments with lymphoblastoid cell lines, we used radiosensitivity, radioresistant DNA synthesis, and irradiation-induced autophosphorylation of ATM Ser-1981 to show that the aminoglycoside-induced full-length ATM protein was functional and corrected, to various extents, the phenotype of A-T cells. These results encourage further testing of other compounds in this class, as well as follow up animal studies. Because some A-T patients with 5–20% of normal levels of ATM protein show slower neurological progression, A-T may prove to be a good model for aminoglycoside-induced read-through therapy.
Ataxia-telangiectasia (A-T) is an autosomal recessive neurodegenerative disorder with onset in early childhood, resulting from mutations in the A-T mutated (ATM) gene (1). The ATM protein is a hierarchical serine–threonine kinase, phosphorylating many substrates involved in repair of double-stranded DNA breaks, control of cell cycle checkpoints, and responses to oxidative stress, as well as in radiosensitivity, cancer susceptibility, immune function, and neurological development (2). ATM protein levels are undetectable in the cells of most A-T patients by conventional testing. However a few patients, often with a milder phenotype, have some detectable ATM protein (3, 4). This finding encouraged us to seek compounds, such as aminoglycosides, that have the potential to read through premature termination codons and restore ATM protein function (5, 6).
To test these effects, we selected 13 lymphoblastoid cell lines (LCLs) with primary premature termination codon (PTC) mutations from a repository of >400 lines derived from A-T patients. Herein, we show representative data from 5 of the 13 LCLs. Using protein truncation testing (PTT) driven by plasmid templates containing PTC mutations in the ATM gene (7), we observed in vitro read-through effects of various magnitudes with three of four aminoglycosides tested. Full-length ATM protein was also documented ex vivo by immunoprecipitation. Correction of radioresistant DNA synthesis and radiosensitivity, as well as autophosphorylation of ATM, suggested that this read-through produces functional ATM protein.
Experimental Procedures
Cell Lines. Lymphoblastoid cell lines were maintained in RPMI medium 1640 (Invitrogen) with 15% FBS (HyClone) and 1% penicillin/streptomycin (Invitrogen) at 37°C and 5% CO2. In ex vivo experiments, aminoglycosides were added daily into culture media at the indicated doses and times. Because streptomycin is also an aminoglycoside that can perturb proofreading by binding to another ribosomal site, ex vivo experiments were performed with or without streptomycin; no differences were noted.
Mutations. We selected only PTC mutations that resulted directly from the mutation, i.e., primary PTC mutations, and not those caused indirectly by upstream mutations or aberrant splicing, because the latter would have no potential therapeutic benefits. The LCLs carried the following mutations (www.benaroyaresearch.org/bri_investigators/atm.htm): AT185LA, homozygous 3673C → T (a UAA G stop codon in PTT fragment 4); TAT51, homozygous 5623C → T (a UGA C stop codon in PTT fragment 5); AT187LA, homozygous 5908C → T (a UAA G stop codon in PTT fragment 6); AT153LA, homozygous 8977C → T (a UGA A stop codon in fragment 8); and AT160LA, hemizygous 7792C → T (a UGA A stop codon in PTT fragment 8), with a deletion of this region as the second allele.
PTT. Messenger RNA was isolated from LCLs containing the PTC mutations by RNeasy kit (Qiagen, Valencia, CA). RT-PCR products were screened by PTT (7) in the presence or absence of aminoglycosides. PCR products containing the PTC mutations from four different patients were cloned into plasmids following the manufacturer's protocols (Invitrogen). The PCR products for each were also mutagenized back to normal and used as control plasmids. DNA sequencing confirmed the PTT fragments in all constructs. The coupled transcription and translation 35S-labeled PTT products from RT-PCR and plasmid clones were electrophoresed on 10% or 15% SDS polyacrylamide gels and transferred onto polyvinylidene difluoride) membrane (Bio-Rad). Occasional extra bands are observed in the translation products, presumably initiated from downstream methonines in the starting template. The blots were dried and exposed to film.
Immunoprecipitation of Aminoglycoside-Induced ATM. LCLs were grown in the presence or absence of various concentrations of aminoglycoside. Nuclear extracts were prepared for each sample according to NE-PER protocol (Pierce). Ten micrograms of anti-ATM antibody (ppATM-2C1, Genetex, Boston) was incubated with nuclear lysates overnight at 4°C. Five microliters of Protein-A magnetic beads (NEB, Beverly, MA) was incubated for 1 h at 4°C to capture the ATM–antibody complex. The immunoprecipitated proteins were run on a 6% SDS polyacrylamide gel, and Western blot analysis was performed for ATM protein detection by using a second anti-ATM antibody (ppATM-5C2, Genetex).
Radioresistant DNA Synthesis. After 4 days of routine culture with or without aminoglycoside, the LCLs were incubated with medium containing 0.04 μCi/ml [14C]thymidine (Perkin–Elmer; 1 Ci = 37 GBq) for 24 h. The medium was replaced with fresh media, and the cells were exposed to various doses of gamma rays. The cells were returned to the incubator for 60 min and pulse-labeled in medium containing 4 μCi/ml [3H]thymidine (Perkin–Elmer) for an additional 60 min. The samples were harvested and counted in a Packard 2900TR scintillation counter. The ratio of incorporated 3H to 14C was used for quantification to standardize the variation in DNA recovery. Triplet replicates were used to minimize the standard error of measurements.
Colony Survival Assay (CSA). CSA was performed as described (8). After 4 days of incubation with or without aminoglycosides, LCLs were plated, in duplicate, in 96-well plates at 50, 100, or 200 cells per well. One plate was exposed to 1.0 Gy radiation, whereas the other was left unirradiated. The cells were incubated for 10–13 days, at which time they were stained with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) dye (tetrazolium-based colorimetric assay, Sigma). Each well was checked under the microscope, and viable cells stained dark blue. The presence of a colony of >32 cells was scored as a positive well, and survival fractions were calculated. Normal and bona fide A-T LCLs were included as positive and negative controls, respectively. Each LCL was tested multiple times.
Immunofluoresence. Before immunostaining, ≈5 × 106 cells were treated with 10-Gy irradiation and incubated at 37°C for 45 min. Cells were rinsed with 1× PBS and dropped onto poly(d-lysine)-coated coverslips, followed by fixation in 4% paraformaldehyde for 15 min at room temperature, rinsing, and nuclear permeabilization with 0.5% Triton X-100 for 10 min at room temperature. After three rinses in 1× PBS, cells were blocked for 1 h at 37°C in 10% FBS, incubated for 1 h at room temperature with anti-ATM pS1981 (Rockland Immunochemicals, Gilbertsville, PA) at a dilution of 1:100, and rinsed three times in 0.1% Triton X-100/PBS. After a second incubation in blocking solution for 30 min at 37°C, cells were stained with FITC-conjugated anti-rabbit IgG (Jackson ImmunoResearch) at a dilution of 1:150 for 45 min at room temperature. Coverslips were mounted onto slides by using Vectashield with 4′,6-diamidino-2-phenylindole (Vector Laboratories). Antibody solutions were prepared in 10% FBS and were applied in a volume of 50 μl per coverslip in a humidity chamber to avoid drying. Images were captured by using Vysis fish analysis software connected to a Leica DM RXA automated microscope equipped with Photometrix SenSyn.
Results
In Vitro PTT Read-Through. Initial in vitro PTT experiments showed consistent but weak read-through of full-length PTT fragments of the ATM gene, and we determined that the RT-PCR template was the limiting factor in these reactions. To provide a more accurate concentration of starting template, and thereby improve the reproducibility of PTC read-through, we cloned the PTT fragments containing specific mutations from four different A-T patients into plasmids: AT185LA, TAT51, AT187LA, and AT153LA. Plasmids containing the same fragments with the mutation corrected by site-directed mutagenesis were also constructed, confirmed by sequencing, and used as controls. Plasmids TAT51 and AT153LA had UGA stop codon mutations; however, these had different +4 nucleotides (i.e., the nucleotide immediately downstream of the stop codon). Plasmids AT185LA and AT187LA had UAA G mutations, on PTT fragments 4 and 6, respectively.
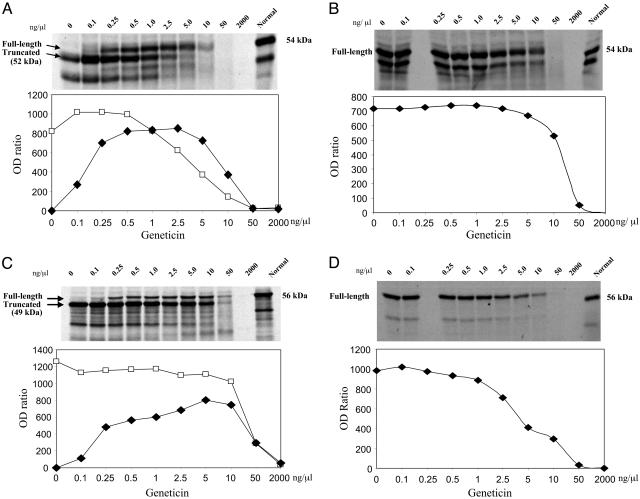
Plasmid TAT51 (UGA C) showed optimal full-length read-through of the PTT fragment with geneticin at a concentration of 2.5 ng/μl (Fig. 1A). Full-length read-through of PTT fragment 5 (54 kDa) was undetectable in the absence of aminoglycoside; this was true for all plasmids and LCLs selected for these experiments. As the band intensity of the full-length product increased, the intensity of the truncated product (52 kDa) diminished, starting at 1.0 ng/μl of geneticin. This inverse relationship was most likely caused by extension of the truncated product beyond the PTC, i.e., to read-through. In addition, a general inhibition of the coupled transcription/translation reaction by the aminoglycoside was noted. Both the full-length and truncated products were completely inhibited above 50 ng/μl. With minor variations, this general inhibition was observed with all four aminoglycosides tested with each of the plasmids.
Fig. 1.
Autoradiograms show in vitro effects of geneticin on read-through of various PTT fragments carrying different PTC mutations in the ATM gene. As the aminoglycoside concentrations increased, the densitometry readings (OD ratio = OD of band/OD of background) for full-length PTT product (filled diamonds) increased from 0 to 800 (A and C). (A) Experiment using plasmid with TAT51 mutation (UGA C) as template. (B) Experiment using plasmid with the corrected TAT51 mutation (CGA C, i.e., normal sequence). (C) Experiment using plasmid with AT153LA mutation (UGA A). (D) Experiment using plasmid with the corrected AT153LA mutation (CGA A). In all experiments, both the full-length and truncated PTT products (open squares) were inhibited by higher concentrations of geneticin.
Using the corrected TAT51 plasmid (CGA C, i.e., normal sequence), only full-length product was observed, as expected (Fig. 1B). Inhibition of full-length protein started at 5 ng/μl. An IC50 (50% inhibitory concentration) was observed at ≈20 ng/μl of geneticin.
Using plasmid AT153LA (UGA A), optimal full-length read-through was observed at a geneticin concentration of 5 ng/μl (Fig. 1C). The corrected plasmid (CGA A) had an IC50 at ≈4 ng/μl (Fig. 1D). Plasmids AT187LA and AT185LA, carrying UAA G mutations, also showed geneticin-induced read-through of full-length protein, though at a somewhat lower level than those of TAT51 and AT153LA (data not shown); this most likely reflects the differing read-through efficiencies of UGA versus UAA stop codons.
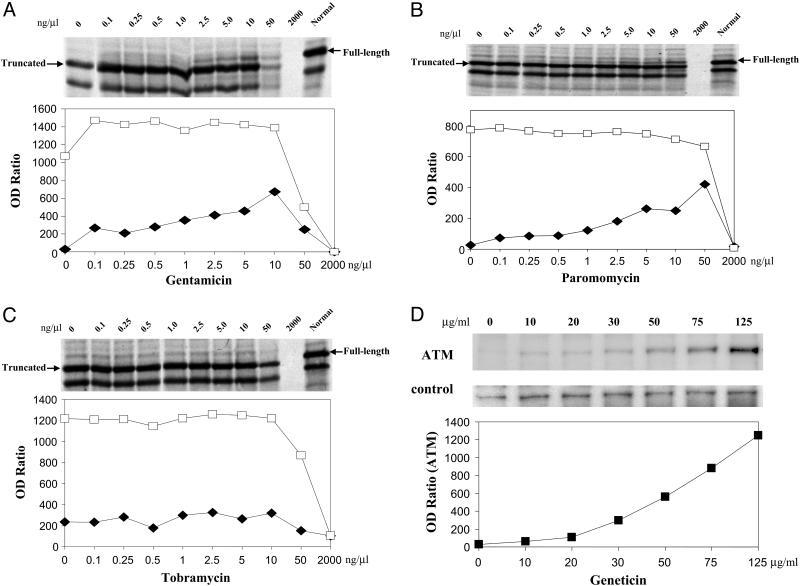
Read-Through with Other Aminoglycosides. We tested three additional aminoglycosides that are commonly used for clinical purposes: gentamicin, paromomycin, and tobramycin. Similar read-through of full-length ATM protein was observed for gentamicin (Fig. 2A) and paromomycin (Fig. 2B); however, as compared to the geneticin results for the same plasmid (TAT51; see Fig. 1A), both compounds showed considerably less read-through activity. IC50 levels showed similar toxicities at high concentrations. In limited testing, we observed no read-through activity with tobramycin at all concentrations tested (Fig. 2C).
Fig. 2.
Read-through effects for aminoglycosides in vitro (A–C) and ex vivo (D). Autoradiograms show in vitro read-through effects of gentamicin (A), paromomycin (B), and tobramycin (C) on the TAT51 (UGA C) mutation. Expression of a truncated 52-kDa PTT product (open squares) was inhibited by all three compounds at highest concentrations. The full-length 54-kDa read-through product (filled diamonds) was observed with gentamicin and paromomycin, but not with tobramycin. (D) Ex vivo geneticin-induced read-through of full-length ATM protein in A-T cells carrying the AT185LA (UAA G) mutation. LCLs were treated with varying concentrations of geneticin for 4 days. Nuclear lysates were immunoprecipitated with antibody to ATM and analyzed by Western blot. Control panel shows a nonspecific band from the same blot to evaluate loading of immunoprecipitates.
Ex Vivo ATM Read-Through. LCLs derived from A-T patients were incubated directly with either geneticin or gentamicin. At 4 days, maximum levels of read-through were observed. However, the level of induced ATM protein, which was negligible in the absence of aminoglycoside, was too low to be detected consistently by conventional immunoblotting (data not shown). To improve the sensitivity of testing, nuclear extracts were first immunoprecipitated with antibody to ATM, and the solubilized precipitate was then tested by immunoblotting using a second antibody to ATM. Both geneticin and gentamicin produced detectable ATM read-through protein, and the concentration was directly proportional to the concentration of aminoglycoside incubated with the LCLs. The highest read-through occurred at 125 μg/ml of geneticin (Fig. 2D). Concentrations higher than 125 μg/ml of geneticin were associated with increasing signs of toxicity and poor cell growth (data not shown).
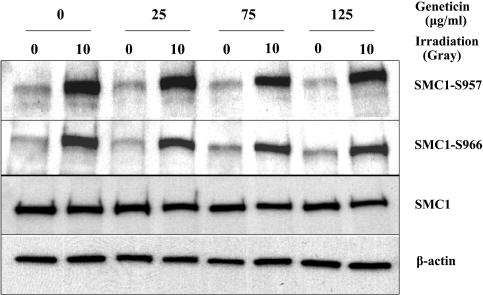
Using normal cells, the dosage of geneticin that induced ATM read-through did not cause any effects on the expression of two control genes, β-actin or SMC1, as measured by immunoblotting (Fig. 3). After 4 days of treatment, we observed normal ATM kinase activity, as demonstrated by phosphorylation levels of SMC1 Ser-957 and -966 after 10 Gy. To date, we have not been able to demonstrate ATM kinase activity in A-T cells treated with optimal doses of aminoglycosides by immunoblotting. We attribute this to the lack of sensitivity of this method. For this reason, we chose other methods to assess the function of read-through ATM protein.
Fig. 3.
Geneticin-induced effects on normal cells. Lanes marked with 0, no irradiation; lanes marked with 10, 10 Gy of irradiation. First two lanes, no drug treatment; second two lanes, 25 μg/ml; third two lanes, 75 μg/ml; fourth two lanes, 125 μg/ml. Two upper blots were developed with antibodies to SMC1 phosphor-Serine S957 or S966. Two lower blots show the same blots developed with antibodies to SMC1 or β-actin.
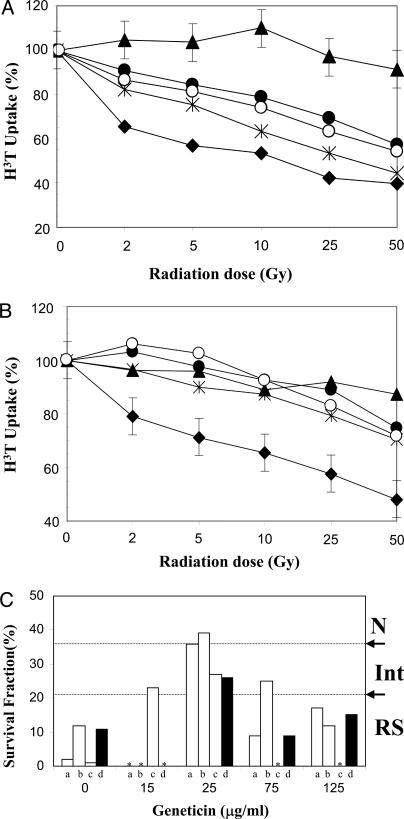
Functional Assessment of Read-Through ATM Protein. To assess whether aminoglycoside-induced read-through resulted in functional ATM protein, we performed radioresistant DNA synthesis (RDS), the CSA, and irradiation-induced autophosphorylation of ATM at Ser-1981, on geneticin-treated LCLs. We found that increasing concentrations of geneticin from 25 to 125 μg/ml produced a consistent pattern of correcting RDS in A-T cells, in response to increasing doses of radiation (Fig. 4A). No effect of geneticin was observed in a similar experiment using another A-T cell that did not carry a PTC mutation (Fig. 4B), indicating that aminoglycoside-induced correction of RDS requires a PTC mutation.
Fig. 4.
Functional analyses of expressed ATM protein. (A) Radioresistant DNA synthesis. A-T cells carrying the AT160LA mutation (UGA A) (filled triangles) were treated with increasing concentrations of geneticin, 25 μg/ml (asterisks), 75 μg/ml (filled circles), and 125 μg/ml (open circles), and were exposed to increasing doses of irradiation. Note that [3H]thymidine uptake of treated cells approached that of normal cells (filled diamonds). (B) Radioresistant DNA synthesis. Absence of geneticin-induced effect on an A-T cell line (BRAT-16.3) that did not carry a PTC mutation is shown. Standard error bars represent typical triplicate results for each curve. (C) Colony survival assay. The graph depicts radiosensitivity of A-T cells after 1-Gy irradiation, after 4 days of treatment with varying concentrations of geneticin. Experiments a, b, and c were performed by using cells with the AT187LA mutation (UAA G) (open bars). Experiment d (filled bars) used cells with the AT160LA mutation (UGA A). Note the correction of radiosensitivity at concentration of 25 μg/ml. Asterisks denote “not tested” for that concentration. N, normal; Int, intermediate; RS, radiosensitive.
Whereas RDS evaluates the S phase checkpoint, the CSA was used to assess the long-term effects of geneticin on A-T cell survival after radiation damage (1 Gy) (8). Fig. 4C shows the survival effects of increasing concentrations of geneticin on LCLs carrying two types of PTC mutations (UAA G and UGA A). Geneticin increased the colony survival fraction of both A-T LCLs from radiosensitive levels (i.e., <21%) to intermediate (21–36%) or normal levels (>37%) (8) at concentrations ranging from 15 to 75 μg/ml; the maximal CSA effect was observed at 25 μg/ml, similar to that seen with RDS. These results were somewhat surprising because the peak ex vivo ATM expression by immunoprecipitation was observed at 125 μg/ml (see Fig. 2D), suggesting that the RSA and CSA data may also reflect a toxic effect of geneticin at higher concentrations. Gentamicin produced similar but weaker functional effects (data not shown).
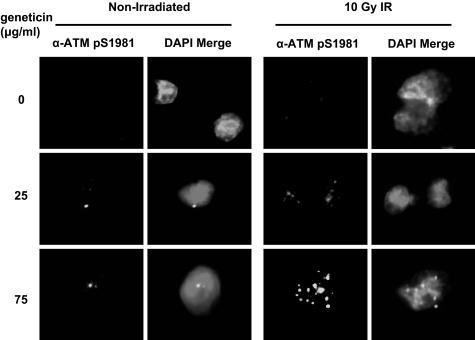
We tested for immunofluoresence (IF) of ATM pS1981 in AT187LA cells after 4 days of geneticin exposure. This assessed both the presence of ATM protein in A-T cells and its ability to autophosphorylate Ser-1981, an ATM kinase function. Forty-five minutes after 10-Gy irradiation, the cells showed irradiation-induced foci (IRIF). Although we observed an increase in intensity of immunostaining with 25 μg/ml of geneticin, as compared to 0 μg/ml geneticin, bright staining of IRIFs was more distinct in cells exposed to 75 μg/ml (Fig. 5); >80% of cells were IRIF positive. At a concentration of 125 μg/ml, most cells were no longer viable (data not shown). The unirradiated AT187LA cells showed only occasional foci (Fig. 5). When IF was performed on another A-T cell that did not carry a PTC mutation, no foci were observed in the unirradiated or irradiated cells in the presence or absence of geneticin (data not shown).
Fig. 5.
ATM pS1981 IRIFs of AT187LA cells after treatment with geneticin. Cells were irradiated with 10 Gy and were incubated for 45 min at 37°C before fixation on coverslips. ATM pS1981 IRIFs were not observed in untreated cells; maximum intensity of staining was seen in the nuclei of cells exposed to 75 μg/ml. Nonirradiated cells, untreated or exposed to comparable concentrations of geneticin, showed only occasional foci.
Discussion
We demonstrate that aminoglycoside-induced read-through of PTC mutations in the ATM gene results in a full-length ATM protein product with functional properties. We further describe a rapid and sensitive in vitro approach for screening of specific aminoglycosides on specific PTC mutations (in their own context) using plasmid-driven PTT; this method provided a semi-quantitative assessment of read-through, one that is easily adaptable to the analysis of PTC mutations in both animal and clinical models.
During four decades of successful development of numerous aminoglycoside antibiotics, extensive industrial and academic research has greatly improved the understanding of PTC read-through (5, 6, 9, 10, 11, 12). Aminoglycosides bind to the internal loop of helix 44 of the 16S ribosomal RNA, a region called the decoding site (11). Comparison of the free and aminoglycoside-bound oligonucleotide structures of this region showed that drug binding to the ribosome induces a local conformational change at the universally conserved 16S rRNA nucleotides A1492 and A1493, displacing these bases toward the minor groove of helix 44. The stability of gentamicin binding to the decoding site reflects the combinatorial effects of two particular hydroxyl functions at opposite poles of the molecule (12). In both prokaryotes and eukaryotes, aminoglycosides induce miscoding by mimicking the conformation change in 16S rRNA that would be induced by a correct codon–anticodon pair, thereby compromising the integrity of codon–anticodon proofreading during translation. As a general rule, glutamine is inserted at nonsense UAG or UAA read-through sites, whereas UGA sites miscode to tryptophan (10). The aminoglycoside binding may also influence the ability of release factors, such as RF1 and RF2, to stabilize the nascent protein strand in the ribosome for further elongation (9).
Aminoglycoside-induced read-through of human PTC mutations has therapeutic potential for nearly one-third of all genetic disorders. This is being critically evaluated for cystic fibrosis (13, 14, 15, 16) and muscular dystrophy (17, 18, 19, 20, 21, 22). After animal studies, gentamicin-induced CFTR protein was recently reported to improve transmembrane conductance across the nasal mucosa in a group of 10 patients with homozygous PTC mutations in the CTRF gene; patients with the more common del508F mutation were used as controls and showed no improvement of conductance (16). Studies treating mdx mice with gentamicin were reported to restore dystrophin function in skeletal muscle (17). However, the muscular dystrophy clinical trials have not been encouraging and a recent report failed to confirm the earlier mouse experiments (18, 20, 21). Aminoglycoside-induced read-through levels have varied from 2% to 10% (21). Aminoglycoside-induced PTC read-through has also been documented in other disorders, such as Hurler's (23), diabetes insipidus (24, 25), cystinosis (26), and X-linked retinitis pigmentosa (27).
Considerable complexity influences these therapeutic outcomes, such as the nature and pathophysiology of a disorder or the pairing of a specific aminoglycoside to a specific PTC mutation. The efficiency of read-through varies inversely to the efficiency of a stop codon, the ranking order for aminoglycoside read-through efficiency generally being UGA > UAA > UAG (28). On the other hand, this ranking varies from one species to another, and is influenced as well by the context within which a stop codon appears, especially with the “wobble” of the +4 nucleotide (22, 29). Other influences included the half-life of the target protein, the availability of biomarkers for documenting clinical improvement, the time that treatment is initiated with respect to the pathogenesis of a disease, the efficiency of drug delivery, and the doses used.
A-T is a promising model for evaluating aminoglycoside-induced read-through. The neurological deterioration is relentlessly progressive, and no treatment is presently available. Approximately 14% of ATM mutations (70 of 485 unique mutations) cause primary stop codons. Although 90% of patients have no detectable ATM protein, the few patients with notable protein levels (5–20%) have milder phenotypes (3, 4). Furthermore, we have documented elsewhere that obligate ATM heterozygotes, who live essentially normal lives, have only 40–50% ATM protein levels (4), suggesting that neurological improvement in A-T patients might be achieved with ATM protein levels somewhere between 5% and 40%. Although we have not yet directly addressed the issue of quantifying ATM read-through in various A-T cells, some of our in vitro and ex vivo data indicate very significant read-through levels, from 0 to 800 OD ratios in vitro, and 80% IRIF-positive cells after a 4-day exposure to 75 μg/ml geneticin.
ATM is a large protein (370 kDa, 150,000 nt), with slow turnover (30). It is thought to exist as an inactive homodimer that autophosphorylates in response to double-strand breaks and chromatin changes (2). The active monomer then functions as a serine–threonine kinase of many important substrates, including H2AX, SMC1, BRCA1, FANCD2, NBS1, MRE11, BLM, p53, p53BP1, MDM2, and CHK2. The ATM protein is found in low concentrations in most cells tested, primarily in the nucleus and, in lesser amounts, in the cytoplasm of terminally differentiated cells, such as Purkinje cells, which are severely depleted in the cerebellum of A-T patients (1). ATM expression is not significantly affected by cell cycle progression. Thus, ATM would appear to have a long half-life, being quickly autophosphorylated as needed for rejoining broken DNA strands and, presumably, quickly deactivated when not needed. Taken together, these facts suggest that A-T is a good candidate for aminoglycoside treatment.
Geneticin is widely used as a selection agent for transfection experiments. We observed no toxic effects of geneticin on expression of β-actin or SMC1. We also showed that the geneticin concentrations used to induce ATM PTC read-through did not inhibit ATM kinase functions in normal cells. On the other hand, our data further suggest that caution should be taken when geneticin is used as a selection agent in complementation experiments involving constructs containing PTC mutations.
The depth of previous research on aminoglycoside read-through encourages the possibility that even more effective compounds in this class can be identified for follow-up animal studies and, eventually, for the treatment of A-T and other genetic disorders. Other compounds that are not aminoglycosides may also influence PTC read-through. Some aminoglycosides also influence splicing, and this holds additional promise for the treatment of patients with genetic defects (31). Furthermore, it may be possible to employ combinations of aminoglycosides and other compounds, similar to how infections are often treated. Thus, high-throughput screening for such compounds should prove rewarding for many patients.
Acknowledgments
We thank David Lawrence for suggesting these experiments. We thank Martin F. Lavin (Queensland Institute of Medical Research, Brisbane, Australia) for his ATM immunoprecipitating antibody and for many valuable discussions. This work was supported by grants from the Ataxia-Telangiectasia Medical Research Foundation (Los Angeles), U.S. Public Health Service Grant NS35322, and the AT Medical Research Trust (United Kingdom).
Author contributions: C.-H.L., H.H.C., K.M.G., L.D., and R.A.G. designed research; C.-H.L., H.H.C., S.A.N., M.M., K.M.G., and L.D. performed research; C.-H.L., S.A.N., and L.D. contributed new reagents/analytic tools; C.-H.L., H.H.C., S.A.N., M.M., K.M.G., L.D., and R.A.G. analyzed data; and C.-H.L., K.M.G., and R.A.G. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: A-T, ataxia-telangiectasia; ATM, A-T mutated; IRIF, irradiation-induced foci; PTC, premature termination codon; PTT, protein truncation test; LCLs, lymphoblastoid cell lines; CSA, colony survival assay; RDS, radioresistant DNA synthesis.
References
- 1.Perlman, S., Becker-Catania, S. & Gatti, R. A. (2003) Semin. Pediat. Neurol. 10, 173-182. [DOI] [PubMed] [Google Scholar]
- 2.Bakkenist, C. J. & Kastan, M. B. (2003) Nature 421, 499-506. [DOI] [PubMed] [Google Scholar]
- 3.Sandoval, N., Platzer, M., Rosenthal, A., Dork, T., Bendix, R., Skawran, B., Stuhrmann, M., Wegner, R. D., Sperling, K., Banin, S., et al. (1999) Hum. Mol. Genet. 8, 69-79. [DOI] [PubMed] [Google Scholar]
- 4.Chun, H. H., Sun, X., Nahas, S. A., Teraoka, S., Lai, C. H., Concannon, P. & Gatt, R. A. (2003) Mol. Genet. Metab. 80, 437-443. [DOI] [PubMed] [Google Scholar]
- 5.Davies, J., Gorini, L. & Davis, B. D. (1965) Mol. Pharmacol. 1, 93-106. [PubMed] [Google Scholar]
- 6.Burke, J. F. & Mogg, A. E. (1985) Nucleic Acids Res. 13, 6265-6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Telatar, M., Wang, Z., Udar, N., Liang, T., Bernatowska-Matuszkiewicz, E., Lavin, M., Shiloh, Y., Concannon, P., Good, R. A. & Gatti, R. A. (1996) Am. J. Hum. Genet. 59, 40-44. [PMC free article] [PubMed] [Google Scholar]
- 8.Sun, X., Becker-Catania, S. G., Chun, H. H., Hwang, M. J., Huo, Y., Wang, Z., Mitui, M., Sanal, O., Chessa, L., Crandall, B., et al. (2002) J. Pediat. 40, 724-731. [DOI] [PubMed] [Google Scholar]
- 9.Karimi, R., Pavlov, M. Y., Buckingham, R. H. & Ehrenberg, M. (1999) Mol. Cell 3, 601-609. [DOI] [PubMed] [Google Scholar]
- 10.Nilsson, M. & Ryden-Aulin, M. (2003) Biochim. Biophys. Acta 1627, 1-6. [DOI] [PubMed] [Google Scholar]
- 11.Lynch, S. R., Gonzalez, R. L. & Puglisi, J. D. (2003) Structure (Cambridge, U.K.). 11, 43-53. [DOI] [PubMed] [Google Scholar]
- 12.Vicens, Q. & Westhof, E. (2003) J. Mol. Biol. 326, 1175-1188. [DOI] [PubMed] [Google Scholar]
- 13.Howard, M., Frizzell, R. A. & Bedwell, D. M. (1996) Nat. Med. 2, 467-469. [DOI] [PubMed] [Google Scholar]
- 14.Bedwell, D. M., Kaenjak, A., Benos, D. J., Bebok, Z., Bubien, J. K., Hong, J., Tousson, A., Clancy, J. P. & Sorscher, E. J. (1997) Nat. Med. 3, 1280-1284. [DOI] [PubMed] [Google Scholar]
- 15.Du, M., Jones, J. R., Lanier, J., Keeling, K. M., Lindsey, J. R., Tousson, A., Bebok, Z., Whitsett, J. A., Dey, C. R., Colledge, W. H., et al. (2002) J. Mol. Med. 80, 595-604. [DOI] [PubMed] [Google Scholar]
- 16.Wilschanski, M., Yahav, Y., Yaacov, Y., Blau, H., Bentur, L., Rivlin, J., Aviram, M., Bdolah-Abram, T., Bebok Z, Shushi, L., et al. (2003) N. Engl. J. Med. 349, 1433-1441. [DOI] [PubMed] [Google Scholar]
- 17.Barton-Davis, E. R., Cordier, L., Shoturma, D. I., Leland, S. E. & Sweeney, H. L. (1999) J. Clin. Invest. 104, 375-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wagner, K. R., Hamed, S., Hadley, D. W., Gropman, A. L., Burstein, A. H., Escolar, D. M., Hoffman, E. P. & Fischbeck, K. H. (2001) Ann. Neurol. 49, 706-711. [PubMed] [Google Scholar]
- 19.Dunant, P., Walter, M. C., Karpati, G. & Lochmuller, H. (2003) Muscle Nerve 27, 624-627. [DOI] [PubMed] [Google Scholar]
- 20.Politano, L., Nigro, G., Nigro, V., Piluso, G., Papparella, S., Paciello, O. & Comi, L. I. (2003) Acta Myol. 22, 15-21. [PubMed] [Google Scholar]
- 21.Howard, M. T., Anderson, C. B., Fass, U., Khatri, S., Gesteland, R. F., Atkins, J. F. & Flanigan, K. M. (2004) Ann. Neurol. 55, 422-426. [DOI] [PubMed] [Google Scholar]
- 22.Bidou, L., Hatin, I., Perez, N., Allamand, V., Panthier, J. J. & Rousset, J. P. (2004) Gene Therapy 11, 619-627. [DOI] [PubMed] [Google Scholar]
- 23.Keeling, K. M. & Bedwell, D. M. (2002) J. Mol. Med. 80, 367-376. [DOI] [PubMed] [Google Scholar]
- 24.Schulz, A., Sangkuhl, K., Lennert, T., Wigger, M., Price, D. A., Nuuja, A., Gruters, A., Schultz, G. & Schoneberg, T. (2002) J. Clin. Endocrinol. Metab. 87, 5247-5257. [DOI] [PubMed] [Google Scholar]
- 25.Sangkuhl, K., Schulz, A., Rompler, H., Yun, J., Wess, J. & Schoneberg, T. (2004) Hum. Mol. Genet. 13, 893-903. [DOI] [PubMed] [Google Scholar]
- 26.Helip-Wooley, A., Park, M. A., Lemons, R. M. & Thoene, J. G. (2002) Mol. Genet. Metab. 75, 128-133. [DOI] [PubMed] [Google Scholar]
- 27.Grayson, C., Chapple, J. P., Willison, K. R., Webster, A. R., Hardcastle, A. J. & Cheetham, M. E. (2002) J. Med. Genet. 39, 62-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin, R., Mogg, A. E., Heywood, L. A., Nitschke, L. & Burke, J. F. (1989) Mol. Gen. Genet. 217, 411-418. [DOI] [PubMed] [Google Scholar]
- 29.Manuvakhova, M., Keeling, K. & Bedwell, D. M. (2000) RNA 6, 1044-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fukao, T., Kaneko, H., Birrell, G., Gatei, M., Tashita, H., Yoshida, T., Cross, S., Kedar, P., Watters, D., Khana, K. K., et al. (1999) Blood 94, 1998-2006. [PubMed] [Google Scholar]
- 31.Schroeder, R., Waldsich, C. & Wank, H. (2000) EMBO J. 19, 1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]