Abstract
Before implantation in the uterus, mammalian embryos set aside trophoblast stem cells that are maintained in the extraembryonic ectoderm (ExE) during gastrulation to generate the fetal portion of the placenta. Their proliferation depends on diffusible signals from neighboring cells in the epiblast, including fibroblast growth factor 4 (Fgf4). Here, we show that Fgf4 expression is induced by the transforming growth factor β-related protein Nodal. Together with Fgf4, Nodal also acts directly on neighboring ExE to sustain a microenvironment that inhibits precocious differentiation of trophoblast stem cells. Because the ExE itself produces the proteases Furin and PACE4 to activate Nodal, it represents the first example, to our knowledge, of a stem cell compartment that actively maintains its own microenvironment.
In eutherian mammals, the exchange of gas and nutrients across the placenta depends on an elaborate vascular network, which forms during embryogenesis with the help of fetal derivatives known as the trophoblast. This tissue arises from the spherical trophectoderm layer of the blastocyst surrounding the inner cell mass (ICM) and the blastocoel. Upon implantation into the uterus, the ICM cavitates and forms the epiblast, while adjacent trophectoderm cells proliferate and form the extraembryonic ectoderm (ExE) and the ectoplacental cone (EPC). Together with a superficial layer of visceral endoderm (VE), these structures constitute the egg cylinder. Throughout the ExE exists a reservoir of self-renewing trophoblast stem cells (TSCs) (1, 2) that provide the EPC with progenitor cells for differentiated spongiotrophoblasts and nondividing polyploid giant cells (3–5). They express essential transcription factors such as the estrogen-related receptor β (Errβ), Eomesodermin, and Cdx2, together with Bmp4, but repress differentiation markers such as Mash2. Their capacity to self-renew and proliferate in the embryo depends on a microenvironment that is established by neighboring cells of the ICM and the epiblast. A critical component of this microenvironment is fibroblast growth factor 4 (Fgf4), but additional, unknown signals are also required (6). Pharmacological inhibition of Errβ blocks the proliferative effect of Fgf4 on TSCs and triggers their differentiation toward the polyploid giant cell fate, substantiating the conclusion that it is an essential stem cell marker (7). At the egg cylinder stage [embryonic day (E) 5.5] and throughout gastrulation, the ExE in addition produces Furin and PACE4, two secreted proteases of the subtilisin-like proprotein convertase (SPC) family also known as SPC1 and SPC4 (8, 9). Recent experiments in mice showed that these proteases act together on neighboring tissues, where they specify anteroposterior asymmetry and stimulate germ layer formation and gastrulation movements (9). Here, we asked whether these proteases also influence the fate of TSCs.
Histological and gene expression analysis of mutant embryos reveals that Furin and PACE4, and a transforming growth factor β-related substrate in the epiblast encoded by Nodal are required to sustain TSCs in the ExE during gastrulation. In part, the role of Nodal is to indure Fgf4 expression in the epiblast. In addition, we use embryo explant culture assays to show that Nodal also acts directly on the ExE, where it is required alongside Fgf4 to sustain the expression of TSC marker genes. Besides identifying Nodal as an essential component of the TSC microenvironment, these findings define a cascade of reciprocal inductive interactions between the ExE and epiblast that are essential for TSCs to retain an undifferentiated character.
Materials and Methods
Mouse Strains. Mice cis heterozygous for null alleles of Furin (10) and PACE4 (8) were maintained on a mixed C57BL/6 × 129 SvEv/SvJ genetic background at the ISREC mouse facility in individually ventilated cages. Timed matings among cis heterozygotes were used to obtain Furin-/-;PACE4-/- double mutants [referred to as double knockout (DKO) embryos]. Mice carrying the NodallacZ reporter allele (11) were maintained on a mixed genetic background of 129SvEV × NMRI. Genotyping by PCR was performed as described (8, 10, 11). Outbred diabetes-resistant NMRI mice were from Harlan (Horst, The Netherlands).
Histology, Whole-Mount in Situ Hybridization, and β-Galactosidase Staining. For histology, paraffin-embedded embryos were sectioned at 7 μm and stained with hematoxylin/eosin. Probes used for RNA whole-mount in situ hybridization were as described (1, 12). LacZ expression was visualized by 5-bromo-4-chloro-3-indolyl β-d-galactoside staining overnight at 37°C after fixation on ice for 30 min (11).
Embryo Explant Cultures. For explant cultures, whole embryos and epiblasts from NMRI mice were dissected during the evening of the sixth day postcoitum (E5.75) between 1700 and 2000 hours and cultured for 20 h in OptiMEM containing 15% (vol/vol) knockout serum replacement factors (Invitrogen), 1% (vol/vol) glutamine, and 100 μg/ml gentamycin sulfate in Millipore filter inserts (pore size, 12 μm) on γ-irradiated STO fibroblasts expressing leukemia inhibitory factor. For factor treatment, epiblasts were freed from VE by using trypsin/pancreatin (13). Mature recombinant Nodal and SPC-resistant precursor were produced in stably transfected 293T cells and applied in equal amounts to embryo explants as described, based on comparative quantitation by Western blot analysis (9). At this concentration, the activity of mature Nodal in 293T cells transfected with the AR3-lux luciferase reporter reached 50–80% of the maximal response and was comparable with that of 20 ng/ml activin A. By comparison, the activity of SPC-resistant precursor reached a plateau and induced 6- to 8-fold less luciferase expression, similar to what has been described after transfection (9). Conditioned medium of untransfected parental cells (mock) was used as a negative control. Human activin A and BMP4 (R & D Systems) were applied at 20 and 50 ng/ml, respectively. FGF4 (Sigma) was used at 40 ng/ml together with 1 μg/ml heparin (Sigma).
Results
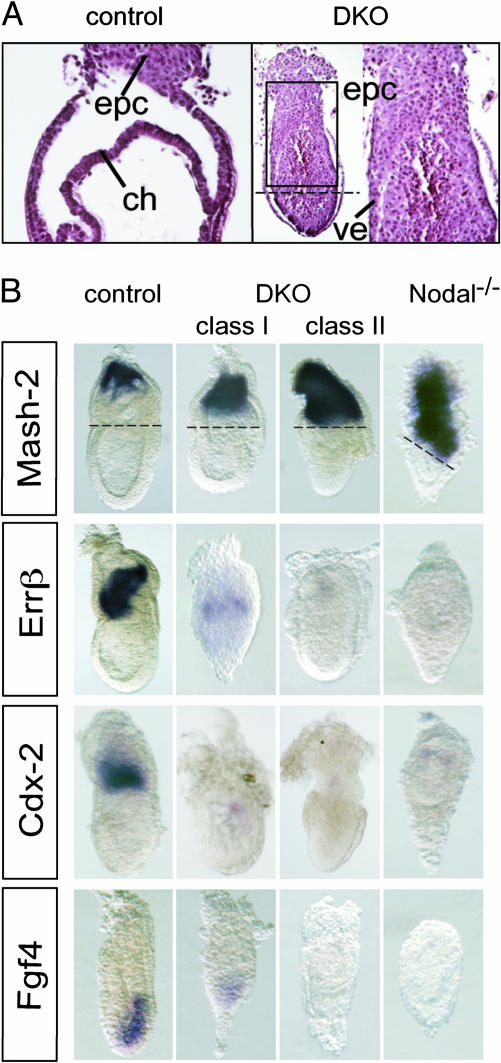
TSC Markers and Fgf4 Are Induced Downstream of Furin, PACE4, and Nodal. Histological analysis of late gastrulation stage Furin-/-;PACE4-/- DKO embryos revealed that virtually all cells in the ExE adopt an ectoplacental morphology (Fig. 1A). Moreover, the expression domain of Mash2, a basic helix–loop–helix (bHLH) transcription factor that promotes the differentiation of spongiotrophoblasts in the EPC (14), was expanded at the expense of the TSC markers Errβ, Cdx2 (Fig. 1B), and Eomesodermin (9). This finding suggests that Furin/PACE4 activities are necessary to inhibit precocious differentiation of TSCs into ectoplacental cell types. It seemed possible, therefore, that Furin and PACE4 are epistatic or act in parallel to Fgf4. Further analysis of DKO embryos revealed that Fgf4 expression was severely attenuated (Fig. 1B, n = 6/8, class I) or undetectable (n = 2/8, class II), indicating that these proteases cleave a substrate involved in the regulation of Fgf4. A known substrate is Nodal, a precursor protein of the transforming growth factor β family expressed in the epiblast and overlying VE (11, 15). Nodal can be cleaved by extracellular forms of Furin and PACE4, which thus convert the precursor to a more active, mature form (9, 16). As shown in Fig. 1B, expression of Fgf4, Errβ, and Cdx2 was also abolished in Nodal mutants. In contrast, Mash2 is ectopically expressed throughout the ExE, as observed in DKO embryos (Fig. 1). These results are consistent with a model wherein Furin and PACE4 up-regulate Fgf4 through the known potentiation of Nodal signaling.
Fig. 1.
Genetic inactivation of Furin and PACE4 or Nodal accelerates the differentiation of TSCs. (A) Sagittal sections through the extraembryonic region on E7.5 shows an expansion of EPC at the expense of the chorion (ch) in Furin/PACE4-deficient DKO embryos (Right), compared with compound heterozygotes (Left). The mutant embryo is also shown at low magnification (the left portion at Right) to indicate the junction between the epiblast and the enlarged EPC (stippled lines). The high magnification (right portion at Right) corresponds to the boxed area. (B) On day E7.5, expression of the EPC marker Mash2 is expanded toward the epiblast boundary (stippled lines) in both DKO (second and third columns) and Nodal mutants (fourth column), whereas progenitor cells marked by Errβ and Cdx2 mRNA in the ExE and chorion are reduced or absent. (Bottom) One day earlier (E6.5), Fgf4 expression in the epiblast was down-regulated compared with control litter mates (first column).
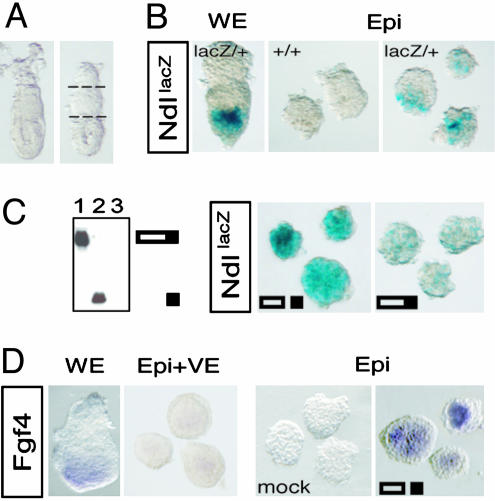
Precursor Processing Stimulates Nodal Autoinduction. The earliest known function of Nodal is to specify a population of anterior VE, which emerges around E5.5 at the apex of the egg cylinder. Concomitantly, Nodal amplifies its own expression in the epiblast and overlying VE by means of autoinduction (12). Furin and PACE4 stimulate both of these processes, possibly by activating the Nodal precursor (9). Intact whole embryos can be cultured in serum-free medium at this stage for up to 20 h without disrupting Nodal signaling, marked for example by the expression of Cripto. By contrast, if the ExE is cut off to remove the source of Furin and PACE4, Cripto and several other target genes downstream of Nodal are silenced (9). The expression of a NodallacZ reporter also appeared to be reduced (9), but to a lesser extent than in epiblast explants stripped of both ExE and VE (Fig. 2 A and B). Therefore, to directly test whether precursor processing stimulates Nodal autoinduction, isolated epiblasts were incubated with recombinant Nodal. As shown in Fig. 2C and Table 1, processed Nodal markedly up-regulated NodallacZ, whereas uncleaved precursor had no effect. Consistent with data from genetic studies (9, 12, 17), this result directly demonstrates that Nodal amplifies its own expression in an autoinductive feedback loop that is positively regulated by SPC-mediated precursor cleavage.
Fig. 2.
Processed Nodal stimulates autoinduction and expression of Fgf4. (A) Intact embryos dissected at E5.75 (Left) were cut twice (stippled lines) before or after enzymatic digestion of the overlying VE layer to culture isolated epiblast (Epi) and ExE explants. (B) Compared with cultured whole embryos (WE), epiblast explants deprived of ExE and VE show reduced expression levels of a NodallacZ reporter allele. Background staining in Ndl+/+ explants lacking the lacZ allele is negligible. Ndl, Nodal. (C) The cleaved, mature form of Nodal (filled rectangle) in conditioned medium of transfected 293T cells (Left, lane 2) amplifies expression of the NodallacZ reporter allele (Center), whereas an SPC-resistant mutant precursor precursor (lane 1) has no effect. (D) Recombinant Nodal can also rescue expression of Fgf4, whereas conditioned medium of untransfected, parental 293T cells (mock) was inactive.
Table 1. Regulation of gene expression in epiblast explants.
| Gene | Staining | None | Mock | Ndl | pre-Ndl |
|---|---|---|---|---|---|
| NodallacZ | + | 12 | 1 | 6 | |
| ++ | 2 | 6 | 0 | ||
| +++ | 2 | 6 | 0 | ||
| Fgf4 | - | 6 | 6 | 11 | 3 |
| + | 0 | 0 | 4 | 4 | |
| ++ | 0 | 0 | 8 | 7 |
Numbers correspond to the total of explants from two to four independent whole-mount in situ stainings that were recovered after treatment with the factors indicated (top row). Processed Nodal (Ndl), uncleaved precursor (pre-Ndl), or conditioned medium of untransfected HEK293Tcells (mock) were applied as described in Materials and Methods. The relative intensities of staining (-, +) were scored visually.
Recombinant Nodal Can Rescue Fgf4 Expression in Epiblast Explants. Because Furin and PACE4 produced in ExE are necessary for Nodal activation, removal of the ExE should also inhibit Fgf4 expression. Confirming this prediction, Fgf4 expression was lost in epiblast explants irrespective of the presence or absence of VE (Fig. 2D). Thus, in both situations, the activity of endogenous Nodal in epiblast explants is below the threshold required for induction of Fgf4. However, upon addition of exogenous recombinant Nodal, expression of Fgf4 was restored. In contrast, BMP4, which can mediate a stimulatory effect of Nodal on Cripto (9), failed to induce Fgf4 (n = 0/6, data not shown). These results demonstrate that Nodal acting directly on epiblast cells is sufficient to maintain Fgf4 expression.
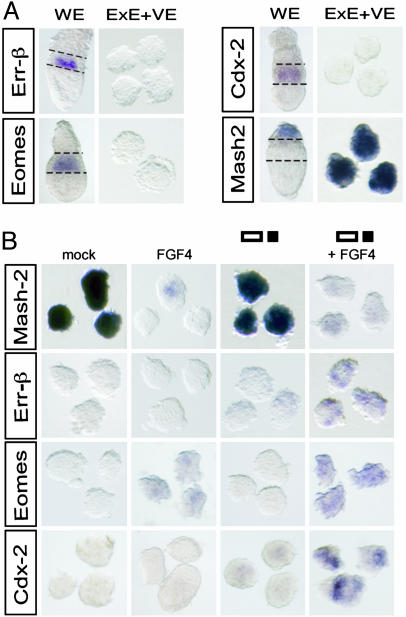
Nodal Acts on ExE Alongside Fgf4 to Sustain TSCs. Fgf4 and additional signals from the epiblast are essential to prevent TSCs from undergoing differentiation (6), but the expression pattern of stem cell markers in isolated ExE explants at early stages has not been examined. As shown in Fig. 3A, ExE explants cultured without the epiblast rapidly down-regulated Errβ, Cdx2, and Eomes mRNA, whereas Mash2 transcripts ectopically accumulated throughout the explants. These results show that isolated ExE explants mimic the molecular defects observed in Furin/PACE4 DKO and Nodal mutants, and rapidly lose their stem cell character. However, the loss of TSCs in a class of Furin/PACE4 DKO embryos appeared to be slightly less severe compared with Nodal mutants and ExE explants, possibly because of weak residual expression of Fgf4 (Fig. 1, class I). Therefore, we wished to determine whether Fgf4 alone might be sufficient to transiently restore normal gene expression in ExE explants if applied in increased amounts. Indeed, FGF4 at a concentration of 40 ng/ml potently inhibited ectopic expression of Mash2. By contrast, recombinant Nodal, activin A (n = 0/4), or BMP4 (n = 0/3) had no effect (Fig. 3B, Table 2, and data not shown). However, although this finding shows that cultured ExE explants respond to FGF4, we did not observe a corresponding up-regulation of Errβ or Cdx2 mRNAs, and Eomes was only weakly induced in 40% of the explants (Fig. 3B and Table 2). Therefore, we asked whether additional signals act in parallel to Fgf4. Interestingly, both Errβ and Eomes were significantly induced if FGF4 was added in combination with Nodal (Fig. 3B), the expression levels being comparable with those observed in whole-embryo cultures (Fig. 3A). This effect was mimicked by recombinant activin A (Errβ, n = 9/16; Eomes, n = 8/11), a related protein sharing its signaling receptors with Nodal, but not by BMP4 (Errβ, n = 0/15; Eomes, n = 0/9). The combination of Fgf4 and Nodal also restored expression of Cdx2 (Fig. 3B). We conclude that Errβ, Cdx2, and Eomes expression during normal development is maintained by Nodal itself, acting together with the epiblast-derived relay signal Fgf4.
Fig. 3.
The combined action of epiblast-derived Nodal and FGF4 inhibits precocious differentiation of the ExE. (A) In ExE explants, removal of the epiblast and EPC results in ectopic expression of Mash2 at the expense of Errβ, Cdx2, and Eomesodermin. For clarity, the boundaries of the ExE in whole embryos (WE) are outlined. (B) FGF4 prevents ectopic expression of Mash2, and, if added together with Nodal, induces expression of Errβ, Eomes, and Cdx2 in ExE explants. Note that Nodal alone has no significant effect on Mash2. Furthermore, in negative control experiments, none of the markers examined is influenced by conditioned medium devoid of Nodal (mock).
Table 2. Regulation of gene expression in ExE explants.
| Gene | Staining | None | Mock | Ndl | pre-Ndl | FGF4 | FGF4 plus Ndl |
|---|---|---|---|---|---|---|---|
| Mash2 | - | 0 | 0 | 0 | 13 | 5 | |
| + | 0 | 0 | 0 | 14 | 17 | ||
| ++ | 0 | 0 | 7 | 1 | 0 | ||
| +++ | 5 | 11 | 0 | 0 | 0 | ||
| Errβ | - | 8 | 8 | 13 | 8 | 9 | 1 |
| + | 0 | 0 | 0 | 0 | 0 | 10 | |
| Eomes | - | 12 | 10 | 12 | 6 | 8 | |
| + | 1 | 0 | 0 | 5 | 9 | ||
| ++ | 0 | 0 | 0 | 0 | 5 | ||
| Cdx2 | - | 8 | 11 | 3 | 6 | 0 | |
| + | 0 | 0 | 2 | 0 | 0 | ||
| ++ | 0 | 0 | 0 | 0 | 8 |
See Table 1 legend for explanation of values.
Discussion
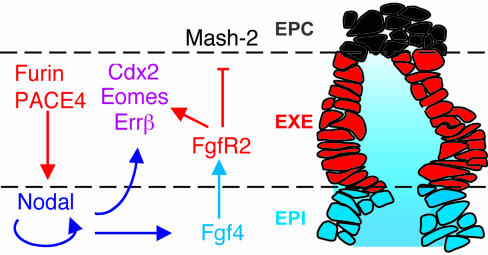
Overall, our results show that the ExE inhibits its own differentiation into ectoplacental tissue through reciprocal interactions with the epiblast that are mediated by Furin, PACE4, and Nodal (Fig. 4B). Previous analysis of Furin;PACE4 DKO embryos suggested that an important function of SPCs is to stimulate Nodal autoinduction in the epiblast (9). By using cultured embryo explants, we here provide direct evidence that Nodal can only amplify its expression after SPC cleavage of the precursor. Furthermore, our results show that Nodal is necessary to maintain the expression of Fgf4. In contrast to Nodal mutants, however, a majority of Furin;PACE4 DKO embryos still weakly expressed Fgf4 throughout the epiblast. The ectopic distribution of Fgf4 transcripts in this class of mutant embryos mirrors the residual expression of Nodal (9). Therefore, it will be interesting to determine in future studies whether Nodal can induce Fgf4 at a lower threshold compared with other target genes.
Fig. 4.
Summary of the reciprocal inductive interactions with the epiblast that are required to prevent precocious differentiation of the ExE into EPC tissue. The cartoon depicts a sagittal section (0.8 μm) of a fixed embryo on E6.5 that served as a template (for clarity, the overlying VE layer is omitted). In this model, epiblast (EPI)-derived Nodal and Fgf4 (blue) establish a microenvironment that sustains TSCs in the adjacent ExE (red). To date, no firm evidence exists for any of the available markers that TSCs represent only a subset of ExE cells. However, the model does not exclude the possibility that some TSCs may become biased toward differentiation already within the ExE, e.g., if their apical surface exposed to amniotic fluid is below a critical size.
Additional experiments show that Fgf4 mediates an inhibitory effect of Nodal on the expression of the early differentiation marker Mash2 in the ExE. However, besides inducing Fgf4 as a secondary signal, Nodal itself also acts directly on the ExE alongside Fgf4 to maintain expression of Errβ, Cdx2, and Eomesodermin. These transcription factors mark undifferentiated TSCs (1), and are required to sustain trophoblast growth (18–20). Thus, our results argue that both Fgf4 and Nodal are essential components of a microenvironment that is required to maintain a population of self-renewing TSCs (Fig. 4).
How do Nodal and Fgf4 signaling synergize in the ExE to induce TSC markers? To address this question, we tested whether Nodal up-regulates the expression of FgfR2, the putative receptor mediating Fgf4 signaling. However, FgfR2 is normally expressed both in Nodal-/- embryos and isolated ExE explants (M.G.-A., unpublished work). Conversely, Fgf4 does not stimulate canonical Nodal signaling via the Smad pathway because it failed to potentiate induction of the luciferase reporter construct AR3-lux in 293T cells (ref. 21 and M.G.-A., unpublished work). Therefore, Nodal and Fgf4 signaling appear to act in parallel and may be integrated directly at the promoter level of target genes as observed for the T box transcription factor brachyury (22). Because all of the TSC markers examined are transcription factors, it is also possible that only one of them directly depends on synergistic activation by Nodal and Fgf4, and subsequently up-regulates the others.
In keeping with a direct role in trophectoderm derivatives, transfection of Nodal cDNA into cultured TSCs was recently found to attenuate their terminal differentiation into giant cells marked by the expression of the late differentiation marker Pl-1. Conversely, in embryos that are homozygous for a hypomorphic Nodal allele, giant cell and spongiotrophoblast formation are accelerated at the expense of labyrinth growth (23). Thus, Nodal was suggested to antagonize the terminal differentiation of trophoblasts during midgestation because its expression is reactivated at that stage specifically in the spongial layer. We propose that a Nodal signaling network in addition already acts before gastrulation to mediate an intricate crosstalk between the ExE and epiblast that enables these tissues to sustain one another, possibly to grow and differentiate in a coordinated fashion. Thus, whereas the ExE normally serves as a reservoir of undifferentiated TSCs to sustain trophoblast growth beyond gastrulation, genetic inactivation of Nodal or its convertases, Furin and PACE4, triggers premature expression of an ectoplacental phenotype. The loss of ExE, in turn, prevents further growth of the epiblast and, hence, functions as a checkpoint to arrest development. Whereas this early developmental arrest precluded a clean and comprehensive analysis of terminal differentiation, we do not rule out the possibility that the ectopic EPC tissue arising in the absence of Nodal signaling in principle could give rise to one or several differentiated lineages, including polyploid giant cells. To address this question, cultured TSC lines may be useful as a model system to characterize in more detail the signaling pathways of Nodal and Fgf4 and their potential to perhaps modulate the fate of TSC derivatives also during later developmental stages that have not been analyzed in the present study.
Acknowledgments
We thank Janet Rossant (Mount Sinai Hospital, Toronto, Canada) for in situ probes of ExE markers and Andreas Trumpp and Anne Grapin-Botton for valuable discussions. This work was supported by the Barmet Foundation (to M.G.-A.) and Swiss National Science Foundation Grant 31-62031.00 (to N.B.-H. and D.B.C.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Fgf4, fibroblast growth factor 4; ExE, extraembryonic ectoderm; EPC, ectoplacental cone; SPC, subtilisin-like proprotein convertase; TSC, trophoblast stem cell; Errβ, estrogen-related receptor β; En, embryonic day n; DKO, double knockout; VE, visceral endoderm.
References
- 1.Tanaka, S., Kunath, T., Hadjantonakis, A. K., Nagy, A. & Rossant, J. (1998) Science 282, 2072-2075. [DOI] [PubMed] [Google Scholar]
- 2.Uy, G. D., Downs, K. M. & Gardner, R. L. (2002) Development (Cambridge, U.K.) 129, 3913-3924. [DOI] [PubMed] [Google Scholar]
- 3.Rossant, J., Gardner, R. L. & Alexandre, H. L. (1978) J. Embryol. Exp. Morphol. 48, 239-247. [PubMed] [Google Scholar]
- 4.Johnson, M. H. & Rossant, J. (1981) J. Embryol. Exp. Morphol. 61, 103-116. [PubMed] [Google Scholar]
- 5.Carney, E. W., Prideaux, V., Lye, S. J. & Rossant, J. (1993) Mol. Reprod. Dev. 34, 357-368. [DOI] [PubMed] [Google Scholar]
- 6.Kunath, T., Strumpf, D. & Rossant, J. (2001) in Stem Cell Biology, eds. Marshak, D. R., Gardner, R. L. & Gottlieb, D. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 267-287.
- 7.Tremblay, G. B., Kunath, T., Bergeron, D., Lapointe, L., Champigny, C., Bader, J. A., Rossant, J. & Giguere, V. (2001) Genes Dev. 15, 833-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Constam, D. B. & Robertson, E. J. (2000) Genes Dev. 14, 1146-1155. [PMC free article] [PubMed] [Google Scholar]
- 9.Beck, S., Le Good, J. A., Guzman, M., Haim, N. B., Roy, K., Beermann, F. & Constam, D. B. (2002) Nat. Cell Biol. 4, 981-985. [DOI] [PubMed] [Google Scholar]
- 10.Roebroek, A. J. M., Umans, L., Pauli, I. G. L., Robertson, E. J., van Leuven, F., Van de Ven, W. J. M. & Constam, D. B. (1998) Development (Cambridge, U.K.) 125, 4863-4876. [DOI] [PubMed] [Google Scholar]
- 11.Collignon, J., Varlet, I. & Robertson, E. J. (1996) Nature 381, 155-158. [DOI] [PubMed] [Google Scholar]
- 12.Brennan, J., Lu, C. C., Norris, D. P., Rodriguez, T. A., Beddington, R. S. & Robertson, E. J. (2001) Nature 411, 965-969. [DOI] [PubMed] [Google Scholar]
- 13.Hogan, B., Beddington, R., Costantini, F. & Lacy, E. (1994) Manipulating the Mouse Embryo: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 14.Guillemot, F., Nagy, A., Auerbach, A., Rossant, J. & Joyner, A. L. (1994) Nature 371, 333-336. [DOI] [PubMed] [Google Scholar]
- 15.Varlet, I., Collignon, J. & Robertson, E. J. (1997) Development (Cambridge, U.K.) 124, 1033-1044. [DOI] [PubMed] [Google Scholar]
- 16.Constam, D. B. & Robertson, E. J. (1999) J. Cell Biol. 144, 139-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Norris, D. P., Brennan, J., Bikoff, E. K. & Robertson, E. J. (2002) Development (Cambridge, U.K.) 129, 3455-3468. [DOI] [PubMed] [Google Scholar]
- 18.Chawengsaksophak, K., James, R., Hammond, V. E., Kontgen, F. & Beck, F. (1997) Nature 386, 84-87. [DOI] [PubMed] [Google Scholar]
- 19.Luo, J., Sladek, R., Bader, J. A., Matthyssen, A., Rossant, J. & Giguere, V. (1997) Nature 388, 778-782. [DOI] [PubMed] [Google Scholar]
- 20.Russ, A. P., Wattler, S., Colledge, W. H., Aparicio, S. A., Carlton, M. B., Pearce, J. J., Barton, S. C., Surani, M. A., Ryan, K., Nehls, M. C., et al. (2000) Nature 404, 95-99. [DOI] [PubMed] [Google Scholar]
- 21.Yan, Y.-T., Liu, J. J., Luo, Y., Chaosu, E., Haltiwanger, R. S., Abate-Shen, C. & Shen, M. M. (2002) Mol. Cell. Biol. 22, 4439-4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Latinkic, B. V., Umbhauer, M., Neal, K. A., Lerchner, W., Smith, J. C. & Cunliffe, V. (1997) Genes Dev. 11, 3265-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma, G. T., Soloveva, V., Tzeng, S. J., Lowe, L. A., Pfendler, K. C., Iannaccone, P. M., Kuehn, M. R. & Linzer, D. I. (2001) Dev. Biol. 236, 124-135. [DOI] [PubMed] [Google Scholar]