Abstract
With few exceptions, the shells (capsids) of sphere-like viruses have the symmetry of an icosahedron and are composed of coat proteins (subunits) assembled in special motifs, the T-number structures. Although the synthesis of artificial protein cages is a rapidly developing area of materials science, the design criteria for self-assembled shells that can reproduce the remarkable properties of viral capsids are only beginning to be understood. We present here a minimal model for equilibrium capsid structure, introducing an explicit interaction between protein multimers (capsomers). Using Monte Carlo simulation we show that the model reproduces the main structures of viruses in vivo (T-number icosahedra) and important nonicosahedral structures (with octahedral and cubic symmetry) observed in vitro. Our model can also predict capsid strength and shed light on genome release mechanisms.
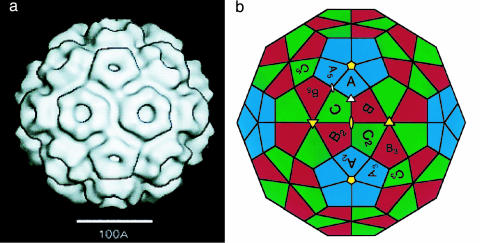
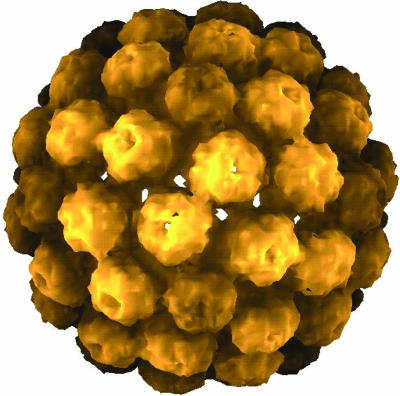
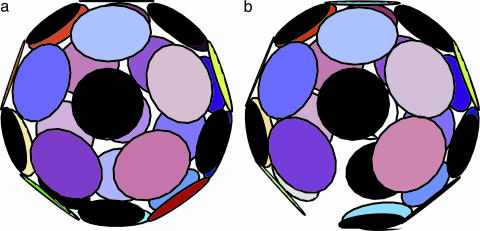
Icosahedral symmetry is ubiquitous among spherical viruses (1). A classic example is the cowpea chlorotic mottle virus (CCMV), a well studied RNA virus with a shell composed of exactly 180 identical proteins (subunits) (2, 3). Fig. 1a is a cryo-transmission electron microscopy reconstruction showing 5- and 6-fold morphological units (capsomers), and Fig. 1b shows the arrangement of the individual subunits within the capsomers. The capsid has 6 5-fold rotation axes, 10 3-fold axes, and 15 2-fold axes, the symmetry elements of an icosahedron. The subunits can be divided into 12 capsomers that contain five subunits (pentamers) and 20 capsomers that contain six subunits (hexamers).
Fig. 1.
Icosahedral symmetry of a viral capsid. (a) Cryo-TEM reconstruction of CCMV. (b) Arrangement of subunits on a truncated icosahedron; A, B, and C denote the three symmetry nonequivalent sites. [Reproduced with permission from ref. 3 (Copyright 1998, Elsevier)].
The current understanding of sphere-like virus structures, like that adopted by CCMV, is based on the Caspar and Klug (CK) “quasi-equivalence” principle (4), which provides the foundation of modern structural virology. CK showed that closed icosahedral shells can be constructed from pentamers and hexamers by minimizing the number T of nonequivalent locations that subunits occupy, with the T-number adopting the particular integer values 1, 3, 4, 7, 12, 13,...(T = h2 + k2 + hk, with h, k equal to nonnegative integers). These CK shells always contain 12 pentamers plus 10 (T-1) hexamers, and these structures have indeed been found to characterize a predominantly large fraction of sphere-like viruses (the CCMV capsid, for example, being a T = 3 structure). Nevertheless, exceptions among WT capsids have been documented (5, 6), including the retroviruses like HIV (7); assembly of subunits with point mutations can also produce breakdown of CK-type capsid icosahedral symmetry (8).
It is significant that the icosahedral point group generates the maximum enclosed volume for shells comprised of a given size subunit (4). But the fact that many viruses, including CCMV, self-assemble spontaneously from their molecular components under in vitro conditions (9) indicates that both icosahedral symmetry and the CK construction should be generic features of the free energy minima of aggregates of viral capsid proteins. Because of current computational limitations, direct evaluation of the capsid free energy by all-atom simulations is not possible; instead one must consider “coarse-grained” model systems to address these questions. The first efforts (10) to construct a minimal model for viral assembly were based on attempts to connect viral icosahedral symmetry with the mathematical problem of obtaining the closest packing of N equal disks (the capsomers) on the surface of a sphere (11). This problem is in turn related to one posed much earlier by Thomson (12), involving the optimal distribution of N mutually repelling points on the surface of a sphere, and to the subsequently posed “covering” problem, where one determines the arrangement of N overlapping disks of minimum diameter that allow complete coverage of the sphere surface (13). In a similar spirit, a self-assembly phase diagram has been formulated for adhering hard disks (14). These models, which all deal with identical capsomers, regularly produced capsid symmetries lower than icosahedral. On the other hand, theoretical studies of viral assembly that assume icosahedral symmetry are able to reproduce the CK motifs (15), and, most recently, it has been demonstrated that the presence of disclinations in CK shells necessarily leads to faceting (the “buckling transition”) for large capsids (16). The most recent approach describes viral structures by using principles of tiling theory (17). Still other approaches based on the assumption of icosahedral symmetry have focused on the pathways and kinetics of the capsid formation process (18, 19). But the origin of icosahedral symmetry in viruses, the validity conditions for the CK construction, and the physical principles underlying the quasi-equivalence principle have yet to be fully elucidated.
Model for the Equilibrium Structure of Viruses
In the present work we provide an answer to the fundamental question: why do viruses adopt icosahedral symmetry? We present a model for equilibrium viral structures that retains the essential features of the process and results in the predominance of icosahedral CK structures as well as the existence of other structures observed in vitro that do not fall into this classification. We start from the observation that, despite the wide range of amino acid sequences and folding structures (20) of viral coat proteins, capsid proteins spontaneously self-assemble into a common viral architecture. The actual kinetic pathways and intermediates involved are quite varied [e.g., CCMV assembles from dimers (21), Polyoma from pentameric capsomers (22), and HK97 from pentamers and hexamers (23)] but the equilibrium structures of viral capsids are invariably made up of the same units (e.g., pentamers and/or hexamers). This finding suggests that, although the interaction potential between subunits is asymmetric and species-specific (24), capsomers interact through a more isotropic and generic interaction potential. The focus of the present work is not on the kinetics process of the assembly but rather on understanding the optimal equilibrium structures.
Based on the ideas noted above we consider a minimal model for the equilibrium structure of molecular shells in which we do not attempt to describe individual subunits but instead focus on the capsomers. The effective capsomercapsomer potential V(r) is assumed to depend only on the separation r between the capsomer centers¶ and captures the essential ingredients of their interaction: a short-range repulsion, representing subunit conformational rigidity, plus a longer-range attraction, corresponding to the driving force (e.g., hydrophobic interaction) for capsomer aggregation. The capsomer-capsomer binding energy ε0 is taken to be 15 kBT (25),∥ a typical value reported from atomistic calculations of subunit binding energies (24). (Here kB is the Boltzmann constant and T the absolute temperature.) Another essential feature of viral capsids is the existence of two different morphological units (pentamers and hexamers). To account for the intrinsic differences between capsomer units we assume that they can adopt two internal states: P(entamer) and H(examer). The potential has the same form for interactions between different capsomer types except that the equilibrium spacing [the minimum of V(r)] includes the geometrical size difference between pentamers and hexamers of the same edge length (size ratio ≈0.85). The energy difference ΔE between a P and an H capsomer, which reflects differences between individual contact interactions and folding conformations of pentamer and hexamer proteins, enters as a Boltzmann factor e-ΔE/kBT that provides the relative thermal probability for a noninteracting unit to be in the P state. For each fixed total number of capsomers N, the number NP of P units (and hence the number NH = N - NP of H units) is permitted to vary and was not fixed to be 12 (as in the CK construction). We carry out Monte Carlo simulations in which N interacting capsomers are allowed to range over a spherical surface while switching between P and H states, thus exploring all possible geometries and conformations. In this way we obtain the optimal structure for a given number N of capsomers and a given capsid radius R.†† The finite-temperature internal energy E(R) is evaluated for each of a range of equilibrated sphere radii R and then minimized with respect to R, leading to a special radius R* for each N. We tested different forms for V(r) and found the conclusions discussed below to be robust.
Results
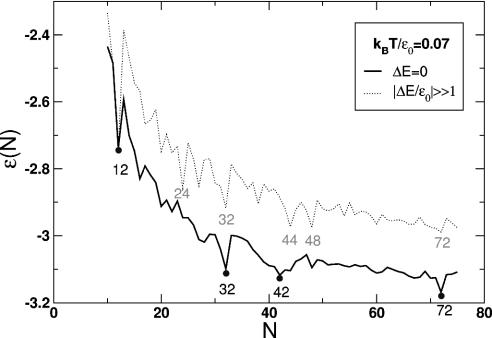
Two Different Morphological Units. The results of our Monte Carlo simulations are shown in Fig. 2 in the form of a plot of the minimized internal energy per capsomer ε(N) (in units of ε0) versus the number N of capsomers.
Fig. 2.
Energy per capsomer for ΔE = 0 (black curve) and |ΔE/εo| large compared to one (dotted curve).
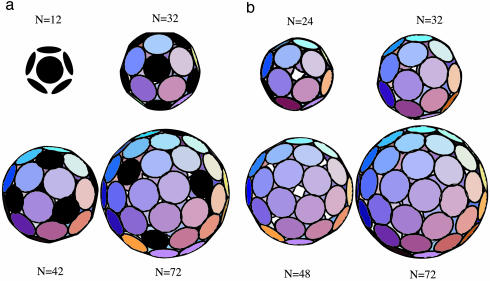
The solid curve shows ε(N) for the case where the energy difference ΔE between the P and H capsomer states equals zero. For large N values, ε(N) is slightly <3ε0, the binding energy per capsomer (including non-nearest-neighbor interactions) for a flat hexagonal array of capsomers. Pronounced minima of ε(N) are seen at N = 12, 32, 42, and 72; the capsid structures associated with these minima are shown in Fig. 3a. All four minimal structures have icosahedral symmetry. Moreover, they correspond precisely to T = 1, T = 3, T = 4, and T = 7 CK structures, respectively. (The N = 32/T = 3 structure should, for instance, be compared with Fig. 1.) Thus, we find that the appearance of both icosahedral symmetry and the T-number organization is indeed a direct consequence of free energy minimization of a very generic interaction that captures the crucial elements of capsid self-assembly: the attraction required for the aggregation, the excluded volume repulsion, and the existence of two different morphological units. In all four cases the equilibrium configuration developed spontaneously, despite the complexity of some of the structures [such as the chiral repeat motif (right-handed, here) of T = 7]. The existence of different possible structures resulting from the same building blocks in our simulations provides insight into polymorphism observed in some animal viruses such as hepatitis B whose capsomers assemble into particles of two different sizes, one with T = 3 symmetry and the other with T = 4 symmetry (26, 27).
Fig. 3.
Minimum energy structures produced by Monte Carlo simulation, with P-state capsomers shown in black. (a) The P and H states here have the same energies. The resulting N = 12, 32, 42, and 72 structures correspond to T = 1, T = 3, T = 4, and T = 7 C-K icosahedra. (b) Minimum energy structures for |ΔE/εo|>>1, i.e., only one size of capsomer. The N = 24 and 48 structures have octahedral symmetry, and N = 32 is icosahedral, whereas N = 72 is highly degenerate, fluctuating over structures with different symmetry, including T = 7.
Capsid self-assembly sets in when the chemical potential of isolated capsomers in solution exceeds -ε(N) (14). Actual size selection of viruses (i.e., the selection of one of the energy minima in Fig. 2) involves certain additional mechanisms that vary among viral species, such as “spontaneous curvature” effects (22), the presence of preformed scaffold structures (28), or the size of the enclosed genome. Assume that these effects restrict the capsid size to a certain range of N values, say N = 70 ± 10, that includes one of the minima of ε(N) (N = 72 in this case). According to Fig. 2, the energy per capsomer for an N = 72 capsid differs from that of its neighbors N = 71 and N = 73 by only ≈0.05 ε0; but the total energy difference (72 times larger) is ≈54 kBT (= 0.05 × 15 kBT × 72), and hence the relative abundance of the N = 71 and N = 73 structures is completely negligible. Because the minimum of ε(N) at N = 72 is sufficiently pronounced, additional size-determining mechanisms only need to provide a weak dependence on N to produce a nearly monodisperse solution of T = 7 capsids.
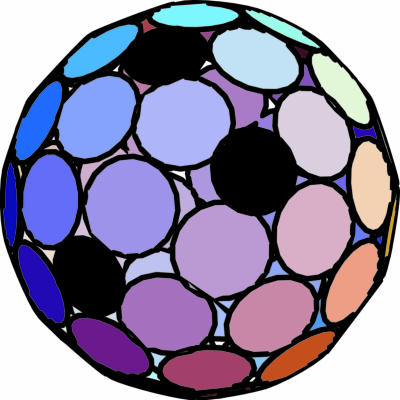
It is notable that in addition to the significant energy differences that exist between N = 72 and N = 71 and 73, there are big holes in N = 71 and N = 73 structures that render them inappropriate for genome protection and delivery. For example, the structure of N = 71 is similar to N = 72 shown in Fig. 3a, except that one of the pentamers is missing. Similarly, Fig. 4 shows one of the many possible nearly degenerate equilibrium structures of N = 73 with 12 pentamers but whose configuration is “spoiled” by sizeable gaps in the capsomer distribution. In fact, most of the other equilibrium, nonicosahedral structures observed in our simulations are characterized similarly by the presence of 12 pentamers, consistent for example with the structures of “spherical” retroviruses that contain 12 pentamers without having the symmetry of an icosahedron (29).
Fig. 4.
Minimum energy structure as in Fig. 3a, but for N = 73; see text.
One Type of Morphological Unit. How essential is reversible P-H switching for icosahedral capsid formation? When we increased the P-to-H switch energy, we encountered a dramatic reorganization of the structure spectrum. The dotted curve in Fig. 2 gives ε(N) for |ΔE/εo| large compared to one; the capsomers are now either all in the H state or all in the P state, depending on the sign of ΔE. The structure spectrum is seen to be significantly more complex. Pronounced minima of ε(N) include those at N = 12, 24, 32, 44, and 48‡‡; the corresponding capsid structures are shown in Fig. 3b.
The suppression of capsomer switching clearly has a profoundly destabilizing effect on icosahedral symmetry. Apart from T = 1, the only surviving T-structure is T = 3 (N = 32). The N = 24 and 48 minima have octahedral symmetry (the N = 24 minimum has the symmetry of a chiral octahedral Archimedean solid known as the “snub-cube”), whereas N = 44 has cubic symmetry. However, we have found that the presence of a small compression of the capsid (caused, for instance, by an external pressure, or a genome size smaller that the preferred size of the capsid protein shell, or a longer-range attractive interaction between capsomers) systematically facilitates the appearance of icosahedral symmetry. For example, for N = 72, consideration of R values only slightly smaller than R* recovers icosahedral symmetry even for the “1-state” case of identical morphological units. Compressed (R < R*) structures favor icosahedral symmetry because the compression stress can be optimally absorbed by the 5-fold sites, allowing the other sites to minimize their overlapping. In fact, genome-free self-assembly studies of capsid proteins of the Polyoma virus, an exceptional “non-CK” virus whose capsomers are all identical (pentamers), report (22) formation of icosahedral N = 72 capsids (see Figs. 3b and 5 and ref. 30) as well as stable N = 24 structures having the symmetry of a left-handed snub-cube.
Fig. 5.
Image reconstitution (30) showing the special structure associated with 72 pentamers (T = 7) in the case of genome-free polyoma capsids.
We conclude that the existence of two different types of morphological units (pentamers and hexamers) is not absolutely required for obtaining capsids with icosahedral symmetry. Nevertheless, the morphological switching, where P and H units are approximately isoenergetic, strongly favors the icosahedral symmetry and dramatically simplifies the spectrum of optimal structures, as is evident in Fig. 2.
Mechanical Properties of Capsids. Our minimal model for capsid structure posits an explicit potential for capsomer interactions, which provides us with a tool to study the mechanical and genome release properties of viral capsids. To address the effect of strain on capsid structure, we repeated our ΔE = 0 simulations at each of successive fixed capsid radii in excess of the optimal radius R*. For fixed N = 32 (T = 3) and NP = 12, the capsid bursts dramatically when R/R* exceeds a critical value (1.107; see Fig. 6b), in the form of a large crack stretching across the capsid surface. The bursting of the capsid is one of several possible gene release scenarios. Just before this point is reached the capsid is uniformly swollen (31) (see Fig. 6a), with all interstitial holes grown larger in size compared to those in the optimized R = R* structure (see N = 32 in Fig. 3a). The appearance of these pores constitutes another mechanism for genome release.
Fig. 6.
Capsid bursting. (a) Expanded N = 32/T = 3 capsid just before bursting (compare with N = 32 in Fig. 3a). (b) Burst capsid with a radius R just exceeding 1.107 times the equilibrium radius R*.
Finally, when we allow the number of capsomers to change during swelling, we find that the bursting scenario competes with still another mechanism, decapsidation; at a critical radius <1.107 R* the capsid energy can be reduced by ejecting one of the 12 pentamers, followed by a decrease in capsid size. These phenomena have been observed, for instance, for the Tymoviruses (32) and a series of Flock House virus mutants (33).
The fact that the minimal model reproduces realistic release mechanisms, in addition to accounting for both the predominant T-structures and the exceptional nonicosahedral structures, suggests that it can be applied as well to studying the mechanical properties of capsids and serve as a guide for the design of artificial viruses.
Acknowledgments
We thank David Nelson for several helpful discussions. D.R. acknowledges support from National Science Foundation Grant CHE-0313563 and the Ministerio de Ciencia y Tecnología of Spain. R.F.B. thanks the National Science Foundation for financial support under DMR Grant 0404507.
Abbreviations: CCMV, cowpea chlorotic mottle virus; CK, Caspar and Klug.
See Commentary on page 15549.
Footnotes
Specifically, we used V(r)= ε0 [(r*/r)12 - 2(r*/r)6] with ε0 the pair binding energy and r* the equilibrium spacing.
A somewhat smaller value of ≈ε0 = 12 kBT has been recently reported by Zlotnick (25), corresponding to a protein–protein interaction of 6 kBT. We have repeated our simulations based on this new estimate. Although the energy value associated with each structure changes because of the change in ε0, we obtain qualitatively the same results as those reported in Figs. 2 and 3.
We have used Metropolis Monte Carlo (MC) simulation with 105 equilibration steps and 105 production steps. An elementary MC step consisted of either an attempt to move a randomly chosen disk over the surface of a sphere in a random direction or an attempt to change its size. The ratio of MC attempts of moving a disk versus switching the size of a disk was set to 10. However, we tested different ratios and the result was robust, independent of the ratio.
This particular sequence of “magic numbers” coincides with maxima of the packing density of N hard disks on a spherical surface. See ref. 11.
References
- 1.Harrison, S. C. (1990) in Fields Virology, eds. Fields, B. N., Howley, P. M., Griffin, D. E., Lamb, R. A., Martin, M. A., Roizman, B., Straus, S. E. & Knipe, D. M. (Raven, New York), Vol. 2, pp. 37-61. [Google Scholar]
- 2.Johnson, J. E. & Speir, J. A. (1997) J. Mol. Biol. 269, 665-675. [DOI] [PubMed] [Google Scholar]
- 3.Fox, J. M., Wang, G., Speir, J. A., Olson, N. H., Johnson, J. E., Baker, T. S. & Young, M. J. (1998) Virology 244, 212-218. [DOI] [PubMed] [Google Scholar]
- 4.Caspar, D. L. D. & Klug, A. (1962) Q. Biol. 27, 1-24. [DOI] [PubMed] [Google Scholar]
- 5.Liddington, R. C., Yan, Y., Moulai, J., Sahli, R., Benjamin, T. L. & Harrison, S. C. (1991) Nature 354, 278-284. [DOI] [PubMed] [Google Scholar]
- 6.Grimes, J. M., Burroughs, J. N., Gouet, P., Diprose, J. M., Zientara, S., Mertens, P. P. C. & Stuart, D. I. (1998) Nature 395, 470-478. [DOI] [PubMed] [Google Scholar]
- 7.Ganser, B. K., Li, S., Klishko, V. Y., Finch, J. T. & Sundquist, W. I. (1999) Science 283, 80-83. [DOI] [PubMed] [Google Scholar]
- 8.Dong, X. F., Natarajan, P., Tihova, M., Johnson, J. E. & Schneemann, A. (1998) J. Virol. 72, 6024-6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adolph, K. W. & Butler, P. J. (1976) Philos. Trans. R. Soc. London B 276, 113-122. [DOI] [PubMed] [Google Scholar]
- 10.Goldberg, M. (1967) J. Mol. Biol. 24, 337-338. [DOI] [PubMed] [Google Scholar]
- 11.Clare, B. W. & Kepert, D. L. (1986) Proc. R. Soc. London Ser. A 405, 329-344. [Google Scholar]
- 12.Thomson, J. J. (1904) Philos. Mag. 7, 237-265. [Google Scholar]
- 13.Tarnai, T. (1991) J. Mol. Biol. 218, 485-488. [DOI] [PubMed] [Google Scholar]
- 14.Bruinsma, R. F., Gelbart, W. M., Reguera, D., Rudnick, J. & Zandi, R. (2003) Phys. Rev. Lett. 90, 248101-248104. [DOI] [PubMed] [Google Scholar]
- 15.Marzec, C. J. & Day, L. A. (1993) Biophys. J. 65, 2559-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lidmar, J., Mirny, L. & Nelson, D. R. (2003) Phys. Rev. E. 68, 051910-051913. [DOI] [PubMed] [Google Scholar]
- 17.Twarock, R. (2004) J. Theor. Biol. 226, 477-482. [DOI] [PubMed] [Google Scholar]
- 18.Berger, B., Shor, P. W., Tucker-Kellogg, L. & King, J. (1994) Proc. Natl. Acad. Sci. USA 91, 7732-7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zlotnick, A. (1994) J. Mol. Biol. 241, 59-67. [DOI] [PubMed] [Google Scholar]
- 20.Brandon, C. & Tooze, J. (1999) Introduction to Protein Structure (Garland, New York), 2nd Ed., pp. 325-344.
- 21.Zlotnick, A., Aldrich, R., Johnson, J. M., Ceres, P. & Young, M. J. (2000) Virology 277, 450-456. [DOI] [PubMed] [Google Scholar]
- 22.Salunke, D. M., Caspar, D. L. D. & Garcea, R. L. (1989) Biophys. J. 56, 887-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie, Z. & Hendrix, R. W. (1995) J. Mol. Biol. 253, 74-85. [DOI] [PubMed] [Google Scholar]
- 24.Reddy, V. S., Giesing, H. A., Morton, R. T., Kumar, A., Post, C. B., Brooks, C. L., III, & Johnson, J. E. (1998) Biophys. J. 74, 546-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zlotnick, A. (2003) Virology 315, 269-274. [DOI] [PubMed] [Google Scholar]
- 26.Zlotnick, A., Cheng, N., Conway, J. F., Booy, F. P., Steven, A. C., Stahl, S. J. & Wingfield, P. T. (1996) Biochemistry 35, 7412-7421. [DOI] [PubMed] [Google Scholar]
- 27.Conway, J. F., Watts, N. R., Belnap, D. M., Cheng, N., Stahl, S. J., Wingfield, P. T. & Steven, A. C. (2003) J. Virol. 77, 6466-6473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thuman-Commike, P. A., Greene, B., Malinski, J. A., King, J. & Chiu, W. (1998) Biophys. J. 74, 559-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ganser-Pornillos, B. K., von Schwedler, U. K., Stray, K. M., Aiken, C. & Sundquist, W. I. (2004) J. Virol. 78, 2545-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reddy, V. S., Natarajan, P., Okerberg, B., Li, K., Damodaran, K. V., Morton, R. T., Brooks, C. & Johnson, J. E. (2001) J. Virol. 75, 11943-11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tama, F. & Brooks, C. L., III (2002) J. Mol. Biol. 318, 733-747. [DOI] [PubMed] [Google Scholar]
- 32.Adrian, M., Timmins, P. A. & Witz, J. (1992) J. Gen. Virol. 73, 2079-2083. [DOI] [PubMed] [Google Scholar]
- 33.Schneemann, A., Dasgupta, R., Johnson, J. E. & Rueckert, R. R. (1993) J. Virol. 67, 2756-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]