Abstract
Epistatic gene interactions are believed to be a major factor in the genetic architecture of evolutionary diversification. In Arabidopsis thaliana, the FRI and FLC genes mechanistically interact to control flowering time, and here we show that this epistatic interaction also contributes to a latitudinal cline in this life history trait within the species. Two major FLC haplogroups (FLCA and FLCB) are associated with flowering time variation in A. thaliana in field conditions, but only in the presence of putatively functional FRI alleles. Significant differences in latitudinal distribution of FLC haplogroups in Eurasia and North Africa also depend on the FRI genotype. There is significant linkage disequilibrium between FRI and FLC despite their location on separate chromosomes. Although no nonsynonymous polymorphisms differentiate FLCA and FLCB, vernalization induces the expression of an alternatively spliced FLC transcript that encodes a variant protein with a radical amino acid replacement associated with the two FLC haplogroups. Numerous polymorphisms differentiating the FLC haplogroups also occur in intronic regions implicated in the regulation of FLC expression. The features of the regulatory gene interaction between FRI and FLC in contributing to the latitudinal cline in A. thaliana flowering time are consistent with the maintenance of this interaction by epistatic selection. These results suggest that developmental genetic pathways and networks provide the molecular basis for epistasis, contributing to ecologically important phenotypic variation in natural populations and to the process of evolutionary diversification.
Keywords: rosette leaf number, alternative splicing, linkage disequilibrium, epistatic selection, candidate gene association
Epistasis has been an enigmatic factor in the conceptualization of the evolutionary process (1, 2). Fisher (1, 3) contended that epistatic gene interactions were of limited relevance to evolutionary diversification, whereas Wright (4) argued for the central role of epistasis in evolution. Recently, renewed interest in the Wrightian view of evolutionary diversification has led to attempts to define and characterize epistasis in a wide range of micro- and macro-evolutionary phenomena. Gene interactions are now widely believed to be crucial features of the maintenance of genetic variation (5), the origins of sex and recombination (6), and the process of speciation (7). The importance of epistasis has also been bolstered by advances in quantitative trait loci (QTL) mapping that reveals examples of epistatic interactions underlying natural phenotypic variation in organisms such as Drosophila (8), Arabidopsis (5, 9), and Caenorhabditis elegans (10).
Concurrent with interest in the evolutionary role of epistasis, molecular geneticists have begun to identify genetic pathways and networks involved in the expression of many phenotypic features (11). These pathways provide frameworks for dissecting the molecular basis of gene interactions underlying ecologically and evolutionarily relevant phenotypic variation. Regulatory genetic pathways that control developmental processes are particularly intriguing, because it is thought that evolution of these regulatory loci underlies key aspects of evolutionary diversification (12).
Despite progress in the characterization of molecular genetic pathways, the study of epistasis continues to rely heavily on characterizing statistical interaction terms in quantitative genetic analysis (13) or defining broad chromosomal regions that contribute nonadditively to phenotypic variation (5). There have been few examples of epistasis resolved at the level of molecular interactions of naturally occurring alleles of specific genes (14-16) and no examples of regulatory gene interactions leading to natural phenotypic variation in ecologically relevant traits. Identifying regulatory gene interactions and determining their molecular mechanisms should provide key insights into the origins and importance of epistasis in evolutionary change.
Here we report on a specific regulatory gene interaction that contributes to natural variation in the timing of flowering in Arabidopsis thaliana. In A. thaliana, flowering time has been associated with size at reproduction and fecundity (17-19). Flowering time is sensitive to climatic signals, including day length (photoperiod) and prolonged cold treatment (vernalization), which serve as ecological cues to ensure that reproductive effort occurs in optimal seasonal environments (20). These signals vary systematically with latitude, and adaptation to these ecological cues would be expected to lead to latitudinal clines in the timing of flowering. Due to its distribution over a wide geographic range, latitude-dependent variation in flowering time in A. thaliana might be expected to be associated with molecular polymorphisms in genes that regulate the transition to flowering in response to environmental cues (21-23).
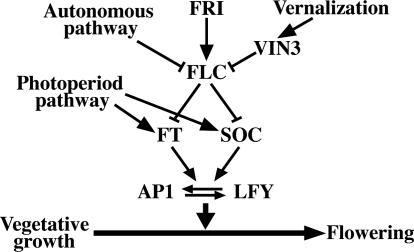
A large number of regulatory genes belonging to several interacting pathways that control flowering time in A. thaliana have been identified (20, 24). Genes in the vernalization pathway, including FRIGIDA (FRI) (25) and FLC (26, 27) (Fig. 1), condition A. thaliana's response to cold temperatures associated with winter, an environmental cue that promotes flowering in the spring. A recent study has demonstrated the presence of a latitudinal cline in flowering time modulated by the FRI gene in A. thaliana (28). FRI encodes a 609-aa coiled-coil protein that represses the transition to flowering, and mutants of this gene display early flowering under long-day controlled-temperature conditions, as well as insensitivity to vernalization (25). Several naturally occurring null FRI alleles have been described, and molecular analyses indicate that these independently derived alleles result from several deletions in the coding region (25, 29).
Fig. 1.
Schematic representation of genetic pathways of flowering time in A. thaliana. Lines with arrows denote up-regulation of gene expression; lines with bars denote repression of gene expression.
In a common garden experiment carried out under over-winter field conditions, a latitudinal cline in bolting time is evident only in ecotypes that are homozygous for a putative functional FRI allele; southern ecotypes in this genotypic class flower significantly earlier than those at northern latitudes (28). In contrast, ecotypes homozygous for a deletion allele of FRI (FRIΔ) flower later under natural over-winter conditions and do not exhibit the cline in flowering time. Thus, it is not latitudinal divergence in FRI functionality that is responsible for establishing the latitudinal cline in A. thaliana flowering time (28). This suggests that functional divergence in another gene, epistatic to FRI, may be responsible for establishing this life history cline.
A possible candidate for this interacting gene is FLC (Fig. 1), which encodes a MADS-box transcriptional activator that represses flowering. Mutants at FLC suppress the late-flowering phenotype of FRI under nonvernalization conditions (30), and molecular analyses indicate that transcription of FLC is up-regulated by FRI (26, 27). Moreover, FLC expression is down-regulated by vernalization, providing a mechanism to ensure flowering after the prolonged cold of winter (26, 27). Our evolutionary genetic analysis of FLC reveals that nucleotide polymorphism in this gene is indeed associated with flowering time variation in A. thaliana and contributes to the latitudinal cline in this trait. The association of FLC variation with flowering time is observed only in epistatic interaction with the upstream regulatory gene FRI. These results demonstrate the importance of epistasis among regulatory genes in generating phenotypic variation in a key life history trait in A. thaliana.
Materials and Methods
Molecular Population Genetic Analyses. A. thaliana ecotypes were obtained from the Arabidopsis Biological Resource Center (Table 2, which is published as supporting information on the PNAS web site). Arabidopsis lyrata seed was provided by O. Savolainen (University of Oulu, Oulu, Finland). Genomic DNA was isolated from young leaves by using a Plant DNeasy Mini kit (Qiagen, Chatsworth, CA). The complete FLC gene, as well as an ≈500-bp upstream region (392 bp of the promoter and the complete 5′ UTR) and ≈34 bp of the 3′ UTR, was amplified in overlapping fragments for 13 ecotypes of A. thaliana with Taq polymerase (Table 3, which is published as supporting information on the PNAS web site). A. lyrata PCRs were carried out on two partially overlapping portions of the gene by using ExTaq DNA polymerase (Takara Shuzo, Kyoto).
A. thaliana PCR products were purified by using QIAquick gel extraction kits (Qiagen) and sequenced directly by using cycle sequencing with BigDye terminators (Applied Biosystems). A. lyrata PCR products were cloned with the TOPO TA PCR cloning kit (Invitrogen); plasmid DNA from three to five independent clones per amplified region was sequenced. Sequencing was carried out at the North Carolina State University Genome Research Laboratory with a Prism 3700 automated sequencer (Applied Biosystems).
All sequences were aligned against the published Col-0 FLC sequence. Molecular population genetic analyses were conducted by using dnasp 3.99 (31). Haplotype trees were constructed by using a maximum parsimony analysis (branch and bound search, stepwise addition) in paup* (32). Insertion/deletion polymorphisms (indels) that were part of long mono- or dinucleotide repeats were excluded from all analyses. Other indels were included in the parsimony analysis of A. thaliana sequences, with each indel treated as a single character. Parsimony analyses with A. lyrata excluded all indels.
Haplotype Tagging and Association Tests. Single-nucleotide polymorphisms (SNPs) distinguishing two major FLC haplotype groups were identified from A. thaliana sequences. Genotyping of two SNPs was carried out by using dcaps (33) in 353 ecotypes (Table 4, which is published as supporting information on the PNAS web site). Previously identified transposon insertions as well as a Col-0 repeat (34, 35) were also genotyped. Association analyses of flowering time under unvernalized growth chamber (long day and short day) and vernalized field conditions was conducted by using sas (Version 9.1, SAS Institute, Cary, NC). Cryptic population structure (stratification) within the sample was controlled by using 92 ecotypes belonging to an unstratified population (36) (see Supporting Text and Fig. 6, which are published as supporting information on the PNAS web site). Sample size in association tests varied slightly depending on ecotypes for which genotypic and phenotypic data were obtained. Conditions and phenotypes measured under growth chamber and over-wintered field conditions are described in refs. 28 and 36.
FLC Expression Analyses. Four A. thaliana ecotypes with nondeletion FRI alleles (Est-0, Pa-1, Le-0, and Po-0) were used to evaluate FLC expression. Seeds were stratified at 4°C for 3 days and planted in a randomized design across two blocks. Planted seeds were placed at 20°C, 12 h per day for 7 days until germination. A block was then moved to 4°C, 12 h per day, whereas the other block remained at 20°C. Two replicates of whole-plant tissue per ecotype were collected from newly germinated plants (V0) and plants grown continuously at 4°C (V15) and 21°C (UV15) for 15 days. Each V0 replicate contained tissue from 10 plants; all others contained tissue from four plants.
RNA was extracted by using an RNeasy kit (Qiagen) and treated with DNA-free DNase (Ambion, Austin, TX). Complementary DNA was synthesized from ≈1 μg of total RNA with the Retroscript reverse transcription kit (Ambion). RT-PCR was performed by using previously described FLC primers (37). Product band intensity in ethidium-stained agarose gels was used as a semiquantitative measure of expression level. Details of the RT-PCR procedure and analysis are included in Supporting Text.
Results
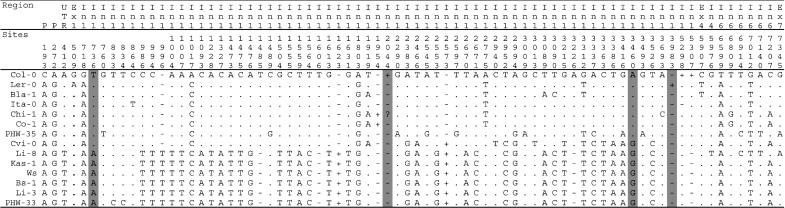
Nucleotide Polymorphisms at the FLC Gene. The latitudinal cline in A. thaliana flowering time modulated by putative functional FRI alleles may arise as a result of the activity of its downstream regulatory target gene, FLC. We thus examined FLC in 13 A. thaliana ecotypes and Col-0 for the presence of genetic variation. Approximately 6.2-7.4 kb were sequenced (length variation is due to the presence of indels) and include the entire coding unit, the 5′ UTR, and portions of the 3′ UTR and the promoter. Sixty-five SNPs and 11 indels were observed across the entire gene (Fig. 2). The silent site nucleotide diversity, π, is 0.004 (Table 1), which is slightly less than the mean of 0.007 observed for previously studied A. thaliana nuclear genes (38).
Fig. 2.
Polymorphisms observed in the sequenced FLC region. Site numbering is based on our alignment and includes indels. Polymorphisms in mononucleotide repeats longer than 4-bp and long dinucleotide repeats are not included. Positions highlighted in gray, along with the Da (1)-12 type insert (not shown), were used in genotyping.
Table 1. Features of FLC sequence variation.
| Sequence length, bp
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Clade | n | Alignment* | Adjusted† | S‡ | A§ | π¶ | θw¶ | Tajima's D |
| FLC | 14 | 7,460 | 5,994 | 65 | 11 | 0.0044 | 0.0036 | 0.8579 |
| FLCA | 8 | 7,460 | 6,017 | 41 | 8 | 0.0022 | 0.0028 | -1,1699 |
| FLCB | 6 | 7,460 | 6,107 | 7 | 3 | 0.0004 | 0.0005 | -1.3903 |
Alignment length
Length excluding indels and missing data
Number of segregating sites
Haplotype number
Estimates based on silent sites
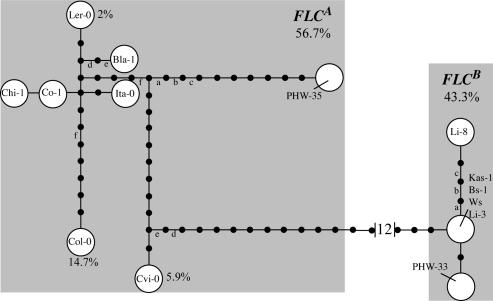
There are 11 distinct FLC haplotypes in our sample, and genealogical relationships among them are shown in Fig. 3. Like several A. thaliana genes (38), FLC exhibits dimorphism; haplotypes are organized into two groups (haplogroups) differentiated by 22 SNPs and six indels (Fig. 3). We refer to these haplogroups as FLCA and FLCB. All but one of these differentiating polymorphisms are in the large ≈3.5-kb first intron of the gene. Using A. lyrata FLC as an outgroup roots the tree between the FLCA and FLCB clades (Fig. 7, which is published as supporting information on the PNAS web site). No exon polymorphisms distinguish FLCA and FLCB, and the protein sequences encoded by the two haplogroups are identical. The only nonsynonymous polymorphism in the sample occurs within FLCA (a conservative R to K replacement found in Ler-0). The two FLC haplogroups differ substantially in the amounts of polymorphism. The eight FLCA alleles have 41 nucleotide polymorphisms, with π = 0.0022. In contrast, the six FLCB alleles have seven SNPs and a 5-fold lower level of nucleotide diversity (π = 0.0004) (Table 1). Patterns with an unbalanced distribution of polymorphism have sometimes been ascribed to partial selective sweeps in other biological systems (39).
Fig. 3.
FLC haplotype network. The tree represents one of four mostparsimonious arrangements; relationships among major haplotype groups do not change with alternative trees. Each line represents a single mutational step corresponding to a SNP or an indel. Mutations marked with lower-case letters are homoplasious. Characters 1919 and 7207, which are highly homoplasious (consistency index ≤ 0.333), were excluded from the tree. Shaded boxes indicate haplogroups. Haplogroup and haplotype frequencies are based on genotyping 353 A. thaliana accessions.
We genotyped a set of 353 primarily Eurasian ecotypes for FLCA and FLCB alleles (Table 2). The FLCA allele is present at slightly higher frequency (57%) than FLCB (43%). Tajima's D for FLC is positive (D = 0.8579), consistent with the presence of two haplotype groups at intermediate frequencies, although it does not differ significantly from predictions of the neutral-equilibrium theory. Previous studies have identified two large transposable element insertions in the first intron of FLC that reduce expression and lead to early flowering (34, 35). These insertions occur at low frequency in our ecotype sample (<3%) and are all nested within the FLCA haplogroup (Fig. 3 and Table 2). A small direct repeat insertion in intron 1, characteristic of the Col-0 ecotype (34, 35), is also nested within FLCA and occurs at moderately low frequency (14.7%).
Epistasis Between FRI and FLC Is Associated with Flowering Time Variation. Field experiments have revealed latitudinal variation in A. thaliana flowering time only in accessions that do not contain inactivating deletions at the FRI gene (28). Molecular genetic studies indicate that FRI up-regulates FLC expression (26, 27); we would thus expect that variation in the latter would show significant association with flowering time, but only in ecotypes that have an active FRI allele.
We tested for effects of FRI and FLC on flowering time using a set of 92 ecotypes shown to behave as an unstratified population appropriate for testing phenotypic associations (36). We genotyped these accessions for FLCA and FLCB alleles and for the two intron 1 transposable element insertions (34, 35); eight ecotypes with transposon insertions were excluded from the analyses, as were five others for which the presence/absence of insertions could not be determined. Together with FRI genotypes classes previously observed for 72 of the remaining ecotypes (28), we determined the two-locus FLC and FRI deletion (FRIΔ) and nondeletion (FRI) genotypes and tested for associations with rosette leaf number at bolting (RLN) and days until bolting, both common indicators of flowering time. There is substantial broad sense heritability (H2)for flowering time under growth chamber and field conditions (36). RLN and days until bolting are highly genetically correlated traits (0.94) (9), and results obtained for both measures were largely congruent. We present the more conservative RLN-based results in the text; results obtained with bolting days are presented in Table 5, which is published as supporting information on the PNAS web site. Only results significant for both traits are discussed.
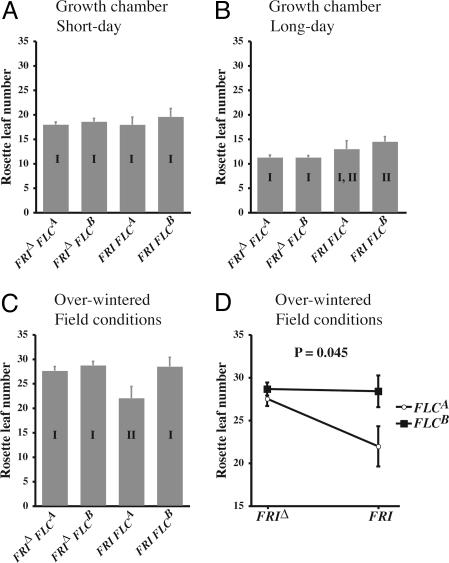
We did not find a significant association of FRI or FLC genotypes with flowering time under short-day controlled environmental conditions (Fig. 4A and Table 6, which is published as supporting information on the PNAS web site). We did find an association of FRI genotype with RLN under long-day controlled environmental conditions (two-way ANOVA: P = 0.0014); ecotypes with FRI deletion alleles exhibit early flowering, as expected (Fig. 4B and Table 6). No association was seen for RLN with FLC or FRI FLC genotype under long-day conditions. We did, however, find significant associations for both genes and RLN under field conditions (FRI genotype P = 0.028; FLC haplogroup P = 0.0049), as well as for the FRI × FLC interaction (P = 0.045) (Fig. 4 C and D and Table 6). This suggests that both genes have an effect on flowering time under ecological field conditions; examination of the variance components reveals that substantial amounts of variance in RLN are explained by FLC, FRI, and their epistatic interaction (11.1%, 4.35%, and 13.43%, respectively; Table 7, which is published as supporting information on the PNAS web site).
Fig. 4.
Associations between FRI FLC genotype and flowering time. Bars that differ in roman numerals are significantly different at P < 0.006 by Fisher's protected least-significant difference test. Error bars indicate standard error. (A) Short-day growth chamber conditions. (B) Long-day growth chamber conditions. (C and D) Vernalized over-wintering field conditions.
Post hoc tests reveal that, under field conditions, there is no significant difference in mean RLN for ecotypes with a FRI deletion allele (FRIΔ FLCA mean RLN = 27.51 ± 0.85, n = 18; FRIΔ FLCB mean RLN = 28.65 ± 0.76, n = 28) (Fig. 4C). In contrast, mean RLN for accessions with a FRI FLCA genotype (21.95 ± 2.35, n = 8) is significantly lower than those that are FRI FLCB (28.84 ± 1.86, n = 9; P = 0.0049). FRI FLCA ecotypes also flower significantly earlier than FRIΔ FLCA or FRIΔ FLCB ecotypes (P = 0.0054 and P = 0.0005, respectively). This indicates that most genotypes flower at approximately the same time in an over-winter field environment, except for the earlier flowering of accessions with an FLCA allele in a FRI nondeletion background. Thus, an association of FLC allele class with flowering time is detectable only in a functional FRI background.
Linkage Disequilibrium Between FLC and FRI. The FRI and FLC genes are not physically linked; FRI is located at the top of chromosome IV, whereas FLC is at the top of chromosome V. Despite their location on different chromosomes, flowering time variation associated with the functional interaction of FRI and FLC could lead to linkage disequilibrium between these two loci. To test for linkage disequilibrium, we obtained FRI genotypes for 321 ecotypes (C. A. Mays and M.D.P., unpublished data) and combined these with observed FLC genotypes. The frequencies of ecotypes with FRIΔ FLCA (29%) and FRIΔ FLCB (35%) are almost equivalent, indicating that FLC alleles are equally distributed in a FRI deletion allele background. The frequency of FRI FLCA (27%) is also similar to that of the former two genotypic classes. In contrast, there is a lower number of accessions with a FRI FLCB genotype (9%). Comparison with expected genotypic frequencies reveals that genotypes FRIΔ FLCB and FRI FLCA are overrepresented in our sample, whereas genotypes FRIΔ FLCA and FRI FLCB are underrepresented. This nonrandom distribution of allelic genotypes is significant in an exact test of gametic disequilibrium (P < 4.7 × 10-7).
FLC Genotypes Display Latitudinal Differentiation. The geographic distribution of FRI FLC genotypes was examined for 298 Eurasian A. thaliana accessions, for which we had site-of-origin information; we tested for FRI and FLC associations with latitude via a two-way analysis of covariance (ANCOVA), including longitude as a covariate. We found it necessary to control for longitude, because it is correlated with latitude in our dataset. There is a significant association of FLC and FRI FLC genotype with latitude (P = 0.0025 and P = 0.001, respectively; Table 8, which is published as supporting information on the PNAS web site). Post hoc tests reveal significant difference in latitudinal distribution of FLCA and FLCB alleles in a FRI nondeletion background (P < 0.001) but not in a FRIΔ background. Plants that have the FRI FLCB genotype are found primarily in northern latitudes (mean latitude = 51.76°N± 0.55, n = 24), whereas accessions with FRI FLCA genotypes have a more southern distribution (mean latitude = 48.37°N± 0.56, n = 78). In contrast, the mean latitudes for FRIΔ FLCA (50.16°N± 0.36, n = 89) and FRIΔ FLCB (50.26°N± 0.26, n = 107) are almost equal. To determine whether the geographic differentiation of FRI FLCA and FRI FLCB genotypes is a consequence of population structure within A. thaliana, we examined the distribution of genotypes in our unstratified dataset. As before, the associations of FLC haplogroup and FRI FLC genotype with latitude are significant (P = 0.013 and P = 0.038, respectively; Table 8). There is significant latitudinal differentiation between FRI FLCA and FRI FLCB genotypes (P = 0.022) and no differentiation between FRIΔ FLCA and FRIΔ FLCB genotypes. This suggests that the distribution of FLC genotypes in a FRI nondeletion background is not random and is not a result of cryptic population structure.
The preferential northern distribution of FRI FLCB genotypes, which are later flowering under field conditions, and southern distribution of earlier-flowering FRI FLCA genotypes could explain the latitudinal cline in flowering time observed for putatively functional FRI genotypes (Fig. 8, which is published as supporting information on the PNAS web site). However, the associations of both genotype and flowering time with latitude could also indicate that other factors correlated with latitude are responsible for the observed variation in flowering time. We tested this hypothesis by running an analysis of covariance (ANCOVA) on our unstratified sample, including FRI and FLC genotypes as main effects and latitude as a covariate. Because flowering time in the common garden has also been shown to be significantly associated with mean January precipitation at site of origin (28), this variable was also included as a covariate. After controlling for the two geographic variables, the effects of FLC and the FRI× FLC interaction on RLN remain significant (P = 0.0053, P = 0.025 respectively; Table 9, which is published as supporting information on the PNAS web site).
Alternative Splicing at FLCA and FLCB. There is no difference in the protein sequences encoded by FLCA and FLCB haplogroup alleles; instead, nearly all polymorphisms that differentiate these two haplogroups are found in the ≈3.5-kb intron 1 of FLC. Large introns in MADS-box regulatory genes are known to contain key cis-regulatory elements (40), and regions of the FLC first intron have been implicated in the control of FLC expression (41, 42). These observations suggest that associations of FLC haplogroups with flowering time may arise from regulatory differences between these two naturally occurring alleles.
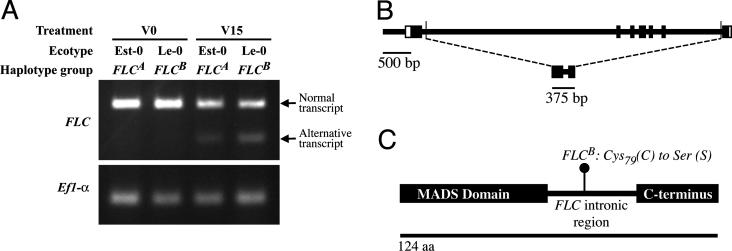
To test this hypothesis, we conducted RT-PCR analysis of FLC transcripts from four ecotypes. Consistent with previous reports, a decrease in the expression levels of FLC is evident after 15 days of vernalization in ecotypes from both FRI FLCA and FRI FLCB genotype classes (Fig. 5A and Fig. 9, which is published as supporting information on the PNAS web site). Moreover, an additional shorter transcript is induced in a vernalization treatment of 15 days (Fig. 5A). This shorter transcript is not observed at high levels in plants grown at normal temperature for 15 days (data not shown). The shorter transcript is an alternatively spliced FLC transcript that uses a nonconsensus cryptic splice donor site in intron 1 (GA) and splice acceptor site in intron 6 (CT) and contains all of the first and last exon of the FLC gene, as well as 73 bp of intron 1 and 15 bp of intron 6 (Fig. 5B). It encodes an in-frame 124-aa protein, which includes the sequence-specific DNA-binding/dimerization MADS domain and the C-terminal region of the normal FLC protein, as well as a novel intervening protein sequence. The alternatively spliced transcript has an A to T transversion at codon 79 of the ORF, resulting in a Cys to Ser radical amino acid replacement between the putative FLCA and FLCB alternate protein products (Fig. 5C).
Fig. 5.
Alternative splicing at FLC. (A) RT-PCR of FLC under different treatments. Plants at V0 were collected soon after germination and were not subjected to vernalization. Plants at V15 were grown at 4°C for 15 days after germination. (B) Structure of the alternative-splice FLC transcript. The upper gene model encompasses the region sequenced for FLC. White boxes represent UTRs, black boxes represent exon regions, and lines represent introns and promoter regions. Slender bars indicate splice sites of the alternative FLC product. The lower gene model represents the alternative transcript after splicing has occurred. (C) Predicted protein of the FLC alternative splice transcript, indicating the amino acid replacement polymorphism between the FLCA and FLCB haplogroups.
We did not detect significant differences in the levels of expression of the full-length or alternative FLC transcripts between the two haplogroups under the different treatments (Fig. 9). However, we did observe a tendency for a higher expression of full-length FLC in FRI FLCA ecotypes under vernalization conditions (Fig. 9B). There is also a tendency for lower levels of the alternativesplice transcript in FRI FLCA ecotypes, but this is partially obscured by the high levels of alternative transcript contributed by one replicate of the Pa-1 ecotype (Fig. 9C). Because the modest size of our assay may have limited power, we cannot rule out the possibility of subtle differences in the levels of the FLC transcripts between FLC haplogroups.
Discussion
Variation in flowering time is a key feature of the life histories of flowering plants and determines crucial aspects of the ecology of plant reproduction; thus, there has been intense interest in determining the genetic architecture of this trait. A recent study has demonstrated that a latitudinal cline in A. thaliana flowering time under field over-winter conditions is modulated by putative functional alleles of the regulatory gene FRI (28). Ecotypes with putatively functional FRI alleles located in southern Eurasian latitudes flower significantly earlier than northern ecotypes. In contrast, there is no correlation of flowering time with latitude among ecotypes that have deletion alleles of FRI. These results suggest a genetic model in which the latitudinal cline in A. thaliana flowering time depends on another gene whose function requires an active FRI allele.
FRI and the MADS-box transcription factor FLC are two genes known to mechanistically interact to control the transition to flowering (26, 27). Two lines of evidence indicate that the latitudinal cline in A. thaliana flowering time is established, in part, by epistasis between these two regulatory genes. First, FRI is known to up-regulate FLC expression, so that ecotypes with deletion alleles of FRI have reduced FLC activity and cannot express phenotypes associated with FLC variation. Our results reveal that variation in flowering time associated with FLC is observed only in plants that have a putative functional FRI allele, in accordance with this model (Fig. 4). Second, we would predict that FLC haplogroups display differences in latitudinal distribution only in ecotypes with putatively functional FRI alleles; this prediction is also borne out by our data. Together, these results demonstrate that FLC, in epistatic interaction with FRI, forms part of the molecular basis for the latitudinal cline in A. thaliana flowering time.
The precise molecular mechanism for differential FLC function associated with flowering time variation requires further investigation. Based on our results, two possible mechanisms could account for functional differences among haplogroups. One mechanism relies on our discovery that vernalization induces expression of an alternatively spliced FLC transcript. The alternate transcript encodes a modified MADS-domain protein, which may act as a dominant negative repressor by competing for target promoter-binding sites or by dimerizing with FLC. Unlike normal FLC, the modified proteins encoded by the FLCA and FLCB alternative transcripts differ by a radical amino acid change, potentially leading to differential activity. A second mechanism is suggested by previous studies of regions in the first intron of FLC involved in the regulation of FLC expression (41, 42). Some of these regions contain polymorphisms differentiating FLCA and FLCB, potentially leading to differential expression. Expression analyses of FLCA and FLCB ecotypes do not reveal significant differences in the expression of either of the FLC transcripts; however, the small scale of the experiments might not be sufficient to reveal subtle differences in levels of expression. Both of the mechanisms for FLC differential function (alternate protein activity or expression differences) provide clear testable hypotheses; these studies are currently in progress.
The finding that the latitudinal cline in A. thaliana flowering time is associated with an epistatic regulatory gene interaction is particularly noteworthy. Although selection on epistatic interactions has been hypothesized, particularly in the maintenance of coadapted gene complexes, it is presumed that recombination would break apart allelic correlations unless selection were sufficiently strong (43). Epistatic selection has been suggested to be a significant evolutionary force among linked loci or in nonrandom mating or highly structured populations (44), although these restrictive features have limited the number of documented cases. Several lines of evidence, however, indicates that the epistatic interaction between the FRI and FLC regulatory genes is maintained by selection. First, the interaction between FRI and FLC clearly contributes to natural phenotypic variation in an ecologically important life history trait. Second, the flowering time variation caused by alternative FLC alleles in a FRI background is distributed along a latitudinal cline; latitudinal clines are classically regarded as strong evidence of selection associated with geographically structured climatic variables (45). Third, linkage disequilibrium among interacting loci has been regarded as a key signature of epistatic selection; the skewed genotypic associations between FRI and FLC alleles suggest that the allele combinations are targeted by selection. Although each of these results alone may be insufficient to conclude the action of selection, together they present a compelling case to suggest that this epistatic regulatory gene interaction is maintained by selection for flowering time variation across the species range of A. thaliana.
The precise nature and direction of selection on the FRI FLC genotypes are unknown, but several possibilities exist. The earlier flowering of FRI FLCA ecotypes under over-wintering conditions suggests that plants carrying this genotype may be more sensitive to vernalization. The differential geographical distribution of FRI FLCA and FRI FLCB genotypes could be due to spatially heterogeneous selection. Sensitivity to vernalization might be favored in southern latitudes, where the vernalization cues of winter are milder; early flowering in response to vernalization could also serve as a mechanism to guarantee flowering before the summer heat (28). Alternatively, less sensitivity to vernalization might be favored in northern latitudes, where colder and longer winters could lead to premature flowering in highly vernalization-sensitive plants; later flowering may also lead to increased fitness associated with larger plant size and greater fruit production (18). The skewed frequencies of FRI FLC genotypes may be due to the availability of habitats where alternative vernalization-sensitive phenotypes are favored. Selection on unknown traits influenced by FLC haplogroups in both FRI backgrounds (deletion and nondeletion) may also be a factor shaping both the geographical distribution and frequencies of these genotypes.
Although the role of epistasis in evolutionary diversification has been the subject of intense debate, there are few known examples of natural phenotypic variation mediated by epistatic interactions between defined genes. Molecular genetic characterization of regulatory gene interactions, such as those observed in the A. thaliana flowering time network, provides evolutionary geneticists with concrete mechanistic frameworks to define epistatic interactions that underlie ecologically and evolutionarily relevant trait variation. We can now begin to bridge the gap between the population epistasis of the evolutionary geneticist and the mechanistic epistasis of molecular and developmental geneticists (46, 47) and to dissect, at the molecular level, the mechanisms and roles of gene interactions in the process of evolutionary change.
Supplementary Material
Acknowledgments
We thank S. Rusza for technical assistance and T. Korves, R. C. Moore, and C. Mays for discussion and suggestions on the manuscript. This work was funded in part by a National Science Foundation Integrated Research Challenges in Environmental Biology grant (to M.D.P., J.S., and T. F. C. Mackay).
Author contributions: A.L.C. and M.D.P. designed research; A.L.C. performed research; A.L.C., J.R.S., K.M.O., J.S., and M.D.P. analyzed data; A.L.C. and M.D.P. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: RLN, rosette leaf number at bolting; SNP, single-nucleotide polymorphism.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY769347-AY769360, AY769865, and AY772670-AY772672).
References
- 1.Fenster, C. B., Galloway, L. F. & Chao, L. (1997) Trends Ecol. Evol. 12, 282-286. [DOI] [PubMed] [Google Scholar]
- 2.Wade, M. J., Winther, R. G., Agrawal, A. F. & Goodnight, C. J. (2001) Trends Ecol. Evol. 16, 498-504. [Google Scholar]
- 3.Fisher, R. A. (1930) The Genetical Theory of Natural Selection (Oxford Univ. Press, Oxford).
- 4.Wright, S. (1931) Genetics 16, 97-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weinig, C., Dorn, L. A., Kane, N. C., German, Z. M., Halldorsdottir, S. S., Ungerer, M. C., Toyonaga, Y., Mackay, T. F., Purugganan, M. D. & Schmitt, J. (2003) Genetics 165, 321-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Charlesworth, B. (1990) Genet. Res. 55, 199-221. [DOI] [PubMed] [Google Scholar]
- 7.Orr, H. A. (1995) Genetics 139, 1805-1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dilda, C. L. & Mackay, T. F. C. (2002) Genetics 162, 1655-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ungerer, M. C., Halldorsdottir, S. S., Modliszewski, J. L., Mackay, T. F. C. & Purugganan, M. D. (2002) Genetics 160, 1133-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shook, D. R. & Johnson, T. E. (1999) Genetics 153, 1233-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cork, J. M. & Purugganan, M. D. (2004) BioEssays 26, 479-484. [DOI] [PubMed] [Google Scholar]
- 12.Doebley, J. & Lukens, L. (1998) Plant Cell 10, 1075-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hard, J. J., Bradshaw, W. E. & Holzapfel, C. M. (1992) Genetics 131, 389-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rawson, P. D. & Burton, R. S. (2002) Proc. Natl. Acad. Sci. USA 99, 12955-12958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Templeton, A. R. (2000) in Epistasis and the Evolutionary Process, eds. Wolf, J. B., Brodie, E. D., III, & Wade, M. J. (Oxford Univ. Press, New York), pp. 41-57.
- 16.McKechnie, S. W. & Geer, B. W. (1988) Genet. Res. 52, 179-184. [DOI] [PubMed] [Google Scholar]
- 17.Mitchell-Olds, T. (1996) Evolution (Lawrence, Kans.) 50, 140-145. [DOI] [PubMed] [Google Scholar]
- 18.Clauss, M. J. & Aarssen, L. W. (1994) J. Ecol. 82, 447-455. [Google Scholar]
- 19.Dorn, L. A., Pyle, E. H. & Schmitt, J. (2000) Evolution (Lawrence, Kans.) 54, 1982-1994. [DOI] [PubMed] [Google Scholar]
- 20.Mouradov, A., Cremer, F. & Coupland, G. (2002) Plant Cell 14, S111-S130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nordborg, M. & Bergelson, J. (1999) Am. J. Bot. 86, 470-475. [PubMed] [Google Scholar]
- 22.Stenoien, H. K., Fenster, C. B., Kuittinen, H. & Savolainen, O. (2002) Am. J. Bot. 89, 1604-1608. [DOI] [PubMed] [Google Scholar]
- 23.Pigliucci, M. & Marlow, E. T. (2001) Oecologia 127, 501-508. [DOI] [PubMed] [Google Scholar]
- 24.Simpson, G. G. & Dean, C. (2002) Science 296, 285-289. [DOI] [PubMed] [Google Scholar]
- 25.Johanson, U., West, J., Lister, C., Michaels, S., Amasino, R. & Dean, C. (2000) Science 290, 344-347. [DOI] [PubMed] [Google Scholar]
- 26.Sheldon, C. C., Burn, J. E., Perez, P. P., Metzger, J., Edwards, J. A., Peacok, W. J. & Dennis, E. S. (1999) Plant Cell 11, 445-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michaels, S. D. & Amasino, R. M. (1999) Plant Cell 11, 949-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stinchcombe, J. R., Weinig, C., Ungerer, M., Olsen, K. M., Mays, C., Halldorsdottir, S. S., Purugganan, M. D. & Schmitt, J. (2004) Proc. Natl. Acad. Sci. USA 101, 4712-4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le Corre, V., Roux, F. & Reboud, X. (2002) Mol. Biol. Evol. 19, 1261-1271. [DOI] [PubMed] [Google Scholar]
- 30.Michaels, S. D. & Amasino, R. M. (2001) Plant Cell 13, 935-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rozas, J. & Rozas, R. (1999) Bioinformatics 15, 174-175. [DOI] [PubMed] [Google Scholar]
- 32.Swofford, D. L. (2000) paup*, Phylogenetic Analysis Using Parsimony (*and Other Methods) (Sinauer, Sunderland, MA).
- 33.Neff, M. M., Turk, E. & Kalishman, M. (2002) Trends Genet. 18, 613-615. [DOI] [PubMed] [Google Scholar]
- 34.Gazzani, S., Gendall, A. R., Lister, C. & Dean, C. (2003) Plant Physiol. 132, 1107-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michaels, S. D., He, Y. H., Scortecci, K. C. & Amasino, R. M. (2003) Proc. Natl. Acad. Sci. USA 100, 10102-10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olsen, K. M., Halldorsdottir, S. S., Stinchcombe, J. R., Weinig, C., Schmitt, J. & Purugganan, M. D. (2004) Genetics 167, 1361-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michaels, S. D., Ditta, G., Gustafson-Brown, C., Pelaz, S., Yanofsky, M. & Amasino, R. M. (2003) Plant J. 33, 867-874. [DOI] [PubMed] [Google Scholar]
- 38.Yoshida, K., Kamiya, T., Kawabe, A. & Miyashita, N. T. (2003) Genes Genet. Syst. 78, 11-21. [DOI] [PubMed] [Google Scholar]
- 39.Yi, S. & Charlesworth, B. (2003) Genet. Res. 82, 101-106. [DOI] [PubMed] [Google Scholar]
- 40.Sieburth, L. E. & Meyerowitz, E. M. (1997) Plant Cell 9, 355-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheldon, C. C., Conn, A. B., Dennis, E. S. & Peacock, W. J. (2002) Plant Cell 14, 2527-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He, Y. H., Michaels, S. D. & Amasino, R. M. (2003) Science 302, 1751-1754. [DOI] [PubMed] [Google Scholar]
- 43.Lewontin, R. (1974) The Genetic Basis of Evolutionary Change (Columbia Univ. Press, New York).
- 44.Clegg, M. T., Allard, R. W. & Kahler, A. L. (1972) Proc. Natl. Acad. Sci. USA 69, 2474-2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Endler, J. A. (1986) Natural Selection in the Wild (Princeton Univ. Press, Princeton, NJ).
- 46.Brodie, E. D., III (2000) in Epistasis and the Evolutionary Process, eds. Wolf, J. B., Brodie, E. D., III, & Wade, M. J. (Oxford Univ. Press, New York), pp. 3-19.
- 47.Phillips, P. C. (1998) Genetics 149, 1167-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.