Abstract
The bacteriostatic natural product enterocin from the marine microbe “Streptomyces maritimus” has an unprecedented carbon skeleton that is derived from an aromatic polyketide biosynthetic pathway. Its caged tricyclic, nonaromatic core is derived from a linear poly-β-ketide precursor that formally undergoes a Favorskii-like oxidative rearrangement. In vivo characterization of the gene encM through mutagenesis and heterologous biosynthesis demonstrated that its protein product not only is solely responsible for the oxidative C—C rearrangement, but also facilitates two aldol condensations plus two heterocycle forming reactions. In total, at least five chiral centers and four rings are generated by this multifaceted flavoprotein. Heterologous expression of the enterocin biosynthesis genes encABCDLMN in Streptomyces lividans resulted in the formation of the rearranged metabolite desmethyl-5-deoxyenterocin and the shunt products wailupemycins D-G. Addition of the methyltransferase gene encK, which was previously proposed through mutagenesis to additionally assist EncM in the Favorskii rearrangement, shifted the production to the O-methyl derivative 5-deoxyenterocin. The O-methyltransferase EncK seems to be specific for the pyrone ring of enterocin, because bicyclic polyketides bearing pyrone rings are not methylated in vivo. Expression of encM with different combinations of homologous actinorhodin biosynthesis genes did not result in the production of oxidatively rearranged enterocin-actinorhodin hybrid compounds as anticipated, suggesting that wild-type EncM may be specific for its endogenous type II polyketide synthase or for benzoyl-primed polyketide precursors.
Keywords: flavoprotein, methyltransferase, polyketide synthase, Streptomyces
Multicyclic aromatic polyketides such as the clinically important tetracyclines, anthracyclines, and angucyclines are biosynthesized in actinomycetes on heterodimeric type II polyketide synthases (PKSs) (1, 2). These dissociable complexes of monofunctional enzymes contain a “minimal” set of proteins [the two ketosynthase subunits KSα and KSβ (also referred to as the chain length factor; ref. 3), acyl carrier protein (ACP), and malonyl-CoA:ACP acyltransferase] that are required for the biosynthesis of the polyketide chain. Additional PKS subunits, including ketoreductases (KR), cyclases, aromatases, oxygenases, etc., are responsible for modification of the nascent poly-β-carbonyl intermediate to form the aromatic product. The context-dependent nature of the type II PKS-encoding gene set thus dictates the metabolic outcome of the pathway, which is dominated by planar, polyaromatic natural products.
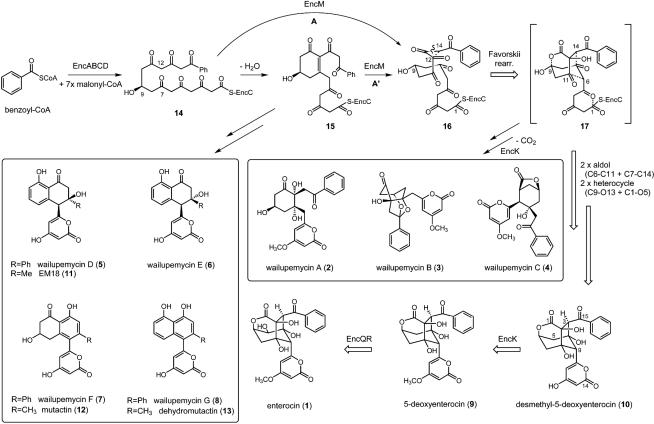
The bacteriostatic agent enterocin (1), however, stands apart from all other type II PKS-derived polyketides (4). Enterocin does not contain a planar polyaromatic core structure, but rather uniquely has a nonaromatic caged core resulting from a branched intermediate. Although the early stages of enterocin biosynthesis have been shown to proceed by means of a typical aromatic polyketide pathway (5) (Fig. 1), labeling (6) and genetic (7) experiments revealed that its biosynthesis involves an unprecedented oxidative rearrangement of its nascent poly-β-carbonyl intermediate. This carbon skeletal Favorskii-like rearrangement prevents successive cyclizations by means of aldol condensations to characteristic multiaromatic end products. The branched intermediate gives rise not only to the tricyclic caged core of enterocin, but also, after decarboxylation, to the structurally diverse wailupemycins A to C (2-4) (8).
Fig. 1.
Proposed biosynthetic pathway to enc-based polyketides (1-10) and structures of analogous act-derived polyketides (11-13). Path A involves monooxidation of 14 to the trione intermediate 16, whereas path A′ involves a dioxygenase role of EncM in which the C7-C12 olefin of the aldol product 15 is oxidatively cleaved to 16. The Favorskii-like rearrangement is speculated to occur on a putative ACP (EncC)-bound intermediate. The relative size of the reaction arrows reflects the flux in the pathway to the major product 1 as can be observed chromatographically (see Fig. 2). Dashed lines in structures 16 and 17 represent newly created bonds. In compounds 14-17, the carbon numbering is based on linear 14 in which priority is given to the carboxyl carbon; the numbering in compounds 1, 9, and 10 is in contrast based on that defined in ref. 18 for 9.
Biosynthetic carbon skeletal rearrangements involving oxidative Favorskii-like chemistry have been proposed in only a few secondary metabolic pathways on the basis of feeding experiments with isotopically labeled biosynthetic intermediates. Additional examples include the fungal polyketide aspyrone (9) and the dinoflagellate polyether okadaic acid (10). The recent cloning and sequencing of the enterocin biosynthetic gene cluster in the marine isolate “Streptomyces maritimus” has now allowed us to examine this biosynthetic reaction in greater detail (5). Mutational analysis revealed that the putative FAD-dependent oxygenase EncM and the methyltransferase EncK jointly catalyze the rearrangement because inactivation of either encoding gene terminated enterocin (1) production and caused the accumulation of the nonrearranged and nonmethylated polyketides wailupemycins D to G (5-8) (Fig. 1) (7). This result suggested that the rearrangement could be radical in nature, with EncK providing two biosynthetic functions, first as a radical source and second as a source of the O-methyl group. In this study, we set out to further examine this highly unusual biosynthetic rearrangement in a series of heterologous expression experiments.
Materials and Methods
General Experimental Details. NMR spectra were recorded on Bruker (Billerica, MA) DRX-300 and DRX-600 spectrometers. 1H and 13C chemical shifts were referenced to the solvent peak (DMSO-d6) δ 2.49 and 39.5 ppm, respectively. Standard parameters were used for 1D and 2D NMR spectra obtained, which included 1H, 13C, heteronuclear multiple quantum correlation (HMQC), and heteronuclear multiple-bond correlation (HMBC). Optical rotation was measured on a Jasco (Easton, MD) P-1010 polarimeter. A Waters 600 pump equipped with a Waters 2487 dual λ absorbance detector was used for semi-preparative HPLC separations. A Waters 996 photodiode array detector was used for analytical HPLC. High-resolution fast atom bombardment MS (HRFABMS) were recorded on a JEOL HX110A high resolution mass spectrometer at the Mass Spectrometry Facility, Department of Chemistry, University of Arizona.
Bacterial Strains and General Techniques for DNA Manipulation. Streptomyces lividans K4-114 was used as a host for transformation of plasmids (11). Escherichia coli XL1-Blue was used for the manipulation of plasmid DNA. Streptomyces coelicolor A3(2) was used to extract genomic DNA for cloning act genes, and the cosmid clone pJP15F11 (5), which harbors the entire enc biosynthesis gene cluster, was used as the source of DNA for cloning enc genes. The E. coli-Streptomyces shuttle pSEK4 and pRM5-based vectors were used for all expression experiments in Streptomyces (3). Recombinant DNA procedures were performed by standard techniques (12, 13). Restriction enzyme-digested DNA fragments were recovered from agarose gels with the QIAquick DNA Purification Kit (Qiagen, Valencia, CA). Oligonucleotides were obtained from Sigma Genosys. PCR was carried out on a PTC-2000 Peltier thermal cycler (MJ Research, Cambridge, MA) with PfuTurbo (Stratagene) DNA polymerase. DNA sequencing by BigDye terminator cycle sequencing reaction using an ABI 377 sequencer (Applied Biosystems) was performed at the Laboratory of Molecular Systematics and Evolution at the University of Arizona.
Construction of Expression Plasmids. pBM43 is a derivative of pMP6 (14) in which the encD gene downfield of the actIII promoter was replaced by a 1.6-kb fragment harboring encDK. The encDK cassette was PCR amplified from the cosmid clone pJP15F11 (5) with the primers 5′-GGTTAATTAACCGGCCGCCCCGACGAAGG-3′ (forward, PacI site underlined) and 5′-GGTGGCCCCGGACGTCATGC-3′ (reverse), and cloned into pCR-Blunt (Invitrogen) to create pBM41. The sequence-verified plasmid was sequentially digested, first with PacI, bluntended with shrimp alkaline phosphatase, and secondly with XbaI. The resulting 1.6-kb fragment was ligated into the XbaI and blunted NdeI sites of pHGF7505 (15) to create pBM42. A 3.2-kb PacI-HindIII fragment containing encDK and the divergent act promoters was cloned into analogous sites of pMP6 to create pBM43.
pBM47 is a pSEK4 derivative in which a 6.1-kb PacI/EcoRI fragment harboring actI-ORFsI,II,III and encMN was cloned into the analogous sites. A 3.0-kb fragment containing actI-ORFsI,II,III was PCR amplified from S. coelicolor A3 (2) genomic DNA by using primers 5′-GCATTAATTAAGCGGCGGTCGAAGGGAGATG-3′ (forward, PacI underlined) and 5′-GCCTCTAGACCACCCGGTGTTCTCCCGG-3′ (reverse, XbaI underlined) and cloned into pCR-Blunt to create pBM44. The 3.0-kb XbaI-HindIII fragment harboring actI-ORFsI,II,III from pBM44 was ligated into the analogous sites of pBM45 to create pBM46. pBM45 is a pCR-Blunt-derived plasmid in which an XbaI/NheI 3.1-kb fragment containing encMN was cloned into the analogous sites. encMN was PCR amplified from pJP15F11 by using primers 5′-GACTCTAGAGCGCCCGTCGTGGCGCCG-3′ (forward, XbaI underlined) and 5′-GCAGCTAGCTGGTCACACGGCGTGGGC-3′ (reverse, NheI underlined).
PacI-HindIII fragments from pMP6 (harboring encD), pBM43 (harboring encDK), and pRM5 (harboring actIII) were separately cloned into the analogous sites of pBM47 to create pBM48, pBM49, and pBM50, respectively.
pBM56 and pBM57 are derivatives of pBM55 in which PacI/HindIII fragments of pMP6 and pBM43 harboring encD and encDK, respectively, were cloned into the analogous sites. pBM55 is a pSEK4-derived plasmid in which the act genes contained within the PacI/EcoRI site were replaced by a PacI/EcoRI cassette harboring actI-ORFsI,II,III-encMNK from pBM54. The actI-ORFsI,II,III-encMNK cassette was constructed from a NheI-EcoRI fragment harboring encK from pBM51, which was ligated into the pCR-Blunt-derived pBM45 (harboring encMN) to create pBM53. An HindIII-XbaI fragment from pBM52 harboring actI-ORFsI,II,III was then cloned into the analogous sites of pBM53 to create pBM54. pBM51 is a pCR-Blunt-derived plasmid harboring encK that was PCR amplified from pJP15F11 by using primers 5′-GTCGCTAGCAGTTACTGACGCTTCGAGTG-3′ (forward, NheI underlined) and 5′-GGTGGCCCCGGACGTCATGC-3′ (reverse).
Culture Conditions and Purification of Polyketides. Plasmids were introduced into S. lividans K4-114, and transformants were selected and grown on solid R2YE with 20 μg/ml thiostrepton as described (14). Spores from single colonies were used as inoculum for liquid fermentations in R2YE containing 20 μg/ml thiostrepton. Cultures were grown at 30°C with shaking (250 rpm) for 4-5 days. Where appropriate, benzoic acid (0.8 mM) was added to the culture after 24 h. Liquid cultures were acidified to pH 5 with 1 M HCl and then extracted with EtOAc. Extracts were dried over anhydrous MgSO4 and concentrated in vacuo. Polyketide production was monitored by analytical HPLC as described (14).
The S. lividans K4-114/pBM43 crude extract (294 mg) from a 1-liter fermentation was subjected to stepwise normal-phase silica gel (Merck, 230-400 mesh) flash column chromatography with 1:1 hexane/EtOAc, EtOAc, 9:1 EtOAc/MeOH, 1:1 EtOAc/MeOH, and MeOH. Each fraction was analyzed by reversed-phase C18 analytical HPLC as described (14). The fraction eluting with 1:1 EtOAc/MeOH was subjected to HPLC purification. Compounds were purified by using a YMC-Pack ODS-A HPLC column (YMC, Kyoto) (250 × 20 mm, 10 μm), employing a gradient from 20% to 70% MeOH in 0.15% trifluoroacetic acid (TFA) over 75 min at a flow rate of 9.5 ml/min with UV detection at 254 nm. Desmethyl-5-deoxyenterocin (10, 9.3 mg) eluted between 12.5 and 14 min.
S. lividans K4-114/pBM57 was similarly grown and extracted, yielding 250 mg of crude extract, which was subjected to flash column chromatography as described above. The EtOAc/MeOH fraction was separated by using the same column as above, employing a gradient from 40% MeOH in 0.15% trifluoroacetic acid to 100% MeOH over 50 min. B26 (20, 14 mg) and dehydroSEK4b (21, 1.5 mg) eluted at 20 and 21.5 min, respectively.
Desmethyl-5-Deoxyenterocin (10. αD -21.4° (c 0.042, MeOH); 1H NMR (DMSO-d6) δ (multiplicity, assignment, coupling constants, HMBCs): 1.63 (brd, H7eq, J = 13.2 Hz, C5, C6, C8, C9), 2.06 (brd, H5eq, J = 14.4 Hz, C3, C4, C6, C7, C9), 2.26 (dd, H7ax, J = 14.1, 3.0 Hz, C2, C8, C9), 2.65 (dd, H5ax, J = 15.0, 4.2 Hz, C3, C4, C9, C15), 4.01 (s, H3, C1, C2, C4, C5, C8, C9, C10, C15), 4.58 (s, H9, C3, C4, C5, C7, C8, C10, C11), 4.83 (brs, H6, C1, C4, C8), 5.29 (d, H13, J = 1.8 Hz, C11, C12, C14), 6.22 (d, H11, J = 1.8 Hz, C9, C10, C12, C13), 7.50 (t, H18/18′, J = 7.8 Hz, C16, C18/18′), 7.60 (t, H19, J = 7.8 Hz, C17/17′), 7.79 (d, H17/17′, J = 7.8 Hz, C15, C17/17′, C19), 11.7 (s, 12-OH, C10, C11, C12, C13); 13C NMR (DMSO-d6) δ: 36.6 (C7), 39.3 (C5), 54.5 (C9), 60.9 (C3), 72.6 (C6), 75.9 (C4), 77.0 (C8), 79.7 (C2), 89.0 (C13), 104.9 (C11), 127.9 (C17/17′), 128.4 (C18/18′), 132.4 (C19), 139.4 (C16), 162.9 (C10), 163.7 (C14), 170.1 (C12), 174.0 (C1), 195.1 (C15); HRFABMS m/z = 415.1025 (C21H19O9 [M+H]+, 415.1029 calculated).
B26 (20). 1H NMR (DMSO-d6) δ (multiplicity, assignment, coupling constants, HMBCs): 2.28 (s, H16, C14, C15), 4.35 (s, H6, C4, C5, C7, C8, C12), 5.17 (d, H2, J = 1.8 Hz, C1, C3, C4), 5.56 (d, H4, J = 1.1 Hz, C2, C3, C5, C6), 6.01 (s, H14, C12, C16), 6.72 (d, H8, J = 2.2 Hz, C6, C9, C10, C12), 6.77 (d, H10, J = 2.6 Hz, C8, C9, C11, C12), 10.8 (s, 9-OH, C7, C8, C9, C10), 11.5 (s, 3-OH, C1, C2, C3, C4, C5); 13C NMR (DMSO-d6) δ: 19.4 (C16), 37.3 (C6), 88.2 (C2), 99.5 (C4), 102.0 (C10), 110.6 (C14), 113.7 (C12), 117.4 (C8), 137.4 (C7), 159.2 (C11), 161.2 (C9), 163.9 (C1), 164.4 (C15), 165.7 (C5), 170.4 (C3), 177.6 (C13); HRFABMS m/z = 301.0712 (C16H13O6 [M+H]+, 301.0712 calculated).
DehydroSEK4b (21). 1H NMR (DMSO-d6) δ (multiplicity, assignment, HMBCs): 2.64 (s, H16, C10, C14, C15), 3.87 (s, H6, C4, C7, C8), 5.28 (s, H2, C1, C3, C4), 6.09 (s, H8, C6, C7, C9, C10), 6.16 (s, H4, C2, C3, C5, C6), 6.60 (s, H12, C10, C11, C13, C14), 6.62 (s, H14, C10, C12, C13, C16), 10.6 (s, 13-OH, C12, C13, C14), 11.8 (s, 3-OH, C2, C3, C4); 13C NMR (DMSO-d6) δ: 22.4 (C16), 37.0 (C6), 89.2 (C2), 100.5 (C12), 101.9 (C4), 112.1 (C8), 114.3 (C10), 116.8 (C14), 141.7 (C15), 159.5 (C11), 160.8 (C5), 161.1 (C7), 161.6 (C13), 163.9 (C1), 170.2 (C3), 178.0 (C9); HRFABMS m/z = 301.0714 (C16H13O6 [M+H]+, 301.0712 calculated).
Bioconversion of Desmethyl-5-Deoxyenterocin to 5-Deoxyenterocin. Desmethyl-5-deoxyenterocin (1 mg) dissolved in DMSO (0.1 ml) was added to a 1-day-old 30-ml culture of S. lividans K4-114/pBM43. After an additional 24 h, the culture was extracted with EtOAc and analyzed by analytical HPLC as described above, showing an ≈30-40% conversion to 5-deoxyenterocin.
Results
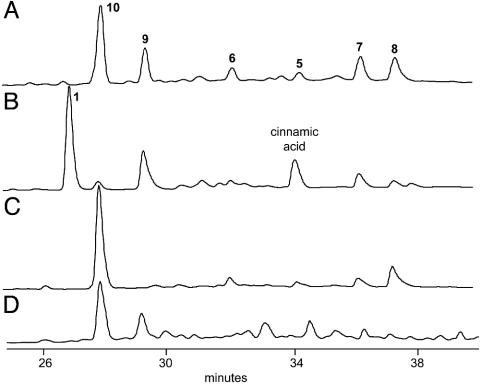
Identification of encM as the Enterocin Favorskiiase-Encoding Gene. On the basis of our earlier report on the identification of the genes encM (FAD-dependent oxygenase) and encK (methyltransferase) as playing a joint role in the enterocin favorskii rearrangement in S. maritimus (7), we set out to heterologously express these genes with those encoding the enterocin minimal PKS to further elucidate this biosynthetic event. We earlier prepared a series of pRM5-based expression plasmids for expression in the engineered host strain S. lividans K4-114 and demonstrated that the minimal enterocin PKS requires its endogenous KR EncD for activity (14). With this in mind, we constructed the plasmid pBM43 by introducing the encDK gene cassette into pMP6 (14) downstream of the actIII promoter (Table 1). This plasmid, which additionally carries the encABCLMN gene cassette downstream of the divergent actI promoter, contains the machinery for the synthesis of the starter unit benzoyl-CoA from exogenous benzoic acid by the benzoate:CoA ligase EncN (16, 17), its loading onto the enterocin ACP EncC, the heterodimeric KSα,β EncA-B, and the acyltransferase EncL. This expression plasmid was introduced by means of transformation into the S. lividans K4-114 (11) and grown in a benzoate-enriched medium. As projected, the S. lividans K4-114/pBM43 transformant produced 5-deoxyenterocin (9) as the major product together with the nonrearranged polyketides wailupemycins D to G (5-8) (Fig. 2A), thereby establishing that no other gene product is required for the rearrangement and subsequent cyclization reactions. In addition to these compounds, a new compound (10) was produced when the transformant was fermented in liquid R2YE media rather than on solid agar plates in ≈100-fold excess.
Table 1. Plasmid constructions and resulting polyketide products in S. lividans K4-114.
| Plasmid | Genes | Major products |
|---|---|---|
| pMP6 | encABCLMN/encD | 5-8, 10* |
| pBM43 | encABCLMN/encDK | 5-10* |
| pBM47 | actl-ORFsI-III-encMN | 18, 19† |
| pBM48 | actl-ORFsI-III-encMN/encD | 11-13‡ |
| pBM49 | actl-ORFsI-III-encMN/encDK | 11-13‡ |
| pBM50 | actl-ORFsI-III-encMN/actIII | 11-13‡ |
| pBM55 | actl-ORFsI-III-encKMN | 18, 19† |
| pBM56 | actl-ORFsI-III-encKMN/encD | 11-13‡ |
| pBM57 | actl-ORFsI-III-encKMN/encDK | 18-21 |
In the presence of 0.8 mM benzoic acid
20 and 21 produced in trace amounts
18 and 19 produced in trace amounts
Fig. 2.
HPLC analysis at 254 nm of crude extracts from S. lividans K4-114/pBM43 plus benzoic acid (trace A), wild-type S. maritimus (trace B), S. lividans K4-114/pMP6 plus benzoic acid (trace C), and the biotransformation of 10 to 9 in S. lividans K4-114/pBM43 (trace D). Cinnamic acid is an intermediate in the biosynthesis of the benzoyl-CoA starter unit from l-phenylalanine (17) as seen in trace A.
Reversed-phase HPLC purification of compound 10 from a 1-liter fermentation provided 9.3 mg for structure characterization. Comparison of the 1H and 13C NMR data with those for 5-deoxyenterocin (18) suggested that 10 was the desmethyl derivative. The absence of the methyl singlet at δ 3.80 in the 1H NMR spectrum supported the loss of the pyrone methyl group. Furthermore, a new, exchangeable proton signal at δ 11.7 showing HMBC correlations into the pyrone confirmed that the pyrone was not methylated, and thus the structure could be assigned as desmethyl-5-deoxyenterocin (10). This assignment was further established by HRFABMS with a molecular formula of C21H19O9 (m/z = 415.1029 [M+H]+, Δ -0.4 millimass units). HPLC analysis of the wild-type S. maritimus strain showed that 10 is a very minor natural constituent that was never characterized (Fig. 2B) (8).
Assignment of EncK as an O-Methyltransferase. In contrast to our earlier in vivo mutagenesis experiments (7), the heterologous expression experiment implies that the oxygenase EncM is solely responsible for catalyzing the Favorskii rearrangement and that EncK may act only as a methyltransferase with rearranged polyketide substrates (Fig. 1). To test this new hypothesis, we recultured the ΔencK mutant S. maritimus KK in liquid media, because this mutant when previously grown on solid media produced only wailupemycins D to G and not desmethyl-5-deoxyenterocin (7). HPLC analysis confirmed our new suspicion, because 10 was produced by liquid cultures of the ΔencK mutant (data not shown). We next turned our attention to the transformant S. lividans K4-114/pMP6 (encABCLMN/encD), which, when grown on solid media, produced only wailupemycins D to G (14). Once again, when fermented in liquid media, 10 was additionally produced (Fig. 2C). These two experiments clearly showed that the influence of fermentation conditions led initially to the incorrect functional assignment of the encK gene product (7).
The role of EncK as an O-methyltransferase was further confirmed through a biotransformation experiment in which 10 was added to a culture of S. lividans K4-114/pBM43 (encABCLMN/encDK). In the absence of supplemental benzoate, this transformant does not produce enc-based polyketides. Thus, when pure 10 was added to the fermentation, we observed by HPLC the production of 5-deoxyenterocin (Fig. 2D), thereby confirming that methylation is the penultimate step in enterocin biosynthesis. EncK is most homologous to AurI, another Streptomycete polyketide pyrone O-methyltransferase (19), with a similarity/identity score of 54/36 and has been shown to complement an aurI genetic mutation in aureothin biosynthesis (J. He, C. Hertweck, L.X., and B.S.M., unpublished observation).
Combinatorial Biosynthesis of Hybrid Enterocin-Actinorhodin Constructs. The antibiotic actinorhodin, whose type II PKS has served as a prototype system in bacteria, is an all malonyl-CoA-derived octaketide (1, 2). Although actinorhodin and enterocin are structurally dissimilar upon initial inspection, their biosynthetic pathways share many early features. The act PKS shunt products EM18 (11), mutactin (12), and dehydromutactin (13) are identical to wailupemycins D (5), F (7), and G (8), respectively, except for the nature of the starter unit, acetate versus benzoate (Fig. 1) (7). Different sets of polyketide tailoring reactions are responsible for taking these compounds down such diverse biosynthetic paths. We thus set out to probe whether the favorskiiase EncM could be used to derail other type II PKSs, such as the act PKS, from producing aromatic polyketides to rather synthesize acetate-primed enterocin-like compounds. A series of hybrid act-enc gene combinations were thus constructed in pRM5 and pSEK4-based act PKS expression vectors (3) and expressed heterologously in S. lividans K4-114 (Table 1).
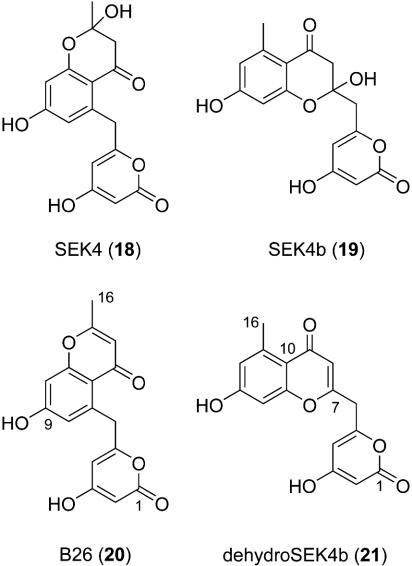
The nonreduced shunt octaketides SEK4 (18) and SEK4b (19) are products of the minimal act PKS ActI-ORFsI-III when expressed in the absence of the KR ActIII (1, 2). We reengineered the plasmid pSEK4 (3) by replacing the actVII (cyclase) and actIV (aromatase) genes with encMN and encKMN to give pBM47 and pBM55, respectively. In both cases, S. lividans K4-114 transformants produced only the known act compounds SEK4 and SEK4b (Fig. 3), suggesting that the enterocin favorskiiase EncM and the O-methyltransferase EncK were not able to use the act minimal PKS products as substrates. To evaluate whether a C9 ketoreduced act substrate is preferred, much like in enc biosynthesis, we next expressed the act minimal PKS with either its endogenous KR ActIII or with the homologous enterocin KR EncD, which has been successfully used before in conjunction with the act PKS (5). Several plasmids containing the act minimal PKS and a KR with different combinations of the genes encKMN (Table 1) all yielded roughly the same polyketide profile when expressed in S. lividans K4-114 (data not shown). In each case, in addition to the KR-reduced act polyketides mutactin, dehydromutactin, and EM18, the nonreduced SEK4 and SEK4b compounds were measured at various levels along with two new compounds. These molecules were isolated by reversed-phase HPLC and characterized by MS and NMR. Proton and carbon NMR data, along with gradient-enhanced heteronuclear multiple quantum correlation (HMQC) and HMBC spectroscopy, quickly revealed that 20 and 21 were dehydrated analogs of SEK4 and SEK4b, respectively, which was supported in each case by HRFABMS. Dehydro SEK4 (aka B26, 20) was previously characterized from the heterologous expression of the naphthocyclinone PKS, although its structure was never assigned by NMR (20). Key HMBC correlations supporting the structure 20 were observed from the H14 olefin at δ 6.01 to the aromatic carbon at δ 113.7 (C12) and the methyl carbon at δ 19.4 (C16). Similarly, the placement of the olefin in dehydro SEK4b (21) was established through HMBC correlations from the H-8 olefin at δ 6.09 into the benzene ring and the C-6 methylene bridge to the pyrone. Interestingly, the levels of the nonreduced polyketides SEK4 and SEK4b and their dehydrated counterparts increased relative to the KR-reduced compounds as the number of enc genes was enlarged. The enc-tailoring proteins EncK and EncM may compete with the KR ActIII or EncD in complexing with the minimal act PKS, thereby perturbing the ketoreduction process. In no case, unfortunately, did we detect oxidatively rearranged or O-methylated act products.
Fig. 3.
Structures of act-derived polyketides 18-21 from the expression of S. lividans K4-114/pBM57.
Discussion
The encM gene encodes a putative 464-aa flavoprotein with remarkable catalytic properties. Sequence analysis suggests that the “favorskiiase” EncM has a covalent attachment of FAD at a conserved histidine residue at position 78, which is reminiscent of many oxidases (21, 22), including those involved in the biosynthesis of the natural products mitomycin (23) and griseorhodin A (24). Flavoenzymes are very versatile catalysts and are involved not only in the activation of oxygen for oxidation and hydroxylation reactions, but also in dehydrogenation reactions, in light emission, and in one- and two-electron transfer reactions (25). Flavoprotein-catalyzed oxidative reactions associated with post-PKS modifications include hydroxylations, epoxidations, Baeyer-Villager rearrangements (26), and now Favorskii rearrangements.
The enterocin flavoprotein EncM is implicated in this study not only to catalyze the Favorskii-like oxidative rearrangement, but also must facilitate the two aldol condensation and two heterocycle-forming reactions in the formation of desmethyl-5-deoxyenterocin (10). In the course of this series of chemical transformations, at least five new chiral centers and four new rings are generated. The presumed EncM precursor is the linear enc PKS C9-reduced octaketide 14 that is oxidized at C12 to form the 11,12,13-trione intermediate 16 (Fig. 1, path A). If EncM functions rather as a dioxygenase, then oxidation of the cyclohexenone intermediate 15 similarly yields 16 (Fig. 1, path A′). Hydration of the C13 carbonyl and scission of the C13-C14 bond with concomitant formation of a new C12-C14 linkage generates the hypothetical Favorskii product 17 bearing a chiral α-hydroxy acid center. This branched intermediate formally undergoes two aldol condensations between C6-C11 and C7-C14 and in the process generates chiral centers at each position. Although cyclases catalyze such aldol condensation reactions during aromatic polyketide assembly (1, 2), the enc gene cluster is a notable exception among all other known type II PKS gene sets for its absence of typical cyclase and aromatase encoding genes (5). We hypothesize by virtue of the formation of 10 that EncM likely facilitates these C-C-forming reactions by orienting 17 in a conducive conformation, because linear poly-β-ketides are extremely reactive and spontaneously cyclize (27, 28). The lactone C-O bond between the carboxylate oxygen and C9 is also generated during the course of 10 biosynthesis. On the basis of the presumed 9R-configuration that is inferred from the structures of the wailupemycin A and B shunt products (8, 29), the C-O linkage should be generated with inversion of stereochemistry with displacement of the acetate-derived C9 hydroxyl group. Pyrone formation with hydroysis of the thioester linkage to the EncC ACP completes the biosynthetic transformation to 10. Because the activity of EncM has not yet been reconstituted in vitro, there remains the possibility that other host proteins may contribute to the transformation in vivo.
We previously proposed on the basis of a gene knockout experiment that the encK gene product also participates in the rearrangement reaction (7). Further experimentation in this study, however, conclusively showed this not to be the case with the characterization of 10. Primary sequence analysis of EncK demonstrates that it belongs to a distinct family of S-adenosyl-l-methionine-dependent O-methyltransferases involved in the methylation of macrolide antibiotics and plant phenylpropanoids.
Although the encM gene product has now been shown to solely catalyze a remarkable series of biosynthetic events during enterocin biosynthesis, its use as a combinatorial biosynthetic reagent to generate “unnatural” natural products may unfortunately be limited. This statement is, however, based upon a single observation with the actinorhodin PKS in which the addition of encM did not redirect the post-PKS tailoring reactions of the act pathway, suggesting that wild-type EncM may be specific for its endogenous type II PKS or for benzoyl-primed polyketide precursors. An in-depth biochemical characterization of EncM will be necessary to probe its mechanism and to evolve it into a suitable reagent for the engineered biosynthesis of new chemical entities.
Acknowledgments
S. lividans K4-114 was generously provided by Kosan Biosciences, Inc. (Hayward, CA), and pRM5 and pSEK4 were kindly provided by C. Khosla (Stanford University, Stanford, CA). This work was supported by National Institutes of Health (NIH) Grant AI47818. Partial support for the 600-MHz NMR spectrometer in the College of Pharmacy was provided by NIH National Center for Research Resources Shared Instrumentation Grant 1S10RR16659.
Author contributions: L.X. and B.S.M. designed research; L.X. and J.A.K. performed research; L.X., J.A.K., and B.S.M. analyzed data; and B.S.M. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ACP, acyl carrier protein; KR, ketoreductase; PKS, polyketide synthase; HMBC, heteronuclear multiple-bond correlation; HRFABMS, high-resolution fast atom bombardment MS.
References
- 1.Rawlings, B. J. (1999) Nat. Prod. Rep. 16, 425-484. [DOI] [PubMed] [Google Scholar]
- 2.Shen, B. (2000) Top. Curr. Chem. 209, 1-51. [Google Scholar]
- 3.McDaniel, R., Ebert-Khosla, S., Hopwood, D. A. & Khosla, C. (1993) Science 262, 1546-1550. [DOI] [PubMed] [Google Scholar]
- 4.Piel, J., Hoang, K. & Moore, B. S. (2000) J. Am. Chem. Soc. 122, 5415-5416. [Google Scholar]
- 5.Piel, J., Hertweck, C., Shipley, P. R., Hunt, D. M., Newman, M. S. & Moore, B. S. (2000) Chem. Biol. 7, 943-955. [DOI] [PubMed] [Google Scholar]
- 6.Seto, H., Sato, T., Urano, S., Uzawa, J. & Yonehara, H. (1976) Tetrahedron Lett., 4367-4370.
- 7.Xiang, L., Kalaitzis, J. A., Nilsen, G., Chen, L. & Moore, B. S. (2002) Org. Lett. 4, 957-960. [DOI] [PubMed] [Google Scholar]
- 8.Sitachitta, N., Gadepalli, M. & Davidson, B. S. (1996) Tetrahedron 52, 8073-8080. [Google Scholar]
- 9.Simpson, T. J. & Holker, J. S. E. (1975) Tetrahedron Lett., 4693-4696.
- 10.Wright, J. L. C., Hu, T., McLachlan, J. L., Needham, J. & Walter, J. A. (1996) J. Am. Chem. Soc. 118, 8757-8758. [Google Scholar]
- 11.Ziermann, R. & Betlach, M. C. (1999) BioTechniques 26, 106-110. [DOI] [PubMed] [Google Scholar]
- 12.Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F. & Hopwood, D. A. (2000) Practical Streptomyces Genetics (John Innes Foundation, Norwich, U.K).
- 13.Sambrook, J. & Russell, D. W. (2001) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, New York).
- 14.Hertweck, C., Xiang, L., Kalaitzis, J. A., Cheng, Q., Palzer, M. & Moore, B. S. (2004) Chem. Biol. 11, 461-468. [DOI] [PubMed] [Google Scholar]
- 15.Yu, T.-W., Müller, R., Müller, M., Zhang, X., Draeger, G., Kim, C.-G., Leistner, E. & Floss, H. G. (2001) J. Biol. Chem. 276, 12546-12555. [DOI] [PubMed] [Google Scholar]
- 16.Xiang, L. & Moore, B. S. (2003) J. Bacteriol. 185, 399-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalaitzis, J. A., Izumikawa, M., Xiang, L., Hertweck, C. & Moore, B. S. (2003) J. Am. Chem. Soc. 125, 9290-9291. [DOI] [PubMed] [Google Scholar]
- 18.Kang, H., Jensen, P. R. & Fenical, W. (1996) J. Org. Chem. 61, 1543-1546. [Google Scholar]
- 19.He, J. & Hertweck, C. (2003) Chem. Biol. 10, 1225-1232. [DOI] [PubMed] [Google Scholar]
- 20.Brünker, P., McKinney, K., Sterner, O., Minas, W. & Bailey, J. E. (1999) Gene 227, 125-135. [DOI] [PubMed] [Google Scholar]
- 21.Mewies, M., McIntire, W. S. & Scrutton, N. S. (1998) Protein Sci. 7, 7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hefti, M. H., Vervoort, J. & van Berkel, W. J. H. (2003) Eur. J. Biochem. 270, 4227-4242. [DOI] [PubMed] [Google Scholar]
- 23.Mao, Y. Q., Varoglu, M. & Sherman, D. H. (1999) Chem. Biol. 6, 251-263. [DOI] [PubMed] [Google Scholar]
- 24.Li, A. Y. & Piel, J. (2002) Chem. Biol. 9, 1017-1026. [DOI] [PubMed] [Google Scholar]
- 25.Fraaije, M. W. & Mattevi, A. (2000) Trends Biochem. Sci. 25, 126-132. [DOI] [PubMed] [Google Scholar]
- 26.Rix, U., Fischer, C., Remsing, L. L. & Rohr, J. (2002) Nat. Prod. Rep. 19, 542-580. [DOI] [PubMed] [Google Scholar]
- 27.Harris, T. M. & Harris, C. M. (1986) Pure Appl. Chem. 58, 283-294. [Google Scholar]
- 28.Shen, Y., Yoon, P., Yu, T.-W., Floss, H. G., Hopwood, D. A. & Moore, B. S. (1999) Proc. Natl. Acad. Sci. USA 96, 3622-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirsch, S. & Bach, T. (2003) Angew. Chem. Int. Ed. 42, 4685-4687. [DOI] [PubMed] [Google Scholar]