Abstract
Human fatty acid synthase is a large homodimeric multifunctional enzyme that synthesizes palmitic acid. The unique carboxyl terminal thioesterase domain of fatty acid synthase hydrolyzes the growing fatty acid chain and plays a critical role in regulating the chain length of fatty acid released. Also, the up-regulation of human fatty acid synthase in a variety of cancer makes the thioesterase a candidate target for therapeutic treatment. The 2.6-Å resolution structure of human fatty acid synthase thioesterase domain reported here is comprised of two dissimilar subdomains, A and B. The smaller subdomain B is composed entirely of α-helices arranged in an atypical fold, whereas the A subdomain is a variation of the α/β hydrolase fold. The structure revealed the presence of a hydrophobic groove with a distal pocket at the interface of the two subdomains, which constitutes the candidate substrate binding site. The length and largely hydrophobic nature of the groove and pocket are consistent with the high selectivity of the thioesterase for palmitoyl acyl substrate. The structure also set the identity of the Asp residue of the catalytic triad of Ser, His, and Asp located in subdomain A at the proximal end of the groove.
Human fatty acid synthase (FAS) is a complex homodimeric (552-kDa) enzyme that regulates the de novo biosynthesis of long-chain fatty acids. This cytosolic enzyme catalyzes the formation of 16 carbon (C16) palmitate, from acetyl-coenzyme A (acetyl-CoA) and malonyl-coenzyme A (malonyl-CoA) in the presence of NADPH. This entire reaction is composed of numerous sequential reactions and acyl intermediates, each catalyzed by a specific enzyme activity (1, 2). Mammalian FAS not only is an essential enzyme in fatty acid synthesis, but also plays an important role during embryonic development (3). Moreover, human FAS recently gained prominence due to its role in obesity and cancer biology. Obesity is a major health problem in developed nations affecting >50% of the U.S. population and seems to be increasing both in severity and prevalence (4). The magnitude of this health problem and the recent difficulties with several weight-loss therapies emphasize the need for different approaches to treat this problem. Of all of the lipogenic enzymes in the fatty acid synthesis pathway, FAS provides the best opportunity for therapeutic applications because of its high expression in lipogenic tissues and multistep enzyme reactions.
Human FAS is an attractive target as both a diagnostic and a prognostic marker for cancer cells. FAS is present at abnormally elevated levels in many varieties of common human cancer, including those of the breast (5–8), prostate (9, 10), colon (11), endometrium (12), ovary (13), thyroid (14), and skin (15). As an anticancer drug target, potentially each of the activities of FAS can be exploited for structure-based design of therapeutic agents. For example, the inhibition of FAS thioesterase (TE) was recently found to halt tumor cell proliferation and inhibit the growth of prostrate tumors in mice (16).
The TE carries out the chain-terminating step of fatty acid synthesis, leading to the release of palmitic acid by the hydrolysis of the acyl-S-phosphopantetheine thioester bound to the preceding acyl carrier protein (ACP) domain. It is a serine active site enzyme in which the nucleophilicity of the serine residue is supported by a conserved histidine residue (17). TE is coupled to the preceding ACP domain by a flexible linker and can be easily separated from the synthase by limited proteolysis (18). Much like its activity in the intact FAS, the isolated TE domain shows maximal activity for C16 acyl chain length, with a sharp decline in activity for chain lengths longer than C18 or shorter than C16 (18–20). However, once removed, the domain can no longer interact with the remainder of FAS to hydrolyze newly synthesized fatty acyl-S-phosphopanthetheine thioester (21). Consequently the FAS lacking the TE does not turn over and loses its chain-length specificity for fatty acid synthesis to an extent that longer fatty acyl chains of C20 and even C22 are bound to the ACP domain (21). Thus, the TE domain of FAS serves a key role as a regulator that determines that the major product of FAS is palmitate and is essential for turnover of the enzyme.
Here, we report the 2.6-Å resolution structure of the 32-kDa TE domain of human FAS. The structure sheds light on the TE enzyme activity and fatty acyl chain selectivity of FAS. This structure for the TE domain of FAS serves as the prototype for all other such FAS TEs, which is well conserved across species from insects to human.
Materials and Methods
Protein Expression, Purification, Activity, and Mutagenesis. The cDNA sequence of human FAS TE was cloned into ProEX vector (Invitrogen), incorporating TeV protease cleavable N-terminal hexahistidine-tag, and expressed in Escherichia coli BL21(DE3) cells. After purification using Talon Co2+ affinity chromatography (Clontech), the histidine tag was cleaved. TE was further purified with a UnoQ column (Pharmacia) by using a continuous gradient from 0 M NaCl to 1 M NaCl in 5% glycerol and 50 mM Tris (pH 8.0). The fractions containing purified untagged protein were collected, concentrated to ≈17 mg/ml, and stored. Enzymatic assays on TE were performed as described (20) by using palmitoyl-CoA as the substrate.
The selenomethionine variant of the TE domain was generated similar to the native protein by using media as described (22). Mutants were made by using the QuikChange site-directed mutagenesis kit (Stratagene), and the expression, purification, and activity assay procedures were similar to wild-type TE.
Crystallization and Structure Determination. Crytals were obtained by using the hanging drop method, with mother liquor containing 1.8 M (NH4)2SO4, and 100 mM Mes (pH 6.5). TE crystals were frozen in mother liquor containing 25% glycerol or 1 M LiSO4. Selenomethionine-substituted protein crystals were oxidized by dipping crystals for 10–15 s in cryo-protection solution containing 0.1% H2O2 to enhance multiwavelength anomalous dispersion (MAD) signals (variation of a procedure previously published in ref. 23) before being frozen. Crystal properties are shown in Table 1.
Table 1. Crystallographic structure analysis.
| Selenomethionine MAD
|
||||
|---|---|---|---|---|
| Native | Edge | Peak | Remote | |
| Data Collection | ||||
| Wavelength, Å | 0.96865 | 0.97930 | 0.97910 | 0.96390 |
| Resolution maximum,* Å | 2.6 (2.69-2.6) | 3.5 (3.69-3.5) | 3.5 (3.69-3.5) | 3.5 (3.69-3.5) |
| Unit cell dimensions,† Å | ||||
| a | 104.78 | 104.95 | 104.78 | 104.77 |
| c | 126.46 | 128.05 | 128.32 | 128.14 |
| Total reflections | 265,768 | 172,565 | 174,295 | 128,992 |
| Unique reflections* | 22,088 (2,215) | 9,539 (1,341) | 9,540 (1,357) | 9,129 (1,316) |
| Completeness,* % | 100 (98.5) | 100 (100) | 100 (100) | 98.9 (97.2) |
| 〈I〉/σ〈I〉* | 17.5 (6.3) | 7.4 (3.1) | 7.7 (2.9) | 8.4 (3.2) |
| Rsym*‡ | 4.8 (33.6) | 7.5 (22.0) | 7.8 (23.6) | 6.5 (21.1) |
| Phasing (50- to 3.5-Å resolution range) | ||||
| Figure of merit | 0.66 | |||
| Refinement statistics (50- to 2.6-Å resolution range) | ||||
| Rcryst/Rfree§ | 0.26/0.28 | |||
| rms deviation | ||||
| Bond length, Å | 0.0077 | |||
| Bond angle, ° | 1.2652 | |||
MAD, multiwavelength anomalous dispersion.
Values in parentheses are for outer resolution shell
P122 space group with two molecules in the asymmetric unit
Rsym = Σhkl|Ihkl - 〈Ihkl〉|/Σhkl Ihkl
R factor = Σhkl|Fo| - |Fc|/Σhkl |Fo| where |Fo| and |Fc| are the observed and calculated structure factor amplitudes for reflection hkl, applied to the work (Rcryst) and test (Rfree) (5% omitted from refinement) sets, respectively
The structure was determined by MAD technique. A three-wavelength dataset was collected from an oxidized crystal of selenomethionine-substituted protein at Advanced Light Source Berkeley HHMI beamline 8.2.1. Anomalous data were processed by using program mosflm (24) and merged by using scala, which is a part of the ccp4 Program Suite (25). Phases were calculated by using using cns (26). Eight of the 10 methionine sites corresponding to two monomers were found in the asymmetric unit, and phases were refined by using cns. The resultant map was solvent flattened by cns and yielded a traceable electron density map with defined side-chain density that allowed placement of the sequence by using program o (27). The structure was refined against a native dataset at 2.6-Å resolution (Table 1) collected at the X4A beamline at Brookhaven and processed by using denzo and merged by using scalepack (28).
Groove in the TE Structure. The program proshape (http://csb.stanford.edu/koehl/ProShape/) was used to identify and characterize the presence of a groove in the TE crystal structure.
Illustrations. All of the illustrations were made by using either dino (www.dino3d.org) or molscript (29) and rendered with raster3d (30).
Results and Discussion
Overall TE Domain Structure. Although the structure of the isolated TE domain that was solved represents two independent molecules (1 and 2) in the asymmetric unit (Table 1), the domain was purified and fully active as a monomer (Z.G. and B.C., unpublished data). Native human FAS is arranged as a head-to-tail antiparallel homodimer (1) as visualized recently in the electron cryo-microscopy (cryo-EM) maps (31). This arrangement would place the two TE domains of the FAS monomers at opposite ends from each other, lending no functional relevance to the formation of a dimer. For these reasons, the dimer is an artifact of crystallization. The structure of molecule 1 is considered hereafter as a representative of the structure.
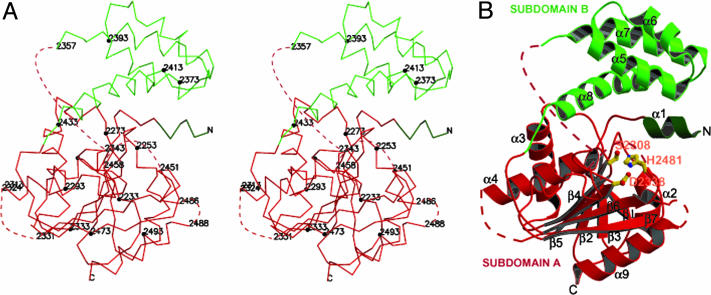
The TE domain of human FAS comprises two subdomains (A and B) (Fig. 1). The larger subdomain A (≈23 kDa) is made up of two noncontiguous segments from the amino- (or N-) and carboxyl- (or C-) terminal ends (Figs. 1, 2, 3). The intervening segment is consigned to the smaller (≈9-kDa) subdomain B. Subdomain A has an overall α/β fold, whereas subdomain B has an all α-helical motif. The entire structure is made up of nine α-helices and eight β strands (Figs. 1 and 2). Most of the entire TE domain structure could be fitted into the electron density, except for the three segments and a glycine residue shown in Figs. 1, 2, 3, with missing or weak density, indicating their high mobility. All of the disordered segments are in solvent-exposed regions and nowhere close to being involved in crystal contacts.
Fig. 1.
Crystal structure of the TE domain. (A) Stereoview of the Cα trace. Every 20 residues (filled circles) and residues at ends of disordered loops are labeled. Dotted lines represent segments that are disordered (also shown in Figs. 2B and 3). Subdomain A is in red and dark green. Subdomain B is in light green. (B) Ribbon diagram of the TE monomer. The catalytic residues (Ser-2308, His-2481, and Asp-2338) are in ball-and-stick representation and are labeled in orange.
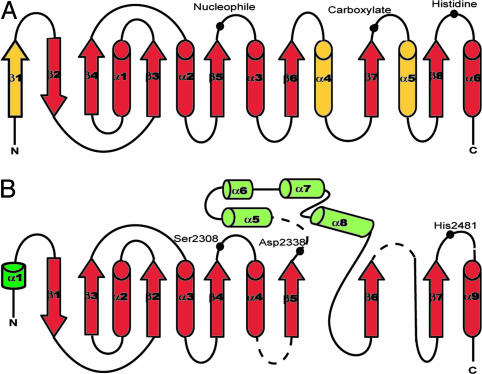
Fig. 2.
Schematic diagram of α/β hydrolase fold. Helices are represented as cylinders, strands as arrows, and random coils as lines. (A) Canonical/core hydrolase fold along with positions of catalytic triad (37). Differences between the core and TE structure are in yellow, whereas similarities are in red. (B) Modified hydrolase fold of TE structure. Subdomain A is in red and dark green. Subdomain B is in light green. Position of the catalytic triad is shown as filled black circles. The core has an extra strand in place of helix α1 in TE structure. Thus, strand β2 in the core corresponds to strand β1 in TE fold, and helix α1 in core corresponds to helix α2 in TE structure. Also helix α4 in the core is replaced by the all-helical subdomain B. Helix α5 in the core also is missing in the corresponding TE structure.
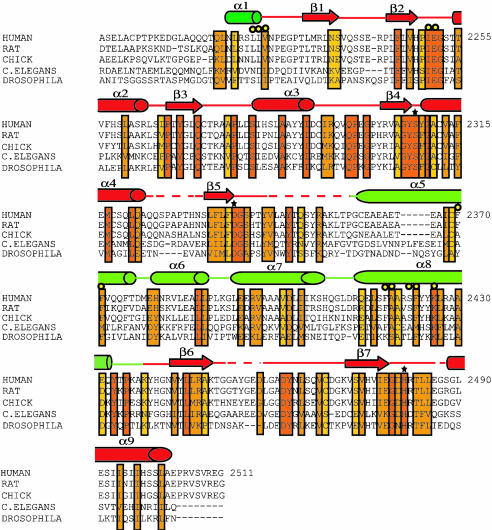
Fig. 3.
Sequence alignment of FAS TE domains of different species. The alignment was carried out by using blast (www.ncbi.nlm.nih.gov/blast) and clustalw (www.ebi.ac.uk/clustalW) (44). The domain originally was described in ref. 20. Conserved residues are boxed. Invariants to partially conserved residues are colored from red to yellow, respectively. The catalytic residues are marked with a star. Yellow circles represent residues forming the candidate palmitoyl binding groove. The elements of the secondary structure from the crystal structure are shown above the amino acid sequence, with helices represented as cylinders, strands as arrows, and random coils as lines. Subdomain A is in red and dark green; subdomain B is in light green.
Subdomains A and B. As shown in Fig. 2, subdomain A of the TE structure does not conform strictly to the α/β hydrolase fold. Also, the position of the catalytic carboxylate residue in this subdomain is atypical (see below). The canonical hydrolase fold is an eight-stranded, mostly parallel β-sheet structure flanked on both sides by α-helices. In comparison, the TE subdomain A lacks the N-terminal β1 strand and the equivalent of helices α4 and α5. In lieu of helix α4, the second subdomain (subdomain B) is inserted between the strands β5 and β6.
A dali search (32) indicated two close structural homologs of the TE domain, the surfactin synthetase (PDB 1jmk) (33) and the 6-deoxyerythronolide B synthase TE (PDB 1kez) (34) with Z scores of 20.2 and 15.7, respectively. However, both surfactin synthetase and DEBS TE were found to overlap with only the subdomain A of the TE structure over 161 and 163 residues, respectively.
The all-α-helical subdomain B is unique to human FAS TE. It is inserted in between strands β5 and β6 (Figs. 1, 2, 3). The subdomain is made up of four α-helices (α5–α8) and is located at an angle to subdomain A, forming either an open or closed cap-like structure. The fold of this subdomain is different from any other TE determined to date as judged by dali. The nearest structural homologs of subdomain B are a domain of phosphonoacetaldehyde hydrolase (35) and β phosphoglucomutase (36) but with significantly low Z scores of ≈6. Both enzymes carry out reactions involving a phosphoryl group. Because TE does not catalyze these types of reactions, the structural homology is considered to have no functional relevance.
Catalytic Triad. One of the important features revealed by TE domain crystal structure analysis is the precise geometry of the catalytic triad of side chain residues, which is present in enzymes with the α/β hydrolase fold. The triad in the hydrolases consists of a highly conserved nucleophile (serine, cysteine, or aspartic acid residue), an acidic residue, and an invariant histidine residue (37). Although two participants of the triad, Ser-2308 and His-2481, had been previously identified in the homologous chicken FAS TE by site-directed mutagenesis experiments (20), only now that the structure of TE has been determined has the third player, Asp-2338, been uncovered. The identity of Asp-2338 is based on its location, where it is the only acidic residue in very close proximity to Ser-2308 and His-2481 (Figs. 1 and 4). The Asp-2338 catalytic residue, along with Ser-2308 and His-2481, is invariant in FASs, emphasizing its importance in the catalytic triad (Fig. 3). To confirm the importance of Asp-2338, its mutation to an Ala residue essentially eliminated TE activity (<0.7% of wild-type activity).
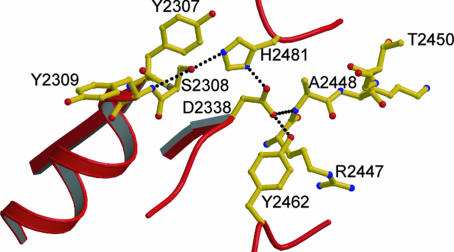
Fig. 4.
Hydrogen bonding network among the various catalytic residues. Dotted lines denote hydrogen bonds connecting the corresponding residues. Hydrogen bond distances range from 2.4 to 3.2 Å.
Unexpectedly, the position of Asp-2338 residue differs significantly from that usually found in the α/β hydrolase fold, which is near the C-terminal end of strand β6 (Fig. 2). The Asp-2338 is instead found at the C-terminal end of strand β5 (Figs. 2 and 3). This unconventional positioning of the acid group of the catalytic triad has been observed previously in only two other enzymes, the human pancreatic lipase (38, 39) and the TE domain of surfactin synthetase (33). The N terminus of the largest disordered loop (residues 2343–2356) is within 6 aa of the Asp catalytic residue and could become ordered on substrate binding and contribute toward binding of the substrate.
In contrast to Asp-2338, the catalytic residues Ser-2308 and His-2481 in FAS TE occupy the general positions in the canonical hydrolase fold. Moreover, Ser-2308 is located in the middle of the signature sequence Gly-X-Ser-X-Gly, where X stands for any amino acid residue, in a very sharp turn called the “nucleophile elbow” (37) between the end of strand β4 and the beginning of helix α4. The tightness of the strand–turn–helix motif induces the Ser residue to adopt energetically unfavorable main chain torsion angles (ϕ = 58° and Ψ = -108°), which is the only residue with a side chain in disallowed region. Interestingly, the tyrosine residue (Tyr-2307) preceding the Ser-2308 is invariant whereas the residue following the serine is always aromatic in TEs ranging from insects to humans (Fig. 3).
The three catalytic residues of the human FAS TE (Ser-2308, His-2481, and Asp-2338) are linked to each other and the neighboring residues by an extensive hydrogen bonding network (Fig. 4). The hydroxyl group of Ser-2308 is involved in a cooperative hydrogen bond, accepting from the adjacent amide backbone of Tyr-2309 and donating to Nε2 of His-2481. This, in turn, would facilitate the activation of Ser-2308 as the nucleophile and help stabilize the oxyanion tetrahedral intermediate expected to form during TE catalytic reaction as seen in serine hydrolases. The His-2481 residue (Nε2) in turn is hydrogen bonded to Asp-2338 (Oδ2). The Asp-2338 (Oδ1 atom) makes additional hydrogen bonds with both the backbone amide of Ala-2448 and the hydroxyl group of Tyr-2462. Tyr-2462 is completely invariant, whereas Ala-2448 is partially conserved from insects to mammals (Fig. 3). Hence the interaction between these residues and the catalytic aspartate residue seems to be important to keep the Asp-2338 fixed in its particular position, further signifying the important role played by this residue. The extensive hydrogen bonding network at the catalytic site keeps the enzyme primed for action by orienting and placing the catalytic residues at their required positions, much like those in serine hydrolases (40).
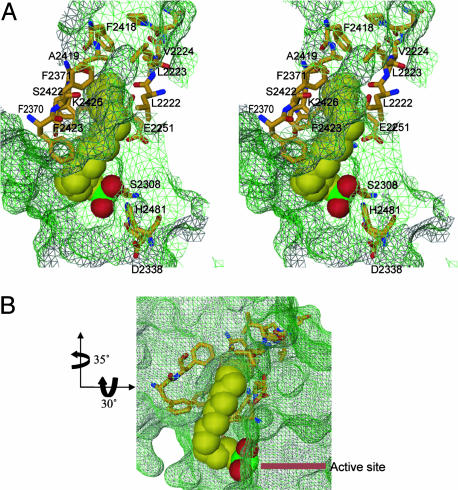
Palmitoyl Chain Binding Site and Chain Length Specificity. To gain an understanding of the mechanism by which the TE domain of FAS regulates chain length specificity, we analyzed the TE structure for the presence of a groove close to the catalytic center that could accommodate long fatty acyl chain substrates. The presence of only one groove is apparent, which is located at the interface between subdomains A and B (Fig. 5). Analysis using proshape (see Materials and Methods) confirmed the presence of the groove with a volume of 186.9 Å3 and a surface area of 140 Å2, the distal end of which forms a pocket (Fig. 5). The groove is close to the amino terminus of the TE domain, which is linked to the ACP domain of FAS that holds the growing acyl chains for scanning and release by the TE domain after reaching the optimum 16 carbon fatty acid chain length. Most of the residues that line the groove originate from the ampiphilic helix α8 of subdomain B. Other residues contributing to it are from helix α5 of subdomain B, the N-terminal helix α1 of subdomain A, and the loop between β2 and α2 of subdomain A. The residues lining the groove, which are mostly hydrophobic in nature, consist of Phe-2418, Ala-2419, Ser-2422, Phe-2423, and Lys-2426 from helix α8, Phe-2370 and Phe-2371 from helix α5, Leu-2222, Leu-2223, and Val-2224 from helix α1, and Ile-2250 and Glu-2251 from the loop between β2 and α2 (Fig. 5A). Of these residues, Ile-2250, Glu-2251, and Lys-2426 are invariant across species, whereas Val-2224, Phe-2371, Ala-2419, and Phe-2423 are mostly conserved (Fig. 3). The rest of the residues are completely conserved in mammals (Fig. 3). All of the above observations taken together suggest the interface between the two subdomains as an excellent fatty acid chain binding region. Two experiments were undertaken to provide credence to the suggestion and to obtain insight into fatty acyl chain selectivity.
Fig. 5.
Lipid binding site. (A) Stereoview of the candidate palmitoyl binding groove for the first 11–12 carbons and the pocket for the last 5–4 carbons. The hexadecyl sulfonyl inhibitor is represented as a Corey–Pauling–Koltun space-filling model (C, yellow; O, red; S, green). Side chains lining the groove are shown as ball-and-stick figures (C, yellow; O, red; N, blue). The side chains making up the active site are also shown as ball-and-stick figures with the color scheme similar to that of the residues. The groove surface map is shown in green wire mesh. The residues making up the groove are labeled except for Ile-2250, which cannot be seen because it falls below the inhibitor. The initial 11–12 carbon atoms from the sulfonyl group are solvent-exposed with the 11th and 12th carbon atoms at the mouth of the pocket. (B) A different view of the groove and pocket. The arrows denote the rotations in going from the orientation of the view in A to B. This view shows that most of the fatty acyl chain of hexadecyl inhibitor is exposed to the solvent.
First, we demonstrated by site-directed mutagenesis that replacements of several of the residues lining the groove and pocket of the TE domain reduced the TE activity, although to differing extents (Table 2).
Table 2. Mutations in the candidate binding groove decrease activity of TE.
| Mutations | Residue location | Relative specific activity,* % |
|---|---|---|
| None | - | 100 ± 1.69 |
| 12250W | Subdomain A, loop between β2 and α2 | 3.1 ± 0.40 |
| F2371W | Subdomain A, α5 | 79.4 ± 5.24 |
| A2419L | Subdomain B, α8 | 3.4 ± 0.53 |
| A2419M | Subdomain B, α8 | 8.1 ± 0.40 |
| S2422A | Subdomain B, α8 | 93 ± 0.83 |
| F2423A | Subdomain B, α8 | 23.2 ± 1.60 |
| F2423W | Subdomain B, α8 | 66.4 ± 5.0 |
| K2426A | Subdomain B, α8 | 0.8 ± 0.02 |
The specific activity (%) of different TE constructs was compared with wild type as measured using the activity assay described in Materials and Methods. Each assay was carried out in duplicate
Second, we were able to model the binding of a C16 palmitoyl chain-containing inhibitor hexadecyl sulfonyl f luoride (HDSF) into the groove. The modeling was guided by the data from the crystal structure of palmitoyl protein TE 1 (PPT1) with covalently bound hexadecyl sulfonate (HDS) (PDB ID code 1exw; ref. 41) and augmented by energy minimization in cns. Like TE, the catalytic site of PPT1 contains a conserved catalytic triad of serine, histidine, and aspartate. The PPT1–HDS complex structure showed the formation of the covalent bond between the sulfur atom of HDS and oxygen atom of the catalytic serine and the presence of the fatty acyl chain in a long mostly hydrophobic surface groove of the protein (41). The modeled covalently bound HDS in the TE structure (shown in Fig. 5) revealed several features of ligand binding with important functional and biochemical implications. Fitting of the HDS caused no major rearrangement of the residues in binding site. The palmitoyl chain makes a sharp bend with respect to the sulfonate group in a similar manner to that seen in the bound PPT1 structure. The 16 carbon atom palmitoyl chain fits well within the groove with the first ≈12 carbon atoms exposed to the solvent and the remaining ≈4 carbon atoms inserted and held in place in the distal pocket. This fit is fully consistent with the observation that mammalian FAS TE catalyses the formation of palmitic acid as its major product (20). The presence of the last ≈4 carbon atoms in the pocket helps tether the fatty acid chain in place for the hydrolysis of the palmitoyl acyl substrate. The nature of the binding site, which combines an exposed groove with a closed pocket, is unique and designed for the specificity of this TE. The finding that the TE domain of mammalian FASs shows no significant activity toward fatty acyl chain lengths shorter than 14 carbon atoms (20) can be explained by nonproductive binding due to the lack of tethering and the resultant greater mobility of the exposed shorter chains. The last ≈4 carbon atoms of the palmitoyl chain partially occupy the distal pocket, leaving enough space to accommodate only 2 additional carbon atoms. This result is consistent with the dramatic loss in activity seen with model substrates longer than 18 carbon atoms (20). It is unclear whether the last ≈4 and ≈6 carbon atoms of the C16 and C18 fatty acyl chains, respectively, will be inserted in a preformed pocket between the two subdomains as seen in the structure of the unbound form, or will be bound initially to an open form of the domain that then subsequently undergoes a hinge-bending motion between the subdomains to enclose the terminal carbon atoms of the fatty acyl chains and create the productive configuration of the active site region.
Considerable efforts were made to crystallize TE with various analogues of long-chain fatty acids (e.g., palmitoyl-CoA, myristoyl-CoA, stearoyl-CoA, palmitic acid, hexadecyl sulfonyl fluoride, and palmitic acid), but no long lived stable complex structures were found. Most of the complexes, as shown by independent enzyme activity or radioactive binding assays in solution, dissociated within 5 min to 1 h, making them unsuitable for crystallization or crystal soaking trials.
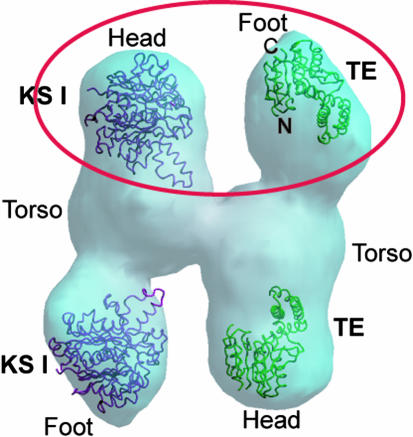
Cryo-EM Fit of TE Structure. Although the 20-Å cryo-EM reconstruction structure of human FAS dimer has been described (31), there was no indication of the N- to C-terminal polarity of the FAS subunit except for a previous indication from antibody labeling that the FAS TE domain was located at one of the ends of the entire structure (42). We attempted a preliminary assessment of the polarity by manually docking the smaller (32-kDa) human FAS TE domain structure, which occupies the C-terminal end of FAS subunit, and the larger (42 kDa) E. coli β-ketoacyl synthase I (or KSI) crystal structure (PDB ID code 1ek4) (43), which is a close homolog of the N-terminal KS domain of mammalian FAS, into each end of the monomer unit designated head and foot of the cryo-EM mass density (Fig. 6). The TE structure fits well into the the tip of the substructure designated foot, whereas the KSI fits best into the tip of the substructure designated head (Fig. 6). In the FAS dimer the ACP domain interacts with both the TE domain of the same monomer as well as the ketoacyl synthase domain in the opposite monomer leading to the formation of two active centers at the two ends of the dimer. The orientation of the TE domain and the KS domain homolog for the best fit at the foot and head, respectively, accommodated for both space and functional interaction of the ACP molecule with both proteins. When the structures were switched the fitting was much poorer, with some parts of KSI falling outside the density and a larger volume of unaccounted density in the TE fit (Fig. 6).
Fig. 6.
Preliminary fit of TE (green backbone trace) and KSI (maroon trace) in the 20-Å resolution cryo-EM mass density. The substructures seen from the density are designated Head, Torso, and Foot. The TE trace fits best in the substructure designated Foot, whereas the KSI backbone trace fit is best in substructure designated Head. The best fit for both KSI and TE is circled in red. The backbone traces of TE and KSI at the bottom of the mass density show less satisfactory fits in the head and foot, respectively. Based on prior proteolytic digestion experiments of intact FAS, three domains were identified: domain I, which comprises the KS-AT/MT-DH enzyme centers and the linker region; domain II, which comprises the ER-KR-ACP region; and domain III, which is assigned to the TE (1, 2). The Head and Torso substructures, which are essentially coalesced, could correspond to domain I and parts of domain II, and the Foot could represent the rest of domain II and domain III.
Conclusions. The principal conclusions that can be drawn from the crystal structure of the TE domain of human FAS are as follows. (i) The TE is composed of two subdomains that differ in size and folding motifs. (ii) The larger A subdomain exhibits an α/β motif, whereas the smaller B subdomain is all helical. (iii) Subdomain A contributes the catalytic triad of Ser, Asp, and His residues. (iv) The long groove with a distal pocket between subdomain B and parts of subdomain A constitutes the fatty acyl chain binding site. The geometry and nature of this site are consistent with the high specificity of the TE toward C16 to C18 fatty acyl substrates. The groove acts like a ruler that measures the correct substrate length. (v) The preliminary fit of the TE into the intact human FAS cryo-EM structure suggests a plausible N to C terminus polarity of the subunits. However, a higher resolution cryo-EM map is needed for further verification of the fit and an in-depth analysis of FAS function.
Acknowledgments
We thank the staff at Berkeley Beamline 8.2.1 and National Synchrotron Light Source Brookhaven X4A for their assistance with x-ray data collection. We also thank Drs. B. Bowman, N. Vyas, A. Nickitenko, C. M. Moure, Y. Mao, and J. L. Fallon for their technical assistance and helpful discussions. This work was supported by National Institutes of Health Grant R01GM068826 and Welch Foundation Grant Q-0581 (to F.A.Q.).
Author contributions: B.C., S.S.C., S.J.W., and F.A.Q. designed research; B.C., Z.G., and S.S.C. performed research; B.C., Z.G., and F.A.Q. analyzed data; and B.C., S.S.C., and F.A.Q. wrote the paper.
Abbreviations: ACP, acyl carrier protein; FAS, fatty acid synthase; TE, thioesterase; KSI, β-ketoacyl synthase I; HDS, hexadecyl sulfonate; Cryo-EM, electron cryo-microscopy.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 1XKT).
References
- 1.Wakil, S. J. (1989) Biochemistry 28, 4523-4530. [DOI] [PubMed] [Google Scholar]
- 2.Smith, S. (1994) FASEB J. 8, 1248-1259. [PubMed] [Google Scholar]
- 3.Chirala, S. S., Chang, H., Matzuk, M., Abu-Elheiga, L., Mao, J., Mahon, K., Finegold, M. & Wakil, S. J. (2003) Proc. Natl. Acad. Sci. USA 100, 6358-6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Must, A., Spadano, J., Coakley, E. H., Field, A. E., Colditz, G. & Dietz, W. H. (1999) J. Am. Med. Assoc. 282, 1523-1529. [DOI] [PubMed] [Google Scholar]
- 5.Alo, P. L., Visca, P., Marci, A., Mangoni, A., Botti, C. & Di Tondo, U. (1996) Cancer 77, 474-482. [DOI] [PubMed] [Google Scholar]
- 6.Bobrow, L. G., Happerfield, L. C., Gregory, W. M., Springall, R. D. & Millis, R. R. (1994) Semin. Diagn. Pathol. 11, 199-207. [PubMed] [Google Scholar]
- 7.Alo, P. L., Visca, P., Trombetta, G., Mangoni, A., Lenti, L., Monaco, S., Botti, C., Serpieri, D. E. & Di Tondo, U. (1999) Tumori 85, 35-40. [DOI] [PubMed] [Google Scholar]
- 8.Jensen, V., Ladekarl, M., Holm-Nielsen, P., Melsen, F. & Soerensen, F. B. (1995) J. Pathol. 176, 343-352. [DOI] [PubMed] [Google Scholar]
- 9.Epstein, J. I., Carmichael, M. & Partin, A. W. (1995) Urology 45, 81-86. [DOI] [PubMed] [Google Scholar]
- 10.Shurbaji, M. S., Kalbfleisch, J. H. & Thurmond, T. S. (1996) Hum. Pathol. 27, 917-921. [DOI] [PubMed] [Google Scholar]
- 11.Rashid, A., Pizer, E. S., Moga, M., Milgraum, L. Z., Zahurak, M., Pasternack, G. R., Kuhajda, F. P. & Hamilton, S. R. (1997) Am. J. Pathol. 150, 201-208. [PMC free article] [PubMed] [Google Scholar]
- 12.Pizer, E. S., Chrest, F. J., DiGiuseppe, J. A. & Han, W. F. (1998) Cancer Res. 58, 4611-4615. [PubMed] [Google Scholar]
- 13.Gansler, T. S., Hardman, W., 3rd, Hunt, D. A., Schaffel, S. & Hennigar, R. A. (1997) Hum. Pathol. 28, 686-692. [DOI] [PubMed] [Google Scholar]
- 14.Sekiguchi, M., Shiroko, Y., Arai, T., Kishino, T., Sugawara, I., Kusakabe, T., Suzuki, T., Yamashita, T., Obara, T., Ito, K. & Hasumi, K. (2001) Biomed. Pharmacother. 55, 466-474. [DOI] [PubMed] [Google Scholar]
- 15.Innocenzi, D., Alo, P. L., Balzani, A., Sebastiani, V., Silipo, V., La Torre, G., Ricciardi, G., Bosman, C. & Calvieri, S. (2003) J. Cutan. Pathol. 30, 23-28. [DOI] [PubMed] [Google Scholar]
- 16.Kridel, S. J., Axelrod, F., Rozenkrantz, N. & Smith, J. W. (2004) Cancer Res. 64, 2070-2075. [DOI] [PubMed] [Google Scholar]
- 17.Pazirandeh, M., Chirala, S. S. & Wakil, S. J. (1991) J. Biol. Chem. 266, 20946-20952. [PubMed] [Google Scholar]
- 18.Mattick, J. S., Nickless, J., Mizugaki, M., Yang, C. Y., Uchiyama, S. & Wakil, S. J. (1983) J. Biol. Chem. 258, 15300-15304. [PubMed] [Google Scholar]
- 19.Lin, C. Y. & Smith, S. (1978) J. Biol. Chem. 253, 1954-1962. [PubMed] [Google Scholar]
- 20.Pazirandeh, M., Chirala, S. S., Huang, W.-Y. & Wakil, S. J. (1989) J. Biol. Chem. 264, 18195-18201. [PubMed] [Google Scholar]
- 21.Singh, N., Wakil, S. J. & Stoops, J. K. (1984) J. Biol. Chem. 259, 3605-3611. [PubMed] [Google Scholar]
- 22.Van Duyne, G. D., Standaert, R. F., Karplus, P. A., Schreiber, S. L. & Clardy, J. (1993) J. Mol. Biol. 229, 105-124. [DOI] [PubMed] [Google Scholar]
- 23.Sharff, A. J., Koronakis, E., Luisi, B. & Koronakis, V. (2000) Acta Crystallogr. D. Biol. Crystallogr. 56, 785-788. [DOI] [PubMed] [Google Scholar]
- 24.Leslie, A. G. W. (1991) in Crystallographic Computing 5: From Chemistry to Biology, eds. Moras, D., Podjanry, A. D. & Thierry, J. C. (Oxford Univ. Press, Oxford, U.K.), pp. 27-38.
- 25.Collaborative Computational Project Number 4 (1994) Acta Crystallogr. D 50, 760-763.15299374 [Google Scholar]
- 26.Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 27.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110-119. [DOI] [PubMed] [Google Scholar]
- 28.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307-326. [DOI] [PubMed] [Google Scholar]
- 29.Kraulis, P. J. (1991) J. Appl. Crystallogr. 24, 946-950. [Google Scholar]
- 30.Merritt, E. A. & Bacon, D. J. (1997) Methods Enzymol. 277, 505-524. [DOI] [PubMed] [Google Scholar]
- 31.Brink, J., Ludtke, S. J., Yang, C. Y., Gu, Z. W., Wakil, S. J. & Chiu, W. (2002) Proc. Natl. Acad. Sci. USA 99, 138-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holm, L. & Sander, C. (1995) Trends Biochem. Sci. 20, 478-480. [DOI] [PubMed] [Google Scholar]
- 33.Bruner, S. D., Weber, T., Kohli, R. M., Schwarzer, D., Marahiel, M. A., Walsh, C. T. & Stubbs, M. T. (2002) Structure 10, 301-310. [DOI] [PubMed] [Google Scholar]
- 34.Tsai, S. C., Miercke, L. J., Krucinski, J., Gokhale, R., Chen, J. C., Foster, P. G., Cane, D. E., Khosla, C. & Stroud, R. M. (2001) Proc. Natl. Acad. Sci. USA 98, 14808-14813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morais, M. C., Zhang, W., Baker, A. S., Zhang, G., Dunaway-Mariano, D. & Allen, K. N. (2000) Biochemistry 39, 10385-10396. [DOI] [PubMed] [Google Scholar]
- 36.Lahiri, S. D., Zhang, G., Dunaway-Mariano, D. & Allen, K. N. (2003) Science 299, 2067-2071. [DOI] [PubMed] [Google Scholar]
- 37.Nardini, M. & Dijkstra, B. W. (1999) Curr. Opin. Struct. Biol. 9, 732-737. [DOI] [PubMed] [Google Scholar]
- 38.Schrag, J. D., Winkler, F. K. & Cygler, M. (1992) J. Biol. Chem. 267, 4300-4303. [PubMed] [Google Scholar]
- 39.Winkler, F. K., D'Arcy, A. & Hunziker, W. (1990) Nature 343, 771-774. [DOI] [PubMed] [Google Scholar]
- 40.Devedjiev, Y., Dauter, Z., Kuznetsov, S. R., Jones, T. L. & Derewenda, Z. S. (2000) Structure Fold. Des. 8, 1137-1146. [DOI] [PubMed] [Google Scholar]
- 41.Das, A. K., Bellizzi, J. J., 3rd, Tandel, S., Biehl, E., Clardy, J. & Hofmann, S. L. (2000) J. Biol. Chem. 275, 23847-23851. [DOI] [PubMed] [Google Scholar]
- 42.Kitamoto, T., Nishigai, M., Sasaki, T. & Ikai, A. (1988) J. Mol. Biol. 203, 183-195. [DOI] [PubMed] [Google Scholar]
- 43.Olsen, J. G., Kadziola, A., von Wettstein-Knowles, P., Siggaard-Andersen, M., Lindquist, Y. & Larsen, S. (1999) FEBS Lett. 460, 46-52. [DOI] [PubMed] [Google Scholar]
- 44.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]