Abstract
Carbohydrate response element (ChRE)-binding protein (ChREBP) is a recently discovered transcription factor that is activated in response to high glucose concentrations in liver independently of insulin. ChREBP was first identified by its ability to bind the ChRE of the liver pyruvate kinase (LPK) gene. We recently reported that the increase in expression of multiple liver lipogenic enzyme mRNAs elicited by feeding a high-carbohydrate diet as well as that of LPK mRNA is markedly reduced in mice lacking ChREBP gene expression (ChREBP-/-) in comparison to WT mice. The present study provides evidence for a direct and dominant role of ChREBP in the glucose regulation of two key liver lipogenic enzymes, acetyl-CoA carboxylase (ACC) and fatty acid synthase (FAS). ACC, FAS, and LPK mRNA levels were higher in WT hepatocytes cultured with high (25 mM) rather than low (5.5 mM) glucose medium, but there was no effect of glucose concentration on these mRNA levels in ChREBP-/- hepatocytes. Similarly, reporter constructs containing ACC, FAS, or LPK gene ChREs were responsive to glucose when transfected into WT but not ChREBP-/- hepatocytes, and glucose transactivation of the constructs in ChREBP-/- hepatocytes was restored by cotransfection with a ChREBP expression plasmid. ChREBP binding to ACC, FAS, and LPK ChRE sequences in vitro was demonstrated by electrophoretic mobility super shift assays. In vivo binding of ChREBP to ACC, FAS, and LPK gene promoters in intact liver nuclei from rats fed a high-carbohydrate diet was demonstrated by using a formaldehyde crosslinking and chromatin immunoprecipitation procedure.
In mammals, simple sugars derived from the digestion of dietary carbohydrates are taken up by the liver and converted to fatty acids for long-term storage when they exceed the amount needed to meet the body's short-term energy requirements. Consumption of high-carbohydrate diets leads to increased mRNA expression, primarily through increased gene transcription, for more than a dozen enzymes in the glycolytic and lipogenic pathways involved in converting glucose to fatty acids (recently reviewed in refs. 1, 2, 3, 4). Included among these enzymes is liver pyruvate kinase (LPK), a regulatory enzyme at the distal end of the glycolytic pathway, acetyl-CoA carboxylase (ACC), which catalyzes the first step of fatty acid biosynthesis, and fatty acid synthase (FAS).
Insulin, which is secreted by the pancreas in response to elevations in blood glucose, is widely recognized to play an essential role in stimulating lipogenesis. SREBP-1c, an insulin-activated isoform of the sterol regulatory element binding family of transcription factors, has been identified as the principal insulin-responsive mediator of increased hepatic lipogenic enzyme gene expression in rodents fed high-carbohydrate diets (5, 6, 7, 8). However, several lines of evidence suggest that a second, glucose-responsive transcription factor contributes to increased lipogenic gene transcription. In vitro in isolated hepatocytes, glucose and insulin display a marked synergism in the transactivation of ACC, FAS, and S14 gene expression (9). S14 is a nuclear protein whose expression is closely associated with lipogenesis (10). The potentiating effects of glucose on ACC, FAS, and S14 gene transactivation were not eliminated by SREBP-1c overexpression (9), suggesting that glucose does not act merely to increase the expression or activation of SREBP-1c. In addition, carbohydrate-responsive promoter regions distinct from known insulin response elements have been identified for a number of lipogenic enzyme genes (recently reviewed in ref. 4). Finally, in SREBP-1c-deficient mice in vivo, the induction of lipogenic enzyme expression by high-carbohydrate diets was reduced but not eliminated (7), providing evidence for an additional physiologically significant mediator of carbohydrate-induced lipogenic enzyme gene expression.
Carbohydrate response element (ChRE)-binding protein (ChREBP) is a recently discovered transcription factor responsible for glucose-induced transcription of LPK (11). ChREBP is expressed constitutively in hepatocytes but, under low glucose conditions, remains in the cytosol and unable to bind DNA as the result of phosphorylation at multiple sites (12). The dephosphorylation of ChREBP in response to high glucose does not require insulin. ChREBP was initially purified based on its ability to bind the LPK ChRE, a palindrome of two modified E boxes separated by five nucleotides (11). Glucose-responsive promoter regions identified for the murine ACC, FAS, and S14 genes contain similar double E box motifs (13, 14, 15). The similarity of these sites to the LPK ChRE suggested to us that ChREBP may directly activate lipogenic gene expression by binding to these sites.
Consistent with this possibility, we recently reported that mRNA levels for all of the major lipogenic enzyme genes, as well as for LPK, were significantly lower in ChREBP-/- mice fed a high-starch diet than those in WT mice (16). Liver triglycerides and total body fat synthesis were decreased in ChREBP-/- mice as a consequence of the decreased lipogenic enzyme expression. These data provide compelling evidence for the physiologically important in vivo role that ChREBP plays in the glucose regulation of LPK and lipogenic enzyme gene transcription. However, it is difficult to determine in intact mice whether the effect of ChREBP deficiency on the transcription of these genes reflects their direct modulation by ChREBP in WT mice or whether indirect mechanisms or secondary deficiencies result in their decreased expression in ChREBP-/- mice.
In the present study, these questions have been addressed by using hepatocytes isolated from CHREBP-/- and WT mice. In addition, specific association of ChREBP with promoter regions of the LPK, ACC, and FAS genes in vivo was investigated by DNA–protein crosslinking followed by chromatin immunoprecipitation (ChIP).
Materials and Methods
Materials. All chemicals were reagent grade and were purchased from Sigma unless otherwise indicated.
Animals. Generation of the ChREBP knockout (ChREBP-/-) mice has been described (16). ChREBP-/- mice and WT littermates from heterozygote ChREBP-/+ breeding pairs were used for these studies and were fed standard rodent chow (Harlan Teklad, Indianapolis). The mice were housed with 12-h light/12-h dark cycles, and hepatocytes were routinely isolated before 9 a.m. All animal experiments were carried out under protocols approved by the University of Texas Southwestern Medical Center Institutional Animal Care and Use Committee and followed the National Institutes of Health guidelines, “Using Animals in Intramural Research.”
Construction of Plasmids. The ChREBP expression vector in pcDNA3.1 has been described (12). Reporter plasmids were constructed with the luciferase expression plasmid pGL-3 Basic vector (Promega). Cloning of the promoter region from -206 to -7 of the rat LPK gene has been described (12). The ACC reporter plasmid contained the promoter region from -514 to +29 of the mouse ACC gene. The FAS reporter plasmid contained a recently identified distal glucose response element of the rat FAS gene (14), from -7221 to -7125, in tandem with the previously characterized proximal ChRE/insulin receptor substrate site from -214 to +27. Additional details on construction of the ACC and FAS reporter plasmids are available in Supporting Text, which is published as supporting information on the PNAS web site.
Primary Hepatocyte Culture and Transfection. Primary hepatocytes were prepared by collagenase perfusion (17) and plated in positively charged tissue culture plates (Corning) in DMEM (Sigma) supplemented with 10 nM insulin, 100 nM dexamethasone, 100 units/ml penicillin, 100 μg/ml streptomycin, and 10% dialyzed FBS (Invitrogen). For investigation of endogenous mRNA expression, the cells were plated at a density of 5 × 106 cells per well in six-well tissue culture plates. Unattached hepatocytes were removed after 2–4 h, fresh culture medium containing the various concentrations of insulin and glucose indicated in the figure legends was added, and the hepatocytes were incubated for an additional 6 h before harvesting and RNA isolation. Relative LPK, ACC, and FAS mRNA levels were determined by real-time RT-PCR using the comparative Ct method as we have described (16). Cyclophilin mRNA was used as the invariant control. For luciferase reporter assays, hepatocytes were plated at a density of 1.25 × 105 cells per well in 24-well tissue culture plates and transfected by using Lipofectamine with reporter plasmid, internal control plasmid pRL-TK, and ChREBP expression vector as indicated in the figure legends. After 4 h, the medium was replaced by DMEM supplemented with 1 nM insulin, 100 nM dexamethasone, 100 units/ml penicillin, 100 μg/ml streptomycin 10% dialyzed FBS, and 5.5 or 27.5 mM glucose. The cells were incubated for an additional 20 h, and luciferase reporter activity determined by using the Dual-Luciferase Reporter Assay System (Promega).
Gel Mobility-Shift Assay. Gel mobility-shift assays (18) were performed as described (11) by using double-stranded oligonucleotides with the following 5′ to 3′ top strand sequences: LPK, ATGGGCGCACGGGGCACTCCCGTGGTTCCTGG; LPK(d), GGGCGCACGGGGCCTCCCGTGGTTCCT; FAS, CTTCCTGCATGTGCCACAGGCGTGTCACCCTC; ACC, CGGTGTCCATGTGAAAACACTGTGGGCAGTCT. ChREBP antiserum (ResGen, Huntsville, AL) was added to the reaction mixtures for supershift assays. The E-box sequences are underlined and denoted in bold. ChIP Assay. Intact nuclei were isolated from either rat or mouse liver, as described before (11), and washed with PBS. Nuclear pellets were suspended in PBS and treated with 1.0% formaldehyde at room temperature to crosslink DNA-binding proteins to cognate cis-acting elements; after 30 min, 125 mM glycine was added to quench the reaction. The nuclei were then suspended in lysis buffer [50 mM Tris·HCl, pH 8.0/1.0% SDS/10 mM EDTA and one Complete Protease Inhibitor minitablet (Roche Diagnostics) per 10 ml], and the nuclear lysate was sonicated to shear chromosomal DNA to a size of ≈500–750 bp. Insoluble material was removed by centrifugation, and the soluble supernatant solution (100 μl) was diluted 10-fold with ChIP dilution buffer (16.7 mM Tris·HCl, pH 8.0/1.1% Triton X-100/1.2 mM EDTA/167 mM NaCl). ChIP was performed as described (19) with some modifications. The diluted chromatin was precleared by incubating for 3 h with 5 μl of normal rabbit serum-coupled protein A-agarose beads saturated with BSA (1 mg/ml) and herring sperm DNA (0.4 mg/ml). The mixture was spun at 2,000 rpm for 2 min in an Eppendorf 5402 microcentrifuge at 4°C, and the supernatant was allowed to bind to either 5 μl of normal rabbit serum or ChREBP antiserum overnight at 4°C. Immunocomplexes were pulled down by protein A-agarose beads and washed extensively as described (19). DNA precipitated in the anti-ChREBP antibody complex was eluted, decrosslinked by incubating at 65°C for 16 h, and treated by Proteinase K. The purified DNA was dissolved in 100 μl of Tris/EDTA (pH 8.0) and used for semiquantitative PCR with gene-specific primers, the sequences of which are provided in Supporting Text. In the semiquantitative PCR, one of the primers (6 pmol per reaction) was labeled with [γ-32P]ATP catalyzed by T4 polynucleotide kinase. DNA purified directly from the chromatin (input DNA) was used as the positive control for PCR. Immunoprecipitated DNA as well as input DNA was subjected to PCR under the following conditions: 94°C for 2 min, 35 cycles of 94°C for 30s, 55°C for 30s, 72°C for 1 min. The reaction products were separated on a polyacrylamide gel (10% acrylamide) with Tris borate-EDTA as running buffer. The dried gel was exposed to x-ray film.
Results
Glucose-Responsive Expression of LPK and Lipogenic Enzyme Genes in Hepatocytes Isolated from ChREBP-/- and WT Mice. Endogenous LPK, ACC, and FAS mRNA levels were significantly higher in WT hepatocytes cultured in high-glucose (25 mM) medium than those cultured in low-glucose (5.5 mM) medium in the absence of insulin (Fig. 1, first bar grouping). Inclusion of insulin (10 nM) in WT hepatocyte cultures containing high but not low concentrations of glucose resulted in even higher levels of ACC and FAS mRNA expression (Fig. 1 B and C, second bar grouping), consistent with the known insulin-dependent, SREBP-1c stimulated transcription of these genes. LPK mRNA expression in the WT cultures was not significantly increased by the addition of insulin irrespective of glucose concentration (Fig. 1A, second bar grouping). In marked contrast to the glucose-responsive gene expression of LPK, ACC, and FAS in WT hepatocytes, no effect of glucose concentration on the mRNA expression of these enzymes was observed in hepatocytes isolated from ChREBP-/- mice in either the absence or presence of insulin (Fig. 1, third and fourth bar groupings).
Fig. 1.
Endogenous mRNA levels of LPK (A), ACC (B), and FAS (C) in hepatocytes isolated from WT but not ChREBP-/- mice are elevated by culturing with high-glucose medium. Adherent hepatocyte cultures prepared as described in Materials and Methods were incubated for an additional 8 h in culture medium containing either a low (5.5 mM, open bars) or high (25 mM, filled bars) glucose (Glc) concentration and in the absence or presence (10 nM) of insulin. RNA was isolated, and relative mRNA levels for LPK, ACC, and FAS were determined by real-time RT-PCR using the comparative Ct method. Enzyme mRNA levels in each sample were normalized to cyclophilin mRNA as the invariant control. The fold change in expression level of each normalized enzyme mRNA level was determined relative to the value in WT, low-glucose hepatocyte cultures containing no insulin, which was arbitrarily defined as one. The values presented are assay means and SD. The experiment shown is representative of two independent experiments.
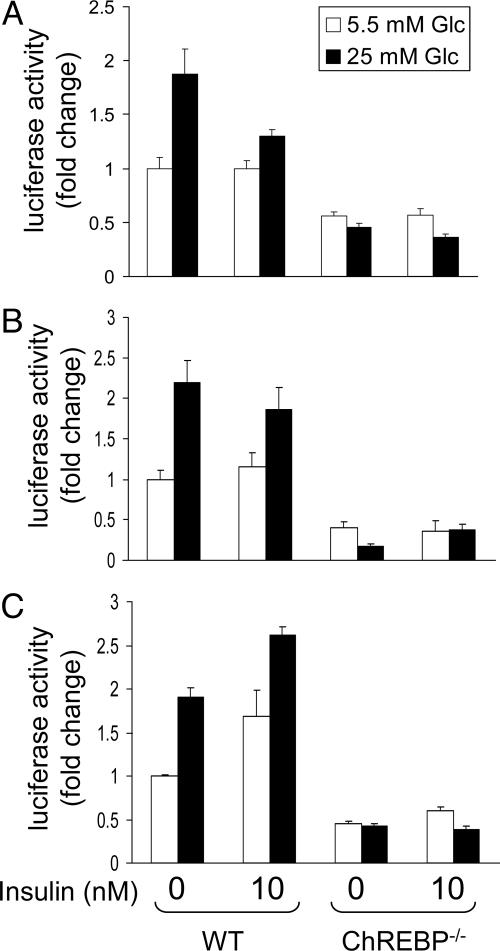
To examine whether previously characterized glucose-responsive promoter regions of the ACC and FAS genes support ChREBP-dependent glucose-stimulated gene transcription, luciferase reporter plasmids containing ChREs of murine ACC, FAS, or LPK genes were transfected into hepatocytes isolated from WT and ChREBP-/- mice. In WT hepatocytes, LPK, ACC, and FAS promoter-driven luciferase expression was significantly higher in the presence of high glucose without additional insulin (Fig. 2, first bar grouping). The glucose-dependent increases in luciferase reporter expression for the three gene promoters were similar in magnitude to the glucose-dependent elevation of the endogenous mRNAs (Fig. 1, first bar grouping). Not surprisingly, FAS promoter-driven luciferase expression in the WT hepatocyte cultures was increased further by the addition of insulin because the cloned FAS gene promoter region contains insulin as well as glucose-responsive elements. In contrast, LPK and ACC promoter-dependent luciferase expression was somewhat lower in WT cultures containing both high glucose and insulin than in those containing high glucose alone. This difference may reflect a modest decrease in glucose concentration over the culture period as the result of insulin-stimulated glycolysis and glycogenesis. In ChREBP-/- hepatocytes, LPK, ACC, and FAS promoter-driven luciferase expression in low glucose cultures (Fig. 2, third bar grouping) was lower than that in WT hepatocytes cultured under the same conditions and, importantly, did not increase in response to higher glucose concentrations in either the absence or presence of insulin (Fig. 2, third and fourth bar groupings). Cotransfection of ChREBP-/- hepatocytes with a plasmid constitutively expressing full-length ChREBP restored glucose transactivation of the LPK, ACC, and FAS promoters to levels similar to those in cotransfected WT hepatocytes (Fig. 3).
Fig. 2.
Luciferase reporter activities of LPK (A), ACC (B), and FAS (C) promoter constructs transfected into ChREBP-/- hepatocytes are not responsive to glucose (Glc). Primary cultured hepatocytes from WT and ChREBP-/- mice were cotransfected with pRL-TK (thymidine kinase promoter driving Renilla luciferase gene) as the internal control and pGL-3 Basic-expressing firefly luciferase under control of the rat LPK promoter region from -206 to -7(A), the mouse ACC promoter from -514 to +29 (B), or the rat FAS promoter region from -7221 to -7125 in tandem with the -214 to +27 region. After transfection, the cells were incubated with 5.5 mM (open bars) or 25 mM (filled bars) glucose-containing medium in the absence or presence of insulin (10 nM) for 12 h. Luciferase activities were measured and corrected for differences in transfection efficiency by Renilla luciferase activity. The values are presented relative to the activity in WT, low-glucose hepatocyte cultures containing no insulin, which was arbitrarily defined as one. The values presented are the mean and SD of replicate (three to five) cultures from a single experiment representative of more than six independent experiments performed with slightly varied incubation times.
Fig. 3.
Cotransfection with a ChREBP expression vector restores glucose (Glc)-responsive reporter expression from LPK (A), ACC (B), and FAS (C) promoters transfected in ChREBP-/- hepatocytes. WT and ChREBP-/- primary hepatocyte cultures were transfected with an expression vector encoding authentic full-length rat ChREBP in addition to the pRL-TK and pGL-3 Basic constructs described in Fig. 2. After transfection, the cells were incubated with 5.5 mM (open bars) or 25 mM (filled bars) glucose-containing medium in the absence of insulin for 20 h. Luciferase activities were measured and corrected for differences in transfection efficiency by Renilla luciferase activity. The values are presented relative to the activity in WT, low-glucose hepatocyte cultures, which was arbitrarily defined as one. Data are shown as the means ± SD (error bars) of six transfections.
ChREBP Binds to the ChRE of Lipogenic Gene Promoters. Gel electrophoretic mobility-shift assays were used to investigate ChREBP binding to ChREs of the ACC and FAS genes. Synthetic oligonucleotides containing the LPK ChRE, double E box motifs from the ACC and FAS gene promoters, and the non-ChREBP binding spacing mutant LPK(d) as a control were incubated with liver nuclear extract from rats fed a high-sucrose diet as a source of activated ChREBP. Complexes that comigrated with the previously identified ChREBP-bound LPK ChRE oligonucleotide were formed with oligonucleotides containing the ACC and FAS ChRE sequences, but not with the LPK(d) oligonucleotide (Fig. 4A). The presence of ChREBP in these complexes was confirmed by incubating the reaction mixtures with ChREBP antibody, which further retarded or “supershifted” the mobility of the ChREBP-oligonucleotide complexes (Fig. 4B). The ability of the ChRE elements of the lipogenic enzyme genes to compete with the LPK ChRE site for ChREBP binding was investigated by incubating ChREBP-bound, 32P-labeled LPK oligonucleotide with unlabeled LPK, ACC, and FAS ChRE sequence oligonucleotides. Both lipogenic ChRE oligonucleotides were able to compete with and release the 32P-labeled LPK oligonucleotide nearly as effectively as unlabeled LPK oligonucleotide (Fig. 4C).
Fig. 4.
ChREBP binds to the ChRE of lopogenic gene promoters. (A) Gel-shift analysis of ChREBP bound to ChREs of LPK, ACC1, and FAS. Nuclear extract from fed rat liver was incubated with radiolabeled olionucleotides corresponding to the ChREs of LPK, ACC1, and FAS. The LPK(d) oligonucleotide, in which the spacing between the two E boxes has been reduced to four nucleotides, was included as a negative control. The position of the DNA–ChREBP complexes is indicated by an arrow. (B) Gel-shift analysis of ChREBP bound to the ChREs of lipogenic gene promoters after immunoreaction with ChREBP antibodies. ChREBP antiserum was added at 1:25 (+) or 1:5 (++) dilutions to binding reactions as described. (C) Competition of ChREBP binding to the radiolabeled ChRE of LPK by unlabeled oligonucleotides of the ChREs of LPK, ACC, and FAS. The unlabeled LPK, ACC, and FAS ChRE oligonucleotides indicated were added in increasing amounts, 10-, 25-, and 150-fold, to binding reactions containing radiolabeled LPK ChRE oligonucleotide.
Anti-ChREBP ChIP Assay. If transcriptional activation of lipogenic enzyme genes and LPK in response to ingestion of a high-carbohydrate meal results in part from ChREBP binding to their ChRE sites, a direct test requires examination of ChREBP promoter occupancy. In vivo interaction of ChREBP with LPK and lipogenic gene promoters was examined by a ChIP assay (Fig. 5). Intact liver nuclei isolated from rodents fed a high-starch diet were treated with formaldehyde to crosslink DNA and bound protein. After nuclear lysis, the chromatin was sheared and subjected to immunoprecipitation by using ChREBP or normal antiserum. DNA sequences in the immunoprecipated complexes were analyzed by PCR using primers designed to amplify the ChRE sites of LPK, ACC, and FAS promoters and the albumin promoter as a negative control. LPK, ACC, and FAS, but not albumin, DNA sequences were detected in crosslinked samples immunoprecipated with anti-ChREBP (Fig. 5, Ch-X) but not in noncrosslinked samples (Ch-NX) or in samples immunoprecipitated with nonimmune serum (NS-X). These findings indicate that ChREBP specifically binds to the glucose response elements of LPK and these lipogenic enzyme genes in vivo.
Fig. 5.
ChIP assay of rat and mouse liver nuclear extract. Intact nuclei isolated from liver of rat and mouse fed a high-starch diet were treated with formaldehyde to crosslink DNA with associated DNA-binding proteins. Chromatin fragments immunoprecipitated with ChREBP antibodies (Ch-X) and purified input DNA were amplified by PCR with primers for rat LPK, rat (r) FAS, rat albumin, mouse (m) ACC, and mouse albumin gene promoters, and the reaction products were separated on a 10% polyacrylamide gel. Immunoprecipitation of noncrosslinked DNA with anti-ChREBP (Ch-NX) and immunoprecipitation with normal serum (NS-X) were performed as controls.
Discussion
The discovery of tandem E box-like sequences similar to the LPK ChRE within carbohydrate-responsive promoter regions of several lipogenic enzyme genes fueled speculation that a single glucose-dependent transcription factor might be responsible for the transcriptional activation of both LPK and carbohydrate-responsive lipogenic enzyme genes in liver. However, previous attempts to identify common nuclear proteins binding to ChRE sites in ACC, FAS, or S14 lipogenic genes and the LPK gene failed to find convincing evidence for any factors conferring appropriate transcriptional activation in response to glucose (20, 21, 22, 23).
ChREBP has been unequivocally established as the dominant positive regulator of LPK gene transcription in vitro and in vivo. In previous studies, we found that in vitro the binding specificity of ChREBP-containing nuclear protein complexes to a series of modified E box palindromes based on the LPK ChRE matched the ability of these sequences to support glucose-responsive transcription of reporter constructs transfected into WT hepatocytes (11). Furthermore, an in vitro-expressed DNA binding domain of the ChREBP protein displays identical sequence specificity. In the present study, in hepatocytes isolated from ChREBP-/- mice, neither LPK mRNA levels nor LPK promoter-driven expression from transfected luciferase reporter constructs were responsive to glucose; the latter activity was restored by cotransfection with a ChREBP expression vector. In vivo, the hormonal and dietary regulation of ChREBP activation in liver (12), which requires dephosphorylation for nuclear transport and DNA binding, is completely consistent with the dietary regulation of LPK mRNA expression. In addition, LPK mRNA levels in ChREBP-/- mice under all dietary conditions thus far examined are 4- to 10-fold lower than in comparably fed WT mice (ref. 16 and unpublished observations).
ChREBP-/- mice provided the first, to our knowledge, in vivo evidence that ChREBP is required for the maximal transcriptional activation of not only LPK gene expression but also all lipogenic enzyme genes including ACC, FAS, S14, ATP citrate lyase, and stearyl CoA desaturase (16). However, the question of whether ChREBP directly functions in the glucose activation of lipogenic enzyme gene transcription still remained. In this study, we provide compelling evidence that ChREBP functions directly in the glucose activation of the key lipogenic enzyme genes, ACC and FAS.
In hepatocytes isolated from ChREBP-/- mouse liver, in contrast to those from WT littermates, ACC and FAS mRNA levels were not elevated when cultured in medium with high concentrations of glucose, even in the presence of elevated levels of insulin. The lack of ACC and FAS mRNA induction in ChREBP-/- hepatocytes cannot be attributed to a deficiency in SREBP-1c. Hepatic SREBP-1c mRNA and protein levels are the same in WT and ChREBP-/- mice (12) and, after hepatocyte isolation, SREBP-1c mRNA levels in both WT and ChREBP-/- hepatocytes in the absence of insulin declined at similar rates to ≈50% of the starting value after 6 h (data not shown). Because transcriptional activation of ACC and FAS genes requires insulin-as well as glucose-dependent signals, these in vitro results are important in providing evidence that the lack of carbohydrate induction of lipogenic gene expression in ChREBP-/- mice, where insulin levels are high, does not result from any unrecognized elevations in counterregulatory hormones.
ACC, FAS, and LPK promoter-driven expression of luciferase reporter constructs also failed to respond to high glucose when transfected into ChREBP-/- but not WT hepatocytes. The restoration of glucose-induced reporter expression by cotransfection of the hepatocytes with a plasmid expressing ChREBP demonstrates that no deficiency other than that of ChREBP prevents the glucose response. Although it is possible that the effect of restoring ChREBP expression on glucose induction of the reporter constructs in isolated hepatocytes is indirect, several lines of evidence argue against this. First, ChREBP binding to putative ChRE double E box motifs of the lipogenic genes was demonstrated in electrophoretic mobility-shift assays with antibodies against ChREBP. Moreover, ChREBP binding to the LPK ChRE sequence was effectively displaced by competition with ACC and FAS ChREs, suggesting that the binding is of similarly high affinity. Finally, if in vivo glucose-dependent stimulation of ACC and FAS genes is mediated by ChREBP binding to these sites, specific ChREBP binding to the promoter regions of the LPK, ACC, and FAS genes should be demonstrable in hepatic nuclei from rodents fed a high-carbohydrate diet. ChIP assays revealed ChREBP binding to the promoter regions of the LPK, ACC, and FAS genes, but not that of the glucose-unresponsive albumin gene.
These results clearly demonstrate that, in the absence of ChREBP, no other factor in liver activates expression of the lipogenic enzyme genes ACC and FAS in response to high glucose. Based on our previous data obtained with ChREBP-/- mice (16) and the results reported here, we conclude that ChREBP directly regulates LPK and the lipogenic enzyme genes in response to high glucose in vivo.
Stoeckman et al. (24) recently reported that, in the human embryonic kidney cell line HEK293, ChREBP overexpression increased reporter expression from a consensus ChRE-containing promoter as well as from hybrid constructs containing two copies of the LPK, ACC, and S14 ChREs, but only when coexpressed with Mlx. Mlx is a member of the c-Myc family of transcription factors that has previously been identified as a potential ChREBP-binding partner (25). Stoeckman et al. (24) also reported that, in primary hepatocytes, coexpression of Mlx had no effect on ChREBP-induced expression of any of the lipogenic gene ChRE constructs, but speculated that high levels of endogenous Mlx in hepatocytes already were sufficient to saturate the glucose response. If true, this evidence would still argue that ChREBP, and not Mlx, is the glucose-responsive factor required for transcriptional activation of LPK and lipogenic gene expression in liver. In addition, ChREBP purified from fed rat liver based on retention of DNA-binding activity (11) contained less than stoichiometric amounts of Mlx as well as other low molecular weight proteins (unpublished results), suggesting that ChREBP may additionally dimerize with other binding partners as well as form functional homodimers. The question of whether ChREBP forms heterodimers or tetramers containing Mlx or potentially other binding partners in liver that could confer distinct regulatory responses is potentially important and warrants further investigation. However, caution should be exercised in extrapolating results from HEK293 cells to the in vivo condition in liver because of both the artificial nature of the reporter constructs investigated and the frequency with which cell lines display anomalous regulatory mechanisms.
A comparison of the mRNA expression patterns of various glycolytic and lipogenic enzymes in SREBP-1c-deficient (7, 8) and ChREBP-/--deficient mice (16) reveals that both transcription factors affect similar enzyme genes. However, the data obtained with hepatocytes in vitro (9) and with ChREBP-/- mice in vivo (16) demonstrate that these transcription factors act independently. Consistent with numerous previous reports that glucose-responsive LPK and lipogenic gene expression requires the metabolism of glucose (see refs. 1, 2, 3, 4), we previously provided evidence that ChREBP activation by dephosphorylation is not directly stimulated by glucose but rather by the glucose metabolite Xu-5P through activation of a Xu-5P-activated protein phosphatase. Thus, ChREBP activation depends on increased glucose flux through the proximal portion of the glycolytic pathway and the pentose shunt. Glucokinase is required for glucose entry into the glycolytic pathway, and it has been reported that insulin can indirectly regulate ChREBP activation through its activation of glucokinase (26). Although the mechanisms of glucokinase activation and induction of gene transcription by insulin are unknown, glucokinase mRNA levels are significantly decreased in liver of SREBP-1c-deficient mice. Thus, the pattern of liver enzyme mRNAs displaying reduced carbohydrate induction in SREBP-1c-/- mice may reflect both the absence of SREBP-1c and a secondary deficiency in ChREBP activity.
Supplementary Material
Author contributions: B.C.M. and K.U. designed research; S.I. and K.I. performed research; S.I. and K.I. analyzed data; and B.C.M. and K.U. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: LPK, liver pyruvate kinase; ACC, acetyl-CoA carboxylase; FAS, fatty acid synthase; ChRE, carbohydrate response element; ChREBP, ChRE-binding protein; ChIP, chromatin immunoprecipitation.
References
- 1.Towle, H. C., Kaytor, E. N. & Shih, H. M. (1997) Annu. Rev. Nutr. 17, 405-433. [DOI] [PubMed] [Google Scholar]
- 2.Girard, J., Ferre, P. & Foufelle, F. (1997) Annu. Rev. Nutr. 17, 325-352. [DOI] [PubMed] [Google Scholar]
- 3.Vaulont, S., Vasseur-Cognet, M. & Kahn, A. (2000) J. Biol. Chem. 275, 31555-315888. [DOI] [PubMed] [Google Scholar]
- 4.Foufelle, F. & Ferre, P. (2002) Biochem. J. 366, 377-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foretz, M., Pacot, C., Dugail, I., Lemarchand, P., Guichard, C., Le Liepvre, X., Berthelier-Lubrano, C., Spiegelman, B., Kim, J. B., Ferre, P. & Foufelle, F. (1999) Mol. Cell. Biol. 19, 3760-3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foretz, M., Guichard, C., Ferre, P. & Foufelle, F. (1999) Proc. Natl. Acad. Sci. USA 96, 12737-12742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang, G., Yang, J., Horton, J. D., Hammer, R. E., Goldstein, J. L. & Brown, M. S. (2002) J. Biol. Chem. 277, 9520-9528. [DOI] [PubMed] [Google Scholar]
- 8.Engelking, L. J., Kuriyama, H., Hammer, R. E., Horton, J. D., Brown, M. S., Goldstein, J. L. & Liang, G. (2004) J. Clin. Invest. 113, 1168-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stoeckman, A. K. & Towle, H. C. (2002) J. Biol. Chem. 277, 27029-20735. [DOI] [PubMed] [Google Scholar]
- 10.Cunningham, B. A., Moncur, J. T., Huntington, J. T. & Kinlaw, W. B. (1998) Thyroid 8, 815-825. [DOI] [PubMed] [Google Scholar]
- 11.Yamashita, H., Takenoshita, M., Sakurai, M., Bruick, R. K., Henzel, W. J., Shillinglaw, W., Arnot, D. & Uyeda, K. (2001) Proc. Natl. Acad. Sci. USA 98, 9116-9121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawaguchi, T., Takenoshita, M., Kabashima, T. & Uyeda, K. (2001) Proc. Natl. Acad. Sci. USA 98, 13710-13715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Callaghan, B. L., Koo, S. H., Wu, Y., Freake, H. C. & Towle, H. C. (2001) J. Biol. Chem. 276, 16033-16039. [DOI] [PubMed] [Google Scholar]
- 14.Rufo, C., Teran-Garcia, M., Nakamura, M. T., Koo, S. H., Towle, H. C., Clarke, S. D. (2001) J. Biol. Chem. 276, 21969-21975. [DOI] [PubMed] [Google Scholar]
- 15.Shih, H. M. & Towle, H. C. (1992) J. Biol. Chem. 267, 13222-13228. [PubMed] [Google Scholar]
- 16.Iizuka, K., Bruick, R. K., Liang, G., Horton, J. D. & Uyeda, K. (2004) Proc. Natl. Acad. Sci. USA. 101, 7281-7286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berry, M. N. & Friend, D. S. (1969) J. Cell Biol. 43, 506-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, Z., Thompson, K. S. & Towle, H. C. (1993) J. Biol. Chem. 268, 12787-12795. [PubMed] [Google Scholar]
- 19.Luo, R. X., Postigo, A. A. & Dean, D. C. (1998) Cell 92, 463-473. [DOI] [PubMed] [Google Scholar]
- 20.Koo, S. H. & Towle, H. C. (2000) J. Biol. Chem. 275, 5200-5207. [DOI] [PubMed] [Google Scholar]
- 21.Kaytor, E. N., Qian, J., Towle, H. C. & Olson, L. K. (2000) Mol. Cell. Biochem. 210, 13-21. [DOI] [PubMed] [Google Scholar]
- 22.Vallet, V. S., Casado, M., Henrion, A. A., Bucchini, D., Raymonjean, M., Kahn, A. & Vaulont, S. (1998) J. Biol. Chem. 273, 20175-20179. [DOI] [PubMed] [Google Scholar]
- 23.Lou, D. Q., Tannour, M., Selig, L., Thomas, D., Kahn, A. & Vasseur-Cognet, M. (1999) J. Biol. Chem. 274, 28285-28394. [DOI] [PubMed] [Google Scholar]
- 24.Stoeckman, A. K., Ma L. & Towle H. C. (2004) J. Biol. Chem. 279, 15662-15669. [DOI] [PubMed] [Google Scholar]
- 25.Meroni, G., Cairo, S., Merla, G., Messali, S., Brent, R., Ballabio, A. & Reymond, A. (2000) Oncogene 19, 3266-3277. [DOI] [PubMed] [Google Scholar]
- 26.Dentin, R., Pegorier, J. P., Benhamed, F., Foufelle, F., Ferre, P., Fauveau, V., Magnuson, M. A., Girard, J. & Postic, C. (2004) J. Biol. Chem. 279, 20314-20326. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.