Abstract
Recent findings have drawn attention to the role of membrane traffic in the signaling of vascular endothelial growth factor (VEGF). The significance of this development stems from the pivotal function of VEGF in vasculogenesis and angiogenesis. The outline of the regulation of VEGF receptor (VEGFR) signaling by membrane traffic is similar to that of the epidermal growth factor receptor (EGFR), a prototype of the intertwining between membrane traffic and signaling. There are, however, unique features in VEGFR signaling that are conferred in part by the involvement of the co-receptor neuropilin (Nrp). Nrp1 and VEGFR2 are integrated into membrane traffic through the adaptor protein synectin, which recruits myosin VI, a molecular motor that drives inward trafficking [17,21,64]. The recent detection of only mild vascular defects in a knockin mouse model that expresses Nrp1 lacking a cytoplasmic domain [104], questions the co-receptor’s role in VEGF signaling and membrane traffic. The regulation of endocytosis by ephrin-B2 is another feature unique to VEGR2/3 [18,19], but it awaits a mechanistic explanation. Current models do not fully explain how membrane traffic bridges between VEGFR and the downstream effectors that produce its functional outcome, such as cell migration. VEGF-A appears to accomplish this task in part by recruiting endocytic vesicles carrying RhoA to internalized active VEGFR2 [58].
Keywords: Angiogenesis, Endocytosis, Membrane traffic, Neuropilin-1, Rho GTPase, VEGFR2
1. Introduction
1.1. Overview: regulation of receptor tyrosine kinase (RTK) signaling by membrane traffic
Membrane traffic has been recognized for some time now as an essential component of RTK signaling in general [1,2], and particularly in the vascular system, [3,4]. That recognition represents a conceptual departure from the previously prevailing view where the sole effect of endocytosis was thought to be the attenuation of cell surface receptors, including that of RTKs [5,6]. To date, endocytosis and membrane traffic have been shown to regulate the signaling pathways of multiple RTKs and their co-receptors. These include EGFR [6–8], the platelet derived growth factor receptor (PDGFR) [9–11], the fibroblast growth factor receptor (FGFR) [12,13] and its co-receptor syndecan-4 [14], VEGFR [15–19] and its co-receptor Nrp [20–22], the insulin and insulin-like growth factor receptors [23–26], the hepatocyte growth factor Met [27], the Eph receptors [28,29], and TrkA, the nerve growth factor receptor [30]. All the RTKs that are internalized after binding their soluble ligands undergo primarily clathrin-mediated endocytosis, and their trafficking is regulated by the same adaptor proteins and small GTPases [3,4]. At the same time, the molecular pathways that intertwine membrane traffic and their downstream phosphorylation cascades appear to differ among RTK types, possibly reflecting the differences between their physiological functions. The mechanism and functional role of membrane traffic is best understood in the signaling pathway of EGFR, and to a lesser degree in VEGFR signaling [31,32]. This review focuses solely on the endocytic pathways that are activated downstream of VEGF in endothelial cells (ECs), in order to discuss the state of knowledge and the impact of this relatively recent aspect of VEGF signaling on the understanding of angiogenesis.
1.2. EGFR as a prototype for the regulation of RTK signaling by membrane traffic
Though the reciprocal relation between EGFR signaling and membrane traffic has been described in a detailed and up-to-date manner elsewhere [31,33], it is worth recapitulating its main components (Fig. 1) because it is known in more detail than that of other RTKs. EGFR undergoes mainly clathrin-dependent endocytosis that is triggered by the binding of its ligand [34]. EGFR dimerization and tyrosine phosphorylation were required for its recruitment to coated pits and clathrin-dependent endocytosis [35]. In the absence of tyrosine phosphorylation, the rates of both coated-pit recruitment and endocytosis of EGFR were reduced [36]. The mechanism that regulates endocytosis of tyrosine-phosphorylated EGFR is complex and still not fully characterized. It appears that there are several redundant and possibly interdependent mechanisms, which involve: (a) EGFR binding to the clathrin endocytosis protein complex AP2, and to the phospho-tyrosine binding adaptor protein growth factor receptor-bound protein 2 (Grb2); (b) ubiquitination of 15 lysines in the EGFR kinase domain by the E3 ubiquitin ligase Casitas B-lineage lymphoma (Cbl), and of 6 other lysines in the carboxy-terminus region; (c) acetylation of 3 of the latter lysines [37]. Knocking down Grb2, a phospho-tyrosine binding protein that interacts with several RTKs, including VEGFR2 [38], had the largest inhibitory effect on EGFR endocytosis. The inhibition resulted from loss of the recruitment of Cbl to EGFR [39], and loss of binding of the Src homology (SH)-3 domain of Grb2 to an unidentified protein(s) containing poly-proline motif(s), possibly subunits of the AP2 complex [40]. The latter was required for the recruitment of EGFR to clathrin-coated pits [41]. Ubiquitination is required for the recruitment of EGFR to clathrin via the adaptor protein epsin-1 [42]. The role of lysine acetylation is not clear – it could have a positive effect, or it could inhibit EGFR endocytosis by competing with the ubiquitination of these lysines.
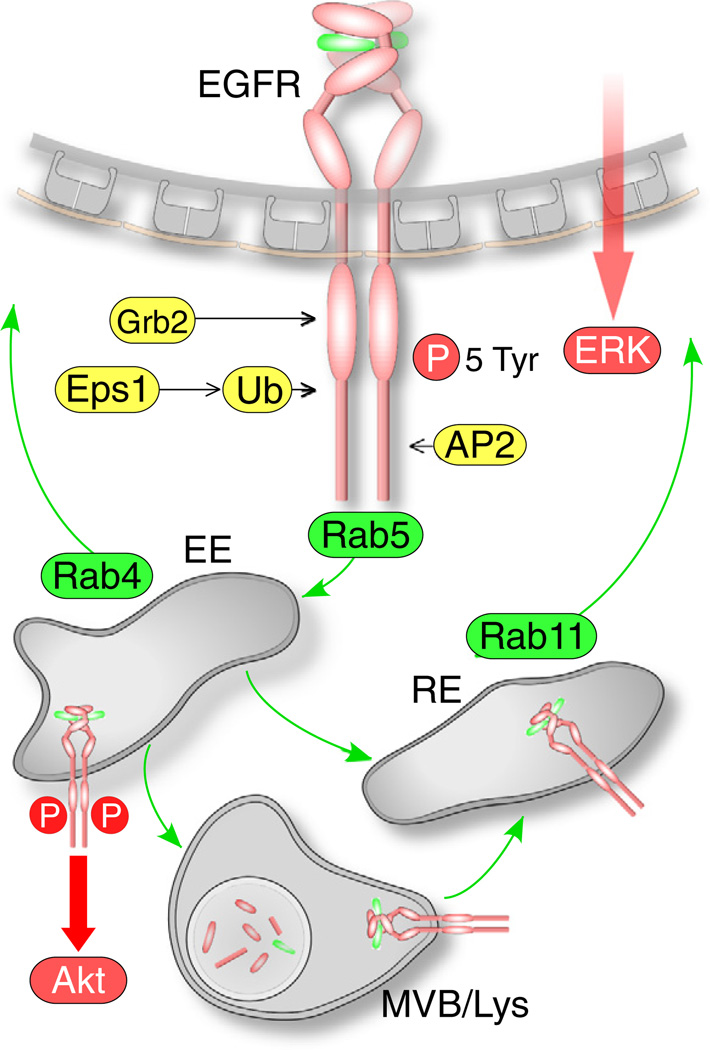
Fig. 1.
Scheme of membrane traffic and signaling of EGFR. Recruitment to clathrin-coated pits requires autophosphorylation (on 5 Tyr residues), association with of Grb2, and the binding of the AP2 complex to Tyr947, Lys1010, and Lys1011. The Cbl ligase ubiquinates (Ub) 21 lysines located in the cytoplasmic domain of the EGFR, which are required for internalization, in part by recruiting the clathrin adaptor protein epsin-1 (Eps1). Phosphorylated EGFR traffics via Rab5 to early endosomes (EE), from where it is recycled to the plasma membrane via Rab4, or to recycling endosomes (RE) from where is recycled to plasma membrane, or to multi-vesicular bodies (MVB). EGFR located in the limiting membrane of the MVB is recycled to the plasma membrane, whereas the EGFR pool in internal vesicles is degraded when these vesicles fuse with the lysosome. Akt activation requires endocytosis and proceeds from EGFR in early endosomes, whereas ERK activation is not endocytosis-dependent.
Internalized EGFR follows the canonical clathrin-dependent endocytic route. EGFR trafficked to early endosomes located in the cell periphery, where it remained dimerized, bound to EGF [43], and phosphorylated [44]. From early endosomes, EGFR either recycled rapidly to the plasma membrane [45], or remained in the same endocytic compartment during its maturation into multi-vesicular bodies (MVB). EGFR in MVBs was partitioned into two populations. The EGFR population on the limiting MVB membrane underwent slow, probably Rab11-dependent recycling that occurred within 15–20 min after the initiation of endocytosis [46]. Another population was located in internal MVB vesicles that fused with lysosomal vesicles, where EGFR was degraded [47]. EGFR regulates concomitantly two major signaling pathways, the phosphoinositide 3-kinase (PI3K)/Akt pathway, and the extracellular signal-regulated kinase (ERK)/mitogen-activated protein kinase (MAPK) pathway [39]. Despite a long-standing convention that optimal EGF-triggered ERK/ MAPK signaling requires EGFR endocytosis, whereas PI3K/Akt signaling does not (reviewed in Ref. [39]), recent studies reversed this view, showing that Akt phosphorylation was severely reduced in cells expressing an internalization-deficient EGFR mutant, but ERK1/2 phosphorylation level was increased [37]. The discrepancy has not been explained. It may arise from the difference between the approaches used to inhibit endocytosis. The studies in the first group used a dominant-negative dynamin mutant that inhibited all clathrin-dependent endocytosis, whereas in the study of Goh et al. only the endocytosis of EGFR was inhibited by mutating ubiquitin and AP2 binding motifs in its cytoplasmic domain [37].
2. Endocytosis and trafficking of VEGFR
The evolving understanding of VEGFR membrane traffic appears to conform in some respects with that of EGFR, though the understanding of VEGFR2 membrane traffic has not reached the level of detail at which EGFR endocytosis is known. Some of the earliest indications for the functional significance of VEGFR2 trafficking came from studies on guided cell migration in Drosophila [15]. The spatial localization of PVR, an ortholog of vertebrate PDGFR and VEGFR, in border cells of the Drosophila embryo, and the capability of the receptor to guide cell migration depended on proteins known to regulate endocytosis. Unlike EGFR, VEGFR signals in cooperation with the Nrp non-catalytic co-receptor, and therefore the membrane traffic of both receptors is reviewed. Recently, the internalization of VEGFR2 and VEGFR3 was found to depend on ephrin signaling, a pathway that was not known before to be linked to VEGFR signaling [18,19]. The dependence on ephrin appears to be unique to VEGFR internalization.
2.1. Endocytic pathway of VEGFR
The response to VEGF, a pivotal agonist of vasculogenesis and angiogenesis [48,49], is mediated in mammals by a 3-member family of RTKs, consisting of VEGFR1 (alternatively named FMS-like tyrosine kinase, flt1), VEGFR2 (alternatively named kinase domain region, KDR, or fetal liver kinase 1, flk1), and VEGFR3 (alternatively named flt4). Since VEGFR2 is the major isoform that regulates vessel growth in the vascular system, we will focus on its endocytic pathway (Fig. 2). Similar to EGFR, VEGFR2 in ECs undergoes clathrin-dependent endocytosis triggered by the binding of its ligand, VEGF-A [16]. The endocytosis of VEGFR1 and VEGFR3 has been studied to a lesser extent than VEGFR2. VEGFR1 also appears to undergo clathrin-dependent endocytosis [50]. Though there is still no direct evidence to determine the endocytic pathway of VEGFR3, it is very likely to be clathrin-dependent.
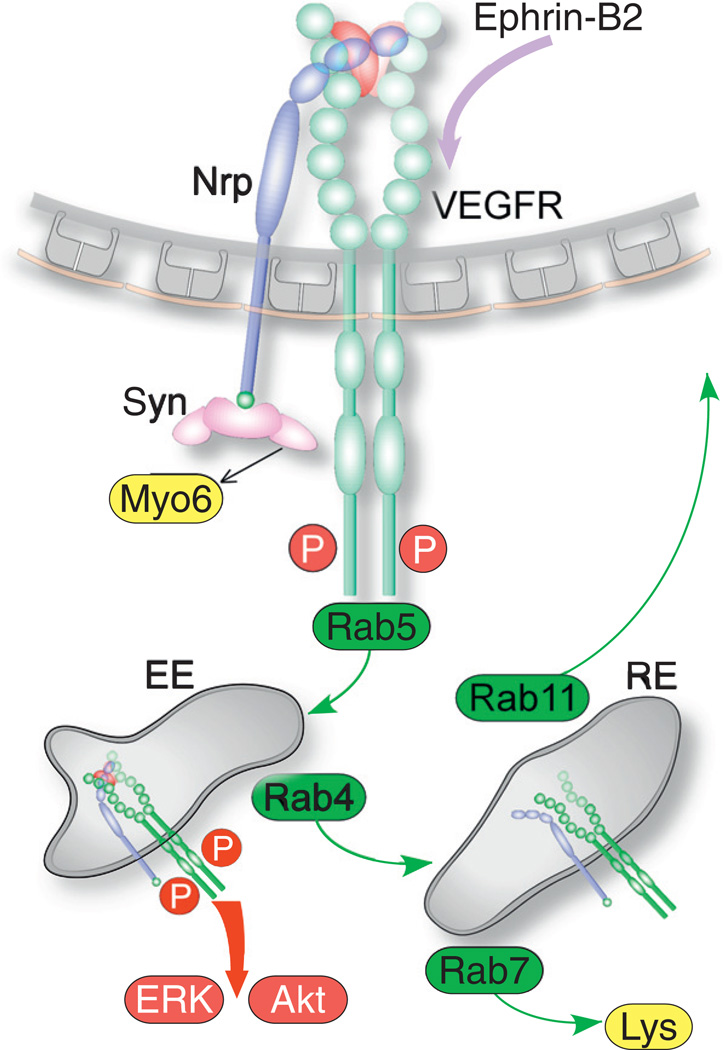
Fig. 2.
Scheme of membrane traffic and signaling of VEGFR and Nrp. Binding of VEGFA induces VEGFR2 auto-phosphorylation on multiple tyrosines and association of Nrp. Engagement of ephrin-B2 by EphB4 is required for VEGFR endocytosis. Endocytosed VEGFR2 is mobilized by myosin VI (myo6) cross-linked to Nrp via synectin, and traffics together with Nrp to early endosomes (EE) via Rab5. From early endosomes, the Nrp-VEGFR complex traffics to recycling endosomes (RE) via Rab4, followed by recycling to the plasma membrane via Rab11, or to lysosomes via Rab7. VEGFR signals from early endosomes and activates Akt and ERK.
Aside from clathrin-dependence, VEGFR2 endocytosis shares several attributes with that of EGFR: (1) In the absence of tyrosine auto-phosphorylation of the kinase region, VEGFR2 internalization rate slowed down significantly [51]. Though most studies concluded that VEGFR2 underwent ligand-triggered endocytosis, it is possible that a lower rate clathrin and AP2-dependent endocytosis persisted even in the absence of VEGF-A and tyrosine phosphorylation of VEGFR2 [52], indicating that VEGFR2 undergoes constitutive recycling. The VEGFR2 sub-population not present on the surface of quiescent cells was localized mostly to early endosomes, from where VEGFR2 underwent rapid Rab4-dependent recycling to the plasma membrane [53,54]. This sub-population is likely to have undergone the VEGF-independent constitutive endocytosis observed by other studies in human umbilical vein ECs (HUVECs) [52,55]. (2) Being clathrin-dependent, VEGFR2 internalization required the AP2 adaptor complex [52]. (3) Grb2 bound VEGFR2 in a VEGF-dependent manner, either directly or through the adaptor proteins Src homology and collagen homology (Shc), which bound auto-phosphorylated Tyr1175 of VEGFR2 [56]. Grb2 was involved also in VEGFR2 downstream signaling [57] and trafficking [58] (see Section 5).
The similarity between the endocytic pathways of EGFR and VEGFR2 also extends to early trafficking, since both receptors underwent Rab5-dependent trafficking to early endosomes [59,60]. The trafficking pattern of VEGF-triggered endocytosis was more complex compared to constitutive VEGFR2 recycling, as it appeared to use each of several routes. In VEGF-stimulated HUVECs, Rab4-dependent trafficking of VEGFR2 ensued from Rab5-associated endosomes [54], similar to the constitutive recycling in quiescent ECs, but the recycling of VEGFR2 to the plasma membrane proceeded from Rab4 to Rab11-associated vesicles, whereas no direct Rab11-dependent recycling from early endosomes to the plasma membrane was observed, in agreement with the canonical model of membrane traffic [61]. In other studies, the bulk of the recycling VEGFR2 population appeared to undergo Rab11-dependent recycling. A recycling feature unique to VEGFR2 is the regulation by its coreceptor, Nrp1, and by the interaction of Nrp1 through its PDZ-binding motif with the adaptor protein synectin [17,54]. In HUVECs treated by the VEGF-A165b isoform that does not bind Nrp1 [62], VEGFR2 was absent from Rab11-associated vesicles [54]. Similarly, VEGFR2 was not targeted to Rab11-associated vesicles in HUVECs expressing a Nrp1 mutant devoid of its carboxy-terminus PDZ binding motif [54]. These observations suggest that Nrp1 and its interaction with PDZ domain-containing proteins are required for Rab11-dependent recycling of VEGFR2. Synectin (also named neuropilin-1 binding protein, NIP, and GAIP-interacting protein, C-terminus, GIPC) is the only known protein that binds to the PDZ-binding motif of Nrp1 [63]. Synectin functions as a universal adaptor of the inward-traffic molecular motor myosin VI [64]. This interpretation counters, however, previous observations that narrowed down the role of both synectin and myosin VI to the inward trafficking of uncoated vesicles up to their fusion with early endosomes [65], where trafficking is regulated by Rab5 rather than by Rab11. Alternatively, other studies attributed the dependence on synectin and myosin VI to VEGFR2 rather than Nrp1 [17]. Though VEGFR2 does not have a PDZ-binding motif, it was suggested that by analogy with the nerve growth factor receptor TrkA, synectin could be recruited via the adaptor protein phosphotyrosine interaction, PH domain, and leucine zipper containing (APPL) [66]. The study of Lanahan et al. [17] ruled out Nrp1-dependent trafficking of the VEGFR2-Nrp1 complex because VEGF-D, an isoform that does not bind Nrp1, produced a similarly lower activation of ERK1/2 as in ECs isolated from synectin−/− mice. The reliance on VEGF-D counters, however, evidence for VEGF-D binding to Nrp1 [67]. Further, the putative absence of Nrp1 in the VEGFR2–VEGF-D complex could not rule out the dependence of VEGFR2 trafficking on Nrp1, because the experiment was carried out in synectin−/− ECs. The absence of synectin would prevent VEGFR2 trafficking even if Nrp1 had been present in the VEGFR2–VEGF-D complex.
Interestingly, the study of Ballmer-Hofer et al. [54] found that once VEGFR2 entered the Rab11-associated trafficking compartment, it was no longer phosphorylated at Tyr1175, a post-translational modification that is required for both MAPK and Akt activation [68]. This indicates that the endosomal compartment from which VEGFR2 signaled preceded Rab11-dependent trafficking. The finding of Ballmer-Hofer et al. [54] concurs with the study of Lanahan et al. [17], where Rab11 knockdown increased MAPK and Akt activation, presumably because it slowed down the departure of VEGFR2 from a preceding compartment. Collectively, these findings suggest that VEGFR2 signals from early or from recycling Rab4/Rab5-associated endosomes. The two studies suggest different sites, however, for the dephosphorylation of VEGR2. Whereas the study of Ballmer-Hofer et al. [54] implies that VEGFR2 was dephosphorylated immediately after exiting the signaling compartment and before Rab11-dependent recycling, the study of Lanahan et al. suggests that VEGFR2 was dephosphorylated by the protein tyrosine phosphatase 1b (PTP1b) near the plasma membrane. The localization of PTP1b activity near the plasma membrane is supported by other studies [69], whereas no PTP was identified in the study of Ballmer-Hofer et al. [54] as a candidate phosphatase of VEGFR2. There is consensus between several studies that part of the VEGFR2 population progressed from either early or recycling endosomes (Rab5 or Rab4-associated, respectively) to Rab7-associated late endosomes and lysosomes, i.e. to degradation [17,54,60]. The lysosome-targeted VEGFR2 fraction was small, however, as long as VEGFR2 co-trafficked with Nrp1. Once the VEGFR2-Nrp1 association was perturbed, a larger fraction of the VEGFR2 population was diverted to Rab7-associated endosomes [54]. While a mechanistic explanation of the dependence of VEGFR2 routing on the association with Nrp1 is lacking, it is possible that the lateral binding of VEGFR2 to Nrp1 interfered with the lysine ubiquitination required for RTK degradation [37,70]. When VEGFR2 degradation was inhibited by knocking down Rab7, MAPK activation increased [17,60]. This observation is in agreement with the premise that VEGFR2 signals from early and/or Rab4-associated recycling endosomes. A caveat that may be considered in regard to the studies on VEGFR2 trafficking discussed above is that their time resolution was relatively low, particularly during the first 30 min after VEGF-A treatment. Since most of the data was sampled at 30 min or later, it is possible that at least part of the data corresponded to the steady state, and did not represent the transient phase of trafficking. In fact, 30 min after endocytosis had been triggered, a large fraction of VEGFR2 in VEGF-A-treated mouse primary ECs had already recycled back to the plasma membrane [21].
The colocalization of internalized VEGFR2 with Rab7, a GTPase required for late endosome fusion with lysosomes [61], indicates that VEGFR2 was targeted to that compartment rather than to the proteasome. However, recent evidence indicates that VEGFR2 was processed by the proteasome prior to undergoing final degradation in the lysosome [32]. Proteolytic cleavage appears to have occurred immediately after VEGFR2 passed through the early endosomes, producing a fragment that corresponds to the transmembrane and ectoplasmic domains of the receptor. Expectedly, inhibiting the proteasome extended the duration of the activation of the downstream effectors Akt and MAPK. The complementary fragment corresponding to the cytoplasmic domain of VEGFR2 could conceivably still function as a tyrosine kinase and activate downstream effectors. Surprisingly, when the cytoplasmic domain was expressed as a chimera fused at its amino-terminus to yellow fluorescent protein (YFP), Tyr1175 was phosphorylated in the absence of VEGF, and Akt and MAPK were activated. However, the putative endogenous cytoplasmic domain was not detected directly, and its phosphorylation state in either the presence or absence of VEGF was not determined.
Similar to EGFR, the main downstream signaling pathways of VEGFR2 regulate either PLC /MAPK or PI3-Kinase/Akt [68]. A confluence of evidence from numerous studies established unambiguously that internalization was required for the optimal tyrosine phosphorylation of VEGFR2 and for the subsequent activation of both pathways [17–19,60,71–73]. Unlike EGFR signaling, the studies on VEGFR2 did not find differential effects of the internalization on Akt versus MAPK activation, concluding, instead, that both signaling pathways were upregulated by endocytosis of VEGFR2. Interestingly, exposure of ECs to matrix-bound, rather than to soluble VEGF-A165 produced an increase in the activation of p38 [72], a MAPK that remodels the actin cytoskeleton and induces cell migration in response to VEGF [74]. The mechanism that accounts for the signaling specificity of matrix-bound VEGF-A165 is unknown. An exception to the similarity between the endocytic mechanisms of EGFR and VEGFR is the involvement of cerebral cavernous malformation 3 (Ccm3) [75], an adaptor/scaffolding protein that is mutated in patients suffering from vascular malformation in the brain. Ccm3 bound to and colocalized with VEGFR2 on the plasma membrane in quiescent ECs. Ccm3 remained colocalized with VEGFR2 in cytoplasmic punctae that seemed to be vesicles, upon VEGF treatment, but the vesicles were not characterized by endocytic markers. Since knockdown of Ccm3 resulted in the internalization of VEGFR2, and reduced the activation of MAPK and Akt, it was deduced that Ccm3 was required for stabilizing VEGFR2 at the cell surface and, therefore, prolonging its signaling. This conclusion is at odds, however, with the recent evidence that endocytosis is required for VEGFR optimal signaling [17–19]. Since Ccm3 internalized and colocalized with VEGFR2, it could hypothetically be involved in VEGFR2 sorting, similar to Ccm1. The latter binds sorting nexin 17 [76], which diverts membrane receptors away from lysosomal degradation by preventing their entry into the internal vesicles of the MVB [77]. This would explain the “destabilization” of VEGFR2 when Ccm3 was knocked down [75].
It was recently observed that as much as a 30% fraction of the cellular VEGFR2 population was localized to the trans-Golgi apparatus in quiescent HUVECs [78]. Upon treatment by VEGF-A165, VEGFR2 exited almost completely from the trans-Golgi to cytoplasmic vesicles. The trans-Golgi VEGFR2 pool was regulated by N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) syntaxin-6 [78]. Since syntaxin-6 regulates transport from the trans-Golgi to the plasma membrane [79], it is likely that the study of Manickam et al. [78] detected the exocytosis of the trans-Golgi VEGFR2 pool to the plasma membrane in response to VEGF.
VEGF signaling is distinct from other RTK ligands in originating from intracellular pools in an autocrine manner [80]. Despite the absence of exogenous VEGF, hypoxia increased the tyrosine phosphorylation of VEGFR2 [80]. Since only a cell-permeable inhibitor of VEGFR2 had an effect on its tyrosine phosphorylation level, it appears that the receptor was activated by the pool of internalized VEGF. The trafficking pathway of the VEGFR2 population activated by intracellular VEGF is unknown.
2.2. Regulation of VEGFR2 endocytosis and sorting by ubiquitination
Aside from the well-known role in marking proteins for degradation, ubiquitination was required for the recruitment of adaptor proteins that sort endocytosed RTKs [81]. This has been demonstrated particularly for EGFR [81]. Although it had been established that VEGFR2 was ubiquitinated in a ligand-dependent manner in response to VEGF [82], the role of ubiquitination in VEGF signaling is less understood in comparison to EGFR. Early studies established that the E3 homologous to E6-AP C terminus (HECT) domain ubiquitin ligase Nedd4 mediated the proteasomal degradation of VEGFR2, and that this degradation could be inhibited by the sequestration of Nedd4 by Grb10 [82], an adaptor protein that bound and was phosphorylated by VEGFR2 [83]. However, Nedd4 was not shown to ubiquinate VEGFR2, or to regulate VEGFR2 sorting during internalization. The E3 ligase Cbl bound phosphorylated Try1052/1057 of VEGFR2 in porcine ECs, and ubiquitinated and inhibited PLC 1 bound to Tyr1173, but it was not implicated in the endocytosis of VEGFR2 [84]. Recent studies established that activated VEGFR2 bound the E3 ligase β-transducin repeats-containing protein (TRCP) via Ser1188/1191 located in a VEGFR2 motif that appears to conform to the PEST (rich in proline [P], glutamic acid [E], serine [S], and threonine [T]) consensus sequence [85]. While TRCP was shown to regulate the polyubiquitination of VEGFR2, it was not linked to VEGFR2 endocytosis [85]. RTK endocytosis is regulated by mono-ubiquitination [86], whereas poly-ubiquitination regulates proteolysis. Similar to EGFR [31], the endocytic sorting of VEGFR2 may be regulated by mono-ubiquitination of yet unidentified lysines that would form binding sites to endocytic adaptor proteins such as epidermal growth factor receptor substrate 15 (EPS15).
2.3. Regulation of VEGFR endocytosis by ephrin
Eph receptors are RTKs that guide neuronal patterning and facilitate the segregation of developing vessels into arteries and veins [87]. Eph signaling is triggered by binding in trans to ephrin receptors on apposing cells. Eph-ephrin signaling is bi-directional, both forward, via a tyrosine kinase phosphorylation cascade (Eph), and in reverse (ephrin), by binding other cell surface receptors in trans (ephrin A subfamily), or by binding cytoplasmic proteins (ephrin B subfamily). The expression patterns of EphB4 and ephrin-B2 in the vascular system are complementary, so that EphB4 is expressed primarily arteries, whereas ephrin-B2 is expressed in veins [88,89]. Though the precise functions of Eph receptors and ephrin in the vasculature have not been determined, their crucial role in vascular development was demonstrated in loss-of-function murine models. Disruption of the genes coding for either protein resulted in severe angiogenesis defects and embryonic lethality [88,89]. The angiogenic defects in the EphB4−/− mouse were symmetric to those in the ephrinB2−/− mouse, mirroring the complementary vein-artery distribution of EphB4 and ephrin-B2, respectively.
Two recent studies revealed an unexpected connection between EphB4-ephrin-B2 signaling, and between the internalization of VGFR2 [18] and VEGFR3 [19]. Engagement of ephrin-B2 by its ligand, EphB4, was followed by internalization of VEGFR3 in lymphatic ECs [19]. Other RTKs, e.g. PDGFR, were not internalized, indicating that the effect of ephrin-B2-EphB4 binding is specific to VEGFR. VEGFR3 failed to undergo endocytosis in ECs from the ephrinB2−/− mouse. VEGF-C-mediated tyrosine phosphorylation of VEGFR3 and the activation of Akt and of MAPK were markedly reduced in the ephrinB2−/− ECs. Ephrin-B2-EphB4 binding caused endocytosis of VEGFR3 in the absence of VEGF-C, but the internalized VEGFR3 was not auto-phosphorylated. VEGFR2 was observed to undergo a similar ephrin-B2-dependent endocytosis in vascular ECs, in the presence of VEGF-A [18].
It is unknown how ephrin-B2-EphB4 signaling induced the inter-nalization of VEGFR2/3, but the above studies provide some insight into the underlying mechanism. VEGFR3 endocytosis appears to have been triggered by transient association in cis between ephrin-B2 and VEGFR3 at the plasma membrane, but once internalized, ephrin-B2 and VEGFR2/3 no longer colocalized [19]. Notably, the interaction of ephrin-B2 with cytoplasmic proteins through its PDZ-binding motif was required for VEGFR2 endocytosis [18]. To date, at least nine PDZ domain-containing proteins that bind ephrin-B2 have been identified (http://www.signaling-gateway.org). One of these, PDZ domain-containing RING finger protein 3 (PDZRN3), is an E3 ubiquitin-protein ligase [90]. Thus, the lateral association of VEGFR2/3 with ephrin-B2 may facilitate the ubiquitination of VEGFR2/3 by ephrin-B2-bound PDZRN3. Though this has not been as well established as for EGFR [37], it is conceivable that Cbl-mediated ubiquitination of VEGFR would be required for its endocytosis, thus accounting for the dependence of VEGFR2 endocytosis on ephrin-B2.
3. Endocytosis and trafficking of Nrp1
3.1. Association between Nrp1 and VEGFR2
The uniqueness of the endocytic pathway of VEGFR is conferred mainly by its association with Nrp1. This is demonstrated by the consequences of perturbing the association between the two receptors on VEGFR2 signaling, as described above. Unlike the transient colocalization between ephrin-B2 and VEGFR at the cell surface [19], Nrp1 and VEGFR were 50–60% colocalized even in quiescent mouse primary ECs, and remained so up to 30 min after stimulation by VEGF-A165 [21]. Once VEGF-A was applied, Nrp1 and VEGFR2 became physically associated [21,91,92], and remained colocalized up to 30 min after the application of VEGF-A165 [21]. It is unknown whether the Nrp1-VEGFR2 association is direct or indirect, and which amino acid sequences and structural motifs of each receptor are required for VEGF-induced association. The dependence of the Nrp1-VEGFR2 association on the binding of the PDZ adaptor protein synectin [93] suggests that the carboxy-terminus of Nrp1 is required for the association. Since VEGFR2 is not known to bind synectin, it is not clear why synectin would be required for the association. If APPL binds VEGFR2, as suggested by Lanahan et al. [17], Nrp1 and APPL-bound synectin dimers could cross link the two receptors [64,94,95], thus explaining the dependence of the Nrp1-VEGFR2 association on synectin.
3.2. Endocytic pathway of Nrp1
Since Nrp1 colocalized extensively with VEGFR2 on the surface of quiescent ECs [21], it could have conceivably accompanied VEGFR2 when the latter underwent constitutive endocytosis [52]. There is direct evidence for VEGF-independent endocytosis and trafficking of Nrp1 together with integrin α5β1 from adhesion complexes, including focal-adhesions, to early endosomes [22]. The ligand-triggered endocytosis of Nrp1 in response to VEGF-A is clearly clathrin dependent, in contrast to semaphorin 3C–induced endocytosis, which is lipid raft-dependent [21]. In agreement with the finding that Nrp1 and VEGFR2 remain largely colocalized throughout endocytosis and trafficking [21], Nrp1 traversed the same endocytic compartments as VEGFR2, i.e. early endosomes [21,54], followed by “slow” recycling via Rab11-dependent trafficking. Only a minor fraction of the VEGFR2-Nrp1 complexes was diverted via Rab7-associated endosomes to lysosomal degradation [54].
3.3. Role of Nrp1 in VEGF signaling
Several lines of evidence suggest that Nrp1 signals independently from VEGFR, or that it is required for transducing aspects of VEGF signaling that are not mediated by VEGFR alone. The putative contribution of Nrp1 to VEGF signaling would obligately be mediated by the cytoplasmic domain. Since the carboxy-terminus of the same domain is used by synectin to cross-link Nrp1 to myosin VI, the molecular motor that drives Nrp1 trafficking [17,21], it is likely that Nrp1-dependent signaling is tightly coupled to Nrp1 trafficking.
The first indications for Nrp1-triggered signaling came from studies where the ectoplasmic domain of Nrp1 was replaced with that of EGFR and expressed in HUVECs lacking endogenous EGFR [96]. EGF treatment of these cells promoted their migration to a similar extent as VEGF-induced migration. Interestingly, when the EGFR-Nrp1 chimera consisted of a Nrp1 cytoplasmic domain lacking the carboxyterminus PDZ-binding motif, VEGF failed to promote cell migration. These results imply that EGF-induced clustering of the cytoplasmic domain of Nrp1 was sufficient to trigger downstream signaling, presumably in a PI3K–dependent manner [96]. However, these conclusions rest on the finding that immunoblotting did not detect EGFR in the HUVECs used by Wang et al. [96]. It is possible that though EGFR was not detectable by immunoblotting, its expression level was sufficient to activate PI3K and elicit cell migration. Moreover, HUVECs are highly variable primary ECs, and are known to express EGFR [97]. Thus, the results of Wang et al. [96] would be valid provided the same EGFR-deficient batch of HUVECs had been used throughout the study.
The requirement for the binding of synectin to the carboxyterminus of Nrp1 was suggested also by studies where nrp1 was knocked down in the zebrafish, inhibiting the angiogenesis of the intersomitic vessels and the sub-intestinal vein [98]. Whereas this vascular phenotype was rescued by the expression of human NRP1 mRNA, expression of a mutant Nrp1 lacking the PDZ-binding was ineffective. The requirement for this motif in vascular development was recapitulated by the similarity between the nrp1 and the synectin morphants.
While the above zebrafish studies suggest that Nrp1 can signal, they do not implicate it directly in transducing VEGF signaling. Evidence to that end was provided by studies in which antibodies specific to the ectoplasmic domain of Nrp1 were used to disrupt its association with VEGFR2 [91]. Strikingly, while the tyrosine phosphorylation level of VEGFR2 and the proliferative and permeability aspects of VEGF signaling were unchanged, the migration of HUVECs treated with Nrp1 antibodies was reduced drastically. The same antibodies strongly inhibited also the VEGF-driven outgrowth of HUVECs from beads encased in a fibrinogen matrix. These results suggest that Nrp1 was required for transducing the motility aspect of VEGF signaling. Similar conclusions could be drawn from a subsequent study from the same group, showing that binding of the VEGF-A121 isoform to Nrp1 was similarly required for sustaining EC migration [99]. Since VEGF-A121 binds Nrp1 but does cross-link it to VEGFR2, this result can be construed to suggest that Nrp1 can signal independently from VEGFR2. Were this scenario correct, how then would a short cytoplasmic domain lacking catalytic activity transduce the binding of VEGF to its ectoplasmic domain? A conceivable mechanism is the recruitment of a cytoplasmic protein complex that can signal once assembled. Aside from synectin, there is evidence for the binding of focal-adhesion kinase (FAK) [100] and the non-receptor tyrosine kinase Fer [101]. Both of these interactions do not appear to be involved in VEGF signaling, but other studies implicated Nrp1 in VEGF-induced tyrosine phosphorylation of FAK and, consequently, in the regulation of focal-adhesion assembly [102]. The potential involvement of Nrp1 in the regulation of focal-adhesions was suggested also by its role in the phosphorylation of p130Cas, an event that appeared to be mediated by PYK2, a tyrosine kinase that is closely related to FAK [103]. Aside from the binding of synectin to the cytoplasmic domain of Nrp1, there is no evidence to implicate endocytosis and membrane trafficking of Nrp1 in any of the putative signaling mechanisms described here.
Given the above convincing evidence for the migration/angiogenesis-specific role of Nrp1 in VEGF signaling, it is surprising that a mouse knockin (KI) model expressing a mutant nrp1 lacking the cytoplasmic domain (nrp1cytoΔ/Δ) was not embryonic lethal, and appeared grossly normal [104]. Phenotyping of the embryonic vascular system of the nrp1cytoΔ/Δ mouse detected no major defects, and analysis of the retinal vasculature in the neonatal nrp1cytoΔ/Δ mouse found a relatively minor defect consisting of excessive artery-vein crossovers. This defect did not interfere with the formation of an intact retinal vasculature. While the analysis of other vascular beds in the nrp1cytoΔ/Δ was not reported, the presence of such minor defects is perplexing given that antibody-mediated disruption of the association between Nrp1 and VEGFR2 interfered with the development of the retinal vasculature [91]. The absence of vascular defects cannot be attributed to compensation by Nrp2, since the retinal vasculature in mice expressing nrp1cytoΔ/Δ on a nrp2-null background was indistinguishable from that in mice expressing nrp1cytoΔ/Δ on a normal genetic background [104]. The discrepancy between the mild vascular phenotype of the nrp1cytoΔ/Δ KI mouse and the abundant evidence for the role of the Nrp1 cytoplasmic domain in angiogenesis remains unreconciled. Further, the apparent failure to rescue the zebrafish nrp1 vascular morphant by expressing human Nrp1 lacking the PDZ binding motif (Nrp1-1ΔSEA) suggests that at least in this model system the cytoplasmic domain, and in particular its PDZ-binding motif, is required for angiogenesis [98]. However, the implications of the nrp1cytoΔ/Δ genetic in-vivo model for the function of Nrp1 in mammalian vascular development should arguably persevere over conclusions drawn from the numerous in-vitro models, or from non-genetic in-vivo models [91]. It appears, therefore, that at least during development, the primary signaling function of the Nrp1 cytoplasmic domain is not the regulation of angiogenesis. Conceivable VEGF-dependent functions would be the regulation of vessel permeability [105,106] or of endothelial cell adhesion via integrin [107]. There is no evidence to suggest that such functions entail Nrp1 endocytosis and trafficking.
4. Endocytosis of adhesion receptors
VEGF signaling induces the endocytosis of two major classes of adhesive receptors: intercellular junction proteins, and ECM receptors. Both adherens and tight junction proteins, i.e. vascular endothelial cadherin (VEcad) and occludin, respectively, are endocytosed in response to VEGF [16,108,109]. Among ECM adhesion receptors, several types of integrin α and β subunits are internalized in response to VEGF [110,111]. Though it is known primarily for its function in the plasminogen activation cascade, the urokinase plasmin activator receptor (uPAR) is also reviewed here, since it interacts with the ECM protein vitronectin and with several integrin β subunits [112].
4.1. Endocytosis of intercellular adhesion receptors
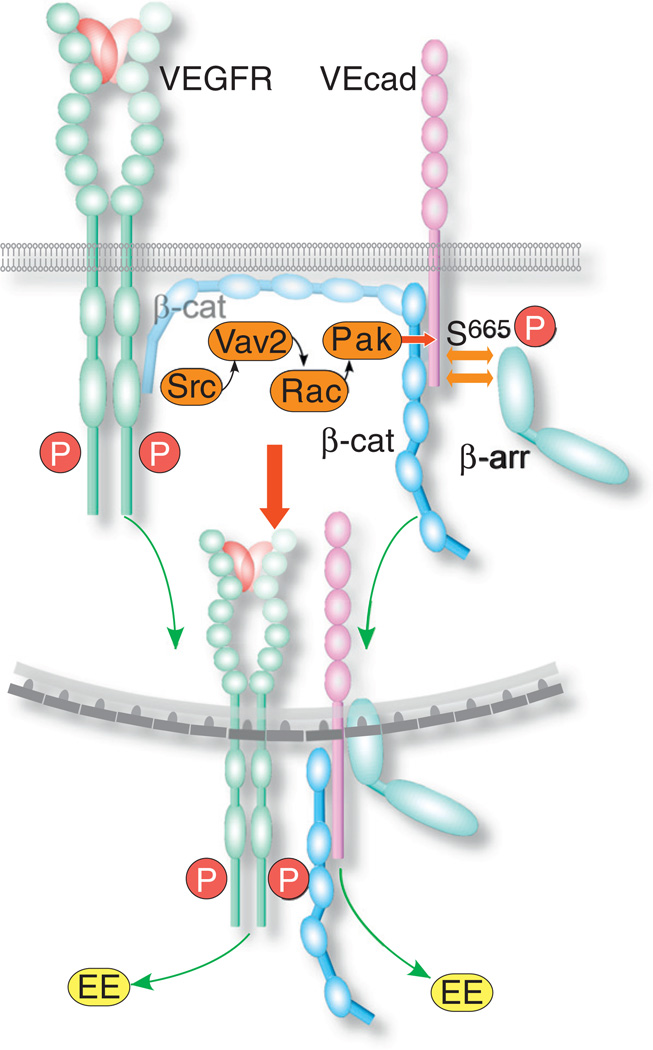
One of the major cell functions regulated by VEGF-A through VEGFR2 is the dissociation of EC junctions and the subsequent increase in vessel permeability. At the cellular level, the loss of vessel patency is caused by the disassembly of EC adherens and tight junctions in a process that involves the Src tyrosine kinase [113]. It appears that in quiescent ECs, VEcad is associated with VEGFR2 via β-catenin [114], and functions to stabilize the receptor at the cell surface [113,115] (Fig. 3). Once VEGFR2 was activated, VEcad dissociated from it and bound β-arrestin [108], an adaptor molecule known primarily for facilitating the endocytosis of G-protein coupled receptors, which facilitates the formation of clathrin-coated vesicles [116]. The recruitment of β-arrestin to VEcad was initiated by Srcdependent phosphorylation and activation of the Rho GEF Vav2. The latter activated Rac, resulting in the activation of p21-activated kinase (PAK), and the phosphorylation of VEcad on Ser665, which served as a β-arrestin-docking site [108]. VEcad underwent clathrin-dependent endocytosis [16,108] and trafficked to early endosomes [108], possibly employing myosin X, an actin-based molecular motor [117]. VEcad did not colocalize after endocytosis with VEGFR2 [16], in agreement with the reported dissociation of VEcad from activated VEGFR2 [118].
Fig. 3.
Scheme of the endocytic mechanism of VEcad. Activated VEGFR induces Src to activate Vav2, which then activates Rac and PAK. The latter phosphorylates the VEcad cytoplasmic domain on Ser665, which serves as a docking site for β-arrestin (β-arr). β-arrestin facilitates the formation of clathrin-coated vesicles. During or after endocytosis, β-catenin (β-cat), which cross-links VEGFR2 and VEcad, dissociates from VEGFR2. VEcad and VEGFR2 proceed to traffic separately to early endosomes (EE).
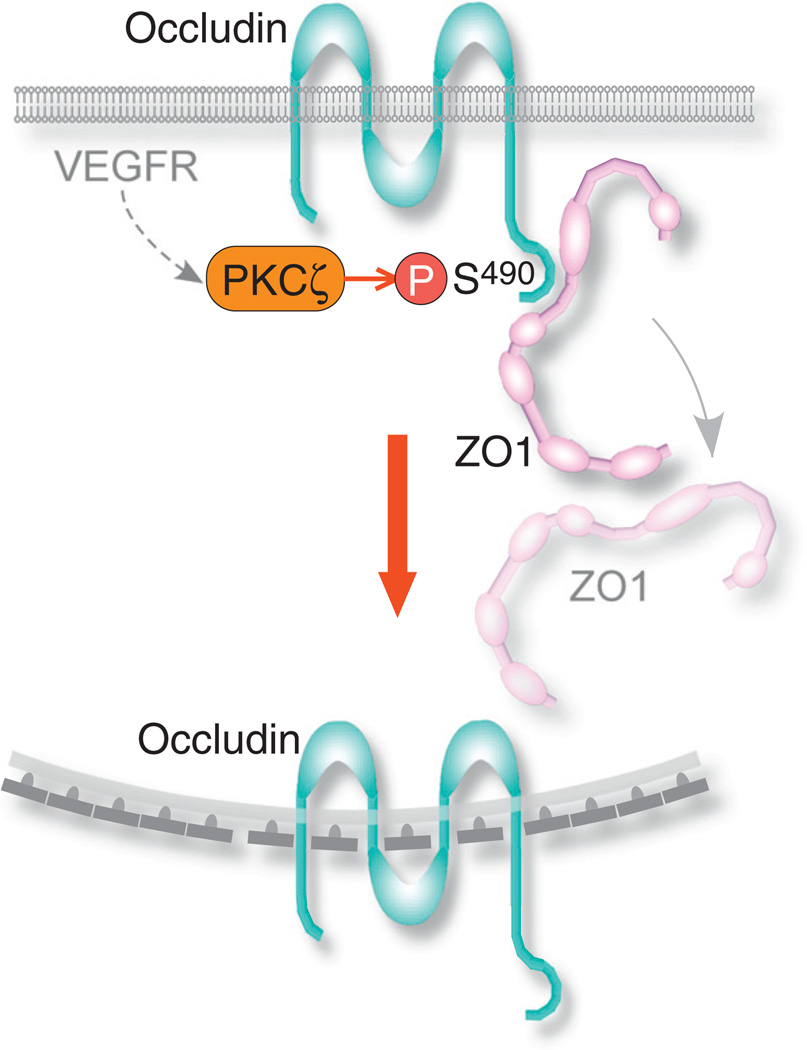
Similar to adherens junctions, the disassembly of tight junctions in response to VEGF is mediated by the endocytosis of its components. Among the integral tight junction proteins, the endocytic mechanism of the 4-pass transmembrane protein occludin has been elucidated in primary bovine retinal endothelial cells [109]. Occludin endocytosis followed the VEGF induced activation of protein kinase C (PKC) ζ, also a tight junction component [119], via an unknown pathway. PKCζ phosphorylated occludin at Ser490, resulting in its removal from the cell borders and its colocalization with clathrin, with clathrin-binding adaptor protein epsin-1, EPS15, and with hepatocyte growth factor-regulated tyrosine kinase substrate (Hrs) (Fig. 4). The latter two proteins contain a ubiquitin-interacting motif and facilitate the ubiquitination and targeting of their ligands to lysosomal degradation [120]. Following endocytosis, occludin reappeared in cytoplasmic punctae that colocalized with early endosome and lysosomal markers [109]. After the Ser490 phosphorylation of occludin, the PDZ domain-containing protein ZO1 dissociated from occludin [121]. Since ZO1 functions as a scaffold that cross-links tight junction transmembrane proteins and the actin cytoskeleton [122], its dissociation from occludin is a key step in junction disassembly. Interestingly, Murakami et al. [109] observed that claudin-5, a 4-pass and ZO1-binding transmembrane protein like occludin, also underwent endocytosis in response to VEGF.
Fig. 4.
Scheme of the endocytic mechanism of occludin. Activated VEGFR induces PKCζ to phosphorylate occludin on Ser490. Following the phosphorylation, ZO1, which was bound to occludin in tight junctions, dissociates from it. Occludin undergoes clathrin-dependent endocytosis.
4.2. Endocytosis of extracellular matrix (ECM) receptors
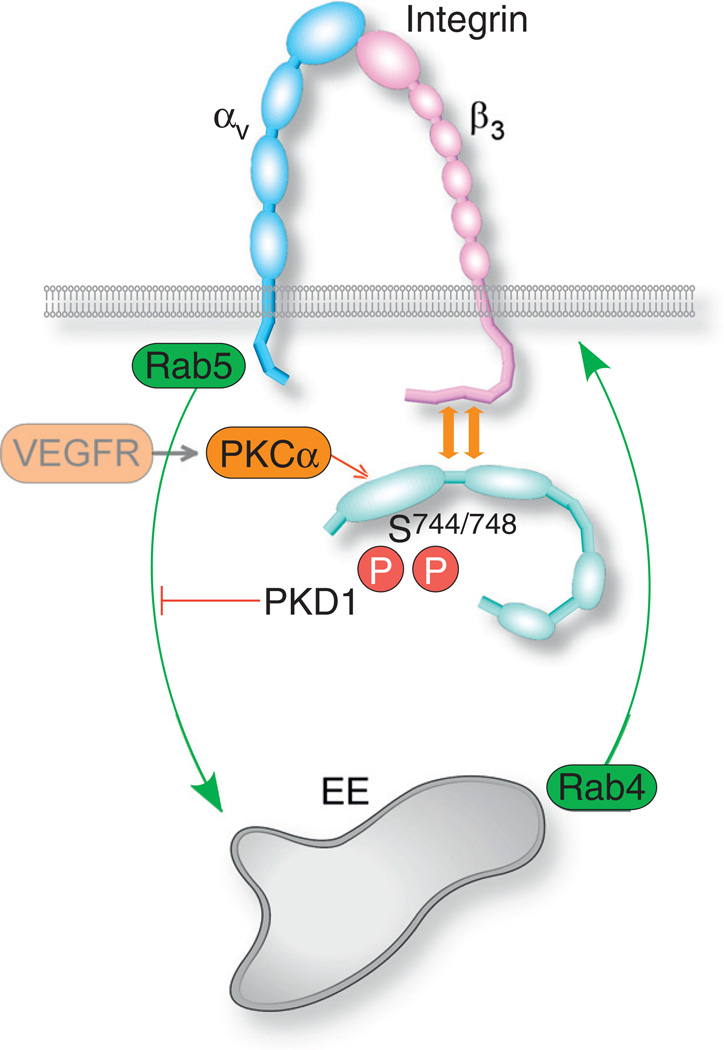
As reviewed elsewhere [123], the endocytosis and trafficking of integrins is required for the regulation of Rho GTPases, and for the regulation of the signaling of integrin-associated growth factor receptors. VEGF-driven endocytosis and trafficking of integrin αvβ3, a fibronectin and vitronectin receptor [124], was mediated in HUVECs by protein kinase D1 (PKD1) [110]. PKD1 associated with integrin αvβ3 constitutively, regardless of VEGF-A signaling [110,125]. VEGF-A activated PKD1 by inducing its phosphorylation of Ser744/748 in the activation loop, most likely by protein kinase Cα [126] (Fig. 5). Additionally, VEGF-A targeted PKD1 to nascent focal-adhesions (focal complexes) on cell protrusions by inducing phosphorylation of Ser916. Knockdown of PKD1 enhanced integrin αvβ3 recycling, while inhibiting the formation of focal-adhesions and the migration of HUVECs [110]. It appears, therefore, that integrin αvβ3 underwent constitutive Rab5 and Rab4-dependent recycling through early endosomes, and that the activation of PKD1 by VEGF-A inhibited the endocytosis of αvβ3 via an unknown mechanism. As a result, PKD1 knockdown tipped the balance maintained by constitutive recycling between inward and outward trafficking towards the latter, resulting in a higher apparent presence of integrin αvβ3 on the cell surface. Though VEGF treatment resulted in a relatively small increase in the recycling rate of integrin αvβ3 in comparison to constitutive recycling, the VEGF-induced relative increase in EC migration rate was far larger [110]. Integrin can undergo either clathrin or caveolin-dependent endocytosis [127], but the study of di Blasio et al. [110] did not positively identify the endocytic pathway.
Fig. 5.
Scheme of the endocytic mechanism of integrin αvβ3. In quiescent ECs, integrin αvβ3 undergoes constitutive recycling to early endosomes. VEGF activation of PKD1 via phosphorylation of Ser744 and Ser748, most likely by PKCα, results in the binding of PKD1 to the cytoplasmic domain of the β3 subunit, and inhibition of integrin αvβ3 endocytosis. This produces an apparent increase in the integrin αvβ3 presence on the cell surface.
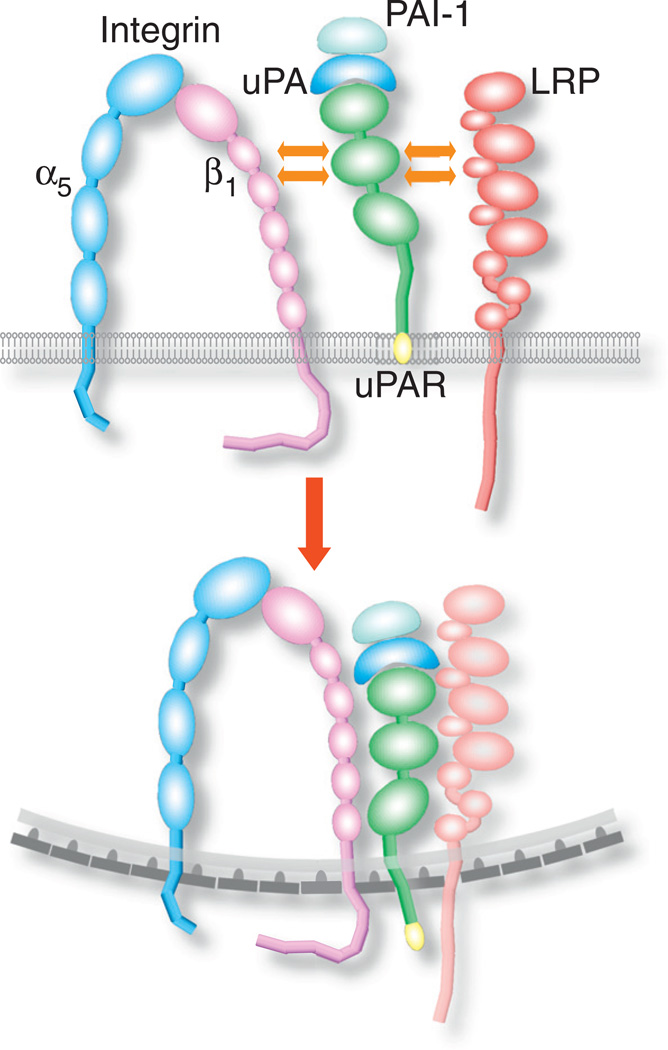
The VEGF-induced endocytosis of the β1 integrin subunit incorporated uPAR, a glycosyl phosphatidylinositol (GPI)-anchored cell surface receptor noted primarily for its role in the plasminogen activation cascade. uPAR binds uPA, which cleaves plasminogen to generate plasmin [128]. Aside from binding uPA, uPAR functions as a receptor for numerous ligands, including the ECM protein vitronectin, and binds in cis several integrin β subunits, including β1 [128,129]. Whereas integrin β1 (in tandem with subunit α5) was localized in quiescent HUVECs mainly at the cell periphery, treatment with VEGF-A165 redistributed it to cytoplasmic vesicular structures, where it colocalized with uPAR [111]. Since previous studies had shown that uPAR underwent clathrin-dependent endocytosis [130], it is likely that VEGF-A165 induced the endocytosis of integrin β1 in the same manner. VEGF-A165 failed to induce clathrin-dependent endocytosis of integrin β1 in ECs of uPAR−/− mice, or in ECs treated with a peptide that inhibited the interaction of uPAR with lipoprotein receptor-related protein (LRP) [130]. These results suggest that integrin β1 endocytosis required coordination between LRP, uPAR, and integrin α5β1, as follows: LRP associated with uPAR that was engaged by uPA bound to PAI-1, followed by the association of uPAR to integrin β1 [130] (Fig. 6). Blocking the integrin β1-uPAR association inhibited EC migration towards a VEGF-A165 gradient [111], indicating that VEGF-induced endocytosis of integrin α5β1 was required for angiogenesis in-vivo. Though VEGF-A165 also induced endocytosis of integrin α3β1, this endocytosis did not require interaction of integrin β1 with uPAR [111].
Fig. 6.
Scheme of the endocytic mechanism of integrin α5β1 and uPAR. Downstream of VEGF, uPA binds together with PAI-1 to uPAR on the EC surface. The binding is followed by lateral association with LRP, after which the uPAR-LRP complex associates laterally with the integrin β1 subunit. Integrin α5β1 undergoes clathrin dependent endocytosis together with uPAR, and possibly also with LRP.
It should be noted that VEGFR2 and integrin β1 [131] or β3 [131–133] signaling were shown to have reciprocal functional relations, but those were not reported to involve endocytosis or trafficking of either receptor.
5. Endocytosis and trafficking of Rho GTPases
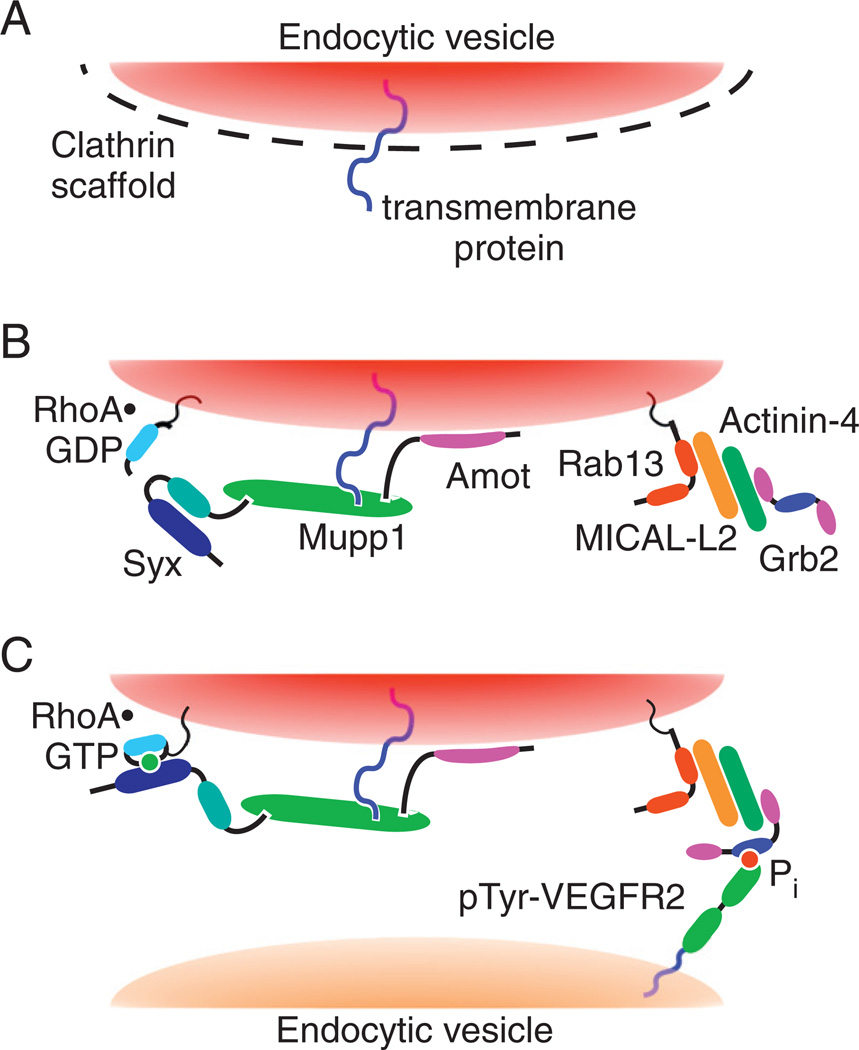
Recent studies found that VEGF signaling can induce the trafficking not only of cell-surface receptors, but also of Rho GTPases [58]. A previous study showed that hepatocyte growth factor, a ligand of the Met RTK, induced membrane trafficking of Rac1, and caused its activation on endosomes by the Tiam1 GEF in a Rab5-dependent manner [134]. Similarly, VEGF-A165 induced the trafficking of RhoA, together with its GEF Syx [58]. Unlike Rac1, the trafficking of RhoA was mediated by Rab13, a GTPase mainly known for regulating the endocytosis of occludin from tight junctions [135]. In primary mouse ECs migrating in response to a VEGF-A165 gradient, Rab13 as well as RhoA and Syx were localized not only to endocytic vesicles, but also at the leading edge of the cells [58]. In agreement with previous studies, it appears that Rab13 is involved not only in the regulation of tight junctions but also in the remodeling of the actin cytoskeleton of migrating cells [136,137]. The hypothetical scenario of VEGF-A-driven trafficking starts with the induction of clathrin-dependent endocytosis of tight junction proteins by VEGF (Fig. 7). Multi PDZ domain protein (MUPP1)-1, which binds the integral tight junction transmembrane proteins junctional adhesion molecule (JAM) and claudin [138,139], would then remain attached to their cytoplasmic domains and would be removed from the tight junctions on vesicles engulfing JAM and claudin. Mupp1 is known to bind Syx [140] and angiomotin (Amot). Amot would facilitate the targeting of Mupp1 and Syx to the lipid bilayer of endocytic vesicles due to its high affinity for phosphatidylinositol 3-phosphate [141], which is enriched in the membranes of vesicles [142]. Rab13 would form a complex with the actin-binding protein actinin-4 (Actn4) through their common binding partner, molecule interacting with CasL-like 2 (Mical-l2) [136]. The Rab13/Mical-l2/Actn4 complex would target the Syx-RhoA carrying vesicles to endocytosed tyrosine-phosphorylated VEGFR2, since Actn4 binds Grb2, which binds VEGFR2 [38]. This mechanism can account for the localization of RhoA, Syx, and Rab13 to the leading edge of migrating ECs [58]. Knockdown of rab13 in the zebrafish inhibited angiogenesis, thus supporting the in-vivo role of VEGF-induced trafficking of RhoA [58].
Fig. 7.
Hypothetical mechanism of Syx and RhoA trafficking from cell junctions to the leading edge. Step A: VEGF-A binding to VEGFR2 triggers clathrin-dependent endocytosis of junctional transmembrane proteins. Step B: Amot targets the Syx-associated complex, containing also Mupp1 and RhoA, to the cytoplasmic leaflet of uncoated endocytic vesicles enclosing the endocytosed junctional transmembrane proteins. Step C: Rab13 associates with and mediates the trafficking of these vesicles. It recruits Grb2, which targets the Rab13-associated vesicles to Tyr-phosphorylated VEGFR2. Syx activates RhoA co-trafficking on the same vesicles, at the leading edge of the migrating cell.
This figure was originally published in the Journal of Biological Chemistry by Wu et al. [58]. The American Society for Biochemistry and Molecular Biology©.
6. Concluding remarks
The unique attributes of VEGFR signaling and its regulation by membrane traffic are conferred largely by the association with the co-receptor Nrp. The phenotyping of the nrp1cytoΔ/Δ mouse ruled out, surprisingly, an obvious signaling role for the cytoplasmic domain of Nrp1 in vasculogenesis and angiogenesis, aside from arteriovenous patterning in the retina. It is likely that there is more to the cytoplasmic domain of Nrp1 than the binding via its PDZ motif to a single protein, synectin. One potential function is the regulation of focal-adhesions in-vitro [102], but it is unclear what is the in-vivo equivalent of this function, since the in-vivo analog of two-dimensional focal adhesions in not known. Another aspect of VEGFR endocytosis that has not yet been determined is the mechanism by which it is triggered by ephrin-B2-EphB4 signaling.
VEGFR and other RTKs exploit the sorting mechanisms of the endosomal system to achieve signaling specificity. The early endosomes appears to be the major sorting center for RTK signaling (reviewed in [81]), and the site wherefrom the receptors signal in the cytoplasm. The sorting tools employed by VEGFR signaling include Rab GTPases [60] and tyrosine phosphorylation [52]. Although there is evidence for the involvement of ubiquitin ligases and ubiquitin-adaptor proteins [82–84,143], this is probably only a part of the role of ubiquitination in regulation of VEGFR membrane traffic and signaling. Similar to EGFR, it is likely that VEGFR is regulated by mono-ubiquitination of lysines in its cytoplasmic domain, and by ubiquitin-binding proteins such as epsin-1 and EPS15. Importantly, the number of animal models that addressed directly the role of membrane traffic in VEGF signaling is relatively small [17–19,58]. Hence, the impact of membrane traffic on the full spectrum of VEGF signaling remains unknown.
Abbreviations
- Actn4
actinin-4
- Amot
angiomotin
- APPL
adaptor protein phosphotyrosine interaction, PH domain, and leucine zipper containing
- Cbl
Casitas B-lineage lymphoma
- EC
endothelial cell
- EGFR
epithelial growth factor receptor
- ECM
extracellular matrix
- EPS15
epidermal growth factor receptor substrate 15
- FGFR
fibroblast growth factor receptor
- FAK
focal-adhesion kinase
- GIPC
GAIP-interacting protein, C-terminus
- GPI
glycosyl phosphatidylinositol
- Grb2
growth factor receptor-bound protein 2
- HECT
E3 homologous to E6-AP C terminus
- Hrs
hepatocyte growth factor-regulated tyrosine kinase substrate
- JAM
junctional adhesion molecule
- KI
knockin
- LRP
lipoprotein receptor-related protein
- NIP
neuropilin-1 binding protein
- Mical-l2
molecule interacting with CasL-like 2
- MUPP1
multi PDZ domain protein 1
- Nrp
neuropilin
- PAK
p21-activated kinase
- PDGFR
platelet derived growth factor receptor
- PDZRN3
PDZ domain-containing RING finger protein 3
- PEST
rich in proline [P], glutamic acid [E], serine [S], and threonine [T]
- PAI
plasminogen activator inhibitor
- PTP1b
protein tyrosine phosphatase 1b
- PKD
protein kinase D
- PKC
protein kinase C
- RTK
receptor tyrosine kinase
- SNARE
N-ethylmaleimide-sensitive factor attachment protein receptor
- SH
Src homology
- Shc
Src homology and collagen homology
- TRCP
β-transducin repeats-containing protein
- uPAR
urokinase plasmin activator receptor
- VEcad
vascular endothelial cadherin
- VEGFR
vascular endothelial growth factor receptor
- YFP
yellow fluorescent protein
References
- 1.Murphy JE, Padilla BE, Hasdemir B, Cottrell GS, Bunnett NW. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(42):17615–17622. doi: 10.1073/pnas.0906541106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zwang Y, Yarden Y. Traffic. 2009;10(4):349–363. doi: 10.1111/j.1600-0854.2008.00870.x. [DOI] [PubMed] [Google Scholar]
- 3.Mukherjee S, Tessema M, Wandinger-Ness A. Circulation Research. 2006;98(6):743–756. doi: 10.1161/01.RES.0000214545.99387.e3. [DOI] [PubMed] [Google Scholar]
- 4.Santambrogio M, Valdembri D, Serini G. Molecular Aspects of Medicine. 2011;32(2):112–122. doi: 10.1016/j.mam.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 5.Haigler HT, McKanna JA, Cohen S. The Journal of Cell Biology. 1979;81(2):382–395. doi: 10.1083/jcb.81.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vieira AV, Lamaze C, Schmid SL. Science. 1996;274(5295):2086–2089. doi: 10.1126/science.274.5295.2086. [DOI] [PubMed] [Google Scholar]
- 7.Di Guglielmo GM, Baass PC, Ou WJ, Posner BI, Bergeron JJ. The EMBO Journal. 1994;13(18):4269–4277. doi: 10.1002/j.1460-2075.1994.tb06747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lai WH, Cameron PH, Doherty II JJ, Posner BI, Bergeron JJ. The Journal of Cell Biology. 1989;109(6 Pt 1):2751–2760. doi: 10.1083/jcb.109.6.2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Donatis A, Comito G, Buricchi F, Vinci MC, Parenti A, Caselli A, Camici G, Manao G, Ramponi G, Cirri P. The Journal of Biological Chemistry. 2008;283(29):19948–19956. doi: 10.1074/jbc.M709428200. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, Pennock SD, Chen X, Kazlauskas A, Wang Z. The Journal of Biological Chemistry. 2004;279(9):8038–8046. doi: 10.1074/jbc.M311494200. [DOI] [PubMed] [Google Scholar]
- 11.Kawada K, Upadhyay G, Ferandon S, Janarthanan S, Hall M, Vilardaga JP, Yajnik V. Molecular and Cellular Biology. 2009;29(16):4508–4518. doi: 10.1128/MCB.00015-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bryant DM, Wylie FG, Stow JL. Molecular Biology of the Cell. 2005;16(1):14–23. doi: 10.1091/mbc.E04-09-0845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hausott B, Rietzler A, Vallant N, Auer M, Haller I, Perkhofer S, Klimaschewski L. Neuroscience. 2011;188:13–22. doi: 10.1016/j.neuroscience.2011.04.064. [DOI] [PubMed] [Google Scholar]
- 14.Tkachenko E, Lutgens E, Stan RV, Simons M. Journal of Cell Science. 2004;117(Pt 15):3189–3199. doi: 10.1242/jcs.01190. [DOI] [PubMed] [Google Scholar]
- 15.Jekely G, Sung HH, Luque CM, Rorth P. Developmental Cell. 2005;9(2):197–207. doi: 10.1016/j.devcel.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 16.Lampugnani MG, Orsenigo F, Gagliani MC, Tacchetti C, Dejana E. The Journal of Cell Biology. 2006;174(4):593–604. doi: 10.1083/jcb.200602080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lanahan AA, Hermans K, Claes F, Kerley-Hamilton JS, Zhuang ZW, Giordano FJ, Carmeliet P, Simons M. Developmental Cell. 2010;18(5):713–724. doi: 10.1016/j.devcel.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sawamiphak S, Seidel S, Essmann CL, Wilkinson GA, Pitulescu ME, Acker T, Acker-Palmer A. Nature. 2010;465(7297):487–491. doi: 10.1038/nature08995. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, Nakayama M, Pitulescu ME, Schmidt TS, Bochenek ML, Sakakibara A, Adams S, Davy A, Deutsch U, Luthi U, Barberis A, Benjamin LE, Makinen T, Nobes CD, Adams RH. Nature. 2010;465(7297):483–486. doi: 10.1038/nature09002. [DOI] [PubMed] [Google Scholar]
- 20.Narazaki M, Tosato G. Blood. 2006;107(10):3892–3901. doi: 10.1182/blood-2005-10-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salikhova A, Wang L, Lanahan AA, Liu M, Simons M, Leenders WP, Mukhopadhyay D, Horowitz A. Circulation Research. 2008;103(6):e71–e79. doi: 10.1161/CIRCRESAHA.108.183327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valdembri D, Caswell PT, Anderson KI, Schwarz JP, Konig I, Astanina E, Caccavari F, Norman JC, Humphries MJ, Bussolino F, Serini G. PLoS Biology. 2009;7(1):e25. doi: 10.1371/journal.pbio.1000025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Backer JM, Kahn CR, White MF. The Journal of Biological Chemistry. 1989;264(3):1694–1701. [PubMed] [Google Scholar]
- 24.Chow JC, Condorelli G, Smith RJ. The Journal of Biological Chemistry. 1998;273(8):4672–4680. doi: 10.1074/jbc.273.8.4672. [DOI] [PubMed] [Google Scholar]
- 25.Kelly KL, Ruderman NB. The Journal of Biological Chemistry. 1993;268(6):4391–4398. [PubMed] [Google Scholar]
- 26.Kublaoui B, Lee J, Pilch PF. The Journal of Biological Chemistry. 1995;270(1):59–65. doi: 10.1074/jbc.270.1.59. [DOI] [PubMed] [Google Scholar]
- 27.Hammond DE, Carter S, McCullough J, Urbe S, Vande Woude G, Clague MJ. Molecular Biology of the Cell. 2003;14(4):1346–1354. doi: 10.1091/mbc.E02-09-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mann F, Miranda E, Weinl C, Harmer E, Holt CE. Journal of Neurobiology. 2003;57(3):323–336. doi: 10.1002/neu.10303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zimmer M, Palmer A, Kohler J, Klein R. Nature Cell Biology. 2003;5(10):869–878. doi: 10.1038/ncb1045. [DOI] [PubMed] [Google Scholar]
- 30.Grimes ML, Zhou J, Beattie EC, Yuen EC, Hall DE, Valletta JS, Topp KS, LaVail JH, Bunnett NW, Mobley WC. The Journal of Neuroscience. 1996;16(24):7950–7964. doi: 10.1523/JNEUROSCI.16-24-07950.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Avraham R, Yarden Y. Nature Reviews. Molecular Cell Biology. 2011;12(2):104–117. doi: 10.1038/nrm3048. [DOI] [PubMed] [Google Scholar]
- 32.Bruns AF, Bao L, Walker JH, Ponnambalam S. Biochemical Society Transactions. 2009;37(Pt 6):1193–1197. doi: 10.1042/BST0371193. [DOI] [PubMed] [Google Scholar]
- 33.Sorkin A, Goh LK. Experimental Cell Research. 2008;314(17):3093–3106. doi: 10.1016/j.yexcr.2008.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamazaki T, Zaal K, Hailey D, Presley J, Lippincott-Schwartz J, Samelson LE. Journal of Cell Science. 2002;115(Pt 9):1791–1802. doi: 10.1242/jcs.115.9.1791. [DOI] [PubMed] [Google Scholar]
- 35.Lamaze C, Schmid SL. The Journal of Cell Biology. 1995;129(1):47–54. doi: 10.1083/jcb.129.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Honegger AM, Dull TJ, Felder S, Van Obberghen E, Bellot F, Szapary D, Schmidt A, Ullrich A, Schlessinger J. Cell. 1987;51(2):199–209. doi: 10.1016/0092-8674(87)90147-4. [DOI] [PubMed] [Google Scholar]
- 37.Goh LK, Huang F, Kim W, Gygi S, Sorkin A. The Journal of Cell Biology. 2010;189(5):871–883. doi: 10.1083/jcb.201001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.D’Angelo G, Martini JF, Iiri T, Fantl WJ, Martial J, Weiner RI. Molecular Endocrinology. 1999;13(5):692–704. doi: 10.1210/mend.13.5.0280. [DOI] [PubMed] [Google Scholar]
- 39.Sorkin A, von Zastrow M. Nature Reviews. Molecular Cell Biology. 2009;10(9):609–622. doi: 10.1038/nrm2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blagoev B, Kratchmarova I, Ong SE, Nielsen M, Foster LJ, Mann M. Nature Biotechnology. 2003;21(3):315–318. doi: 10.1038/nbt790. [DOI] [PubMed] [Google Scholar]
- 41.Johannessen LE, Pedersen NM, Pedersen KW, Madshus IH, Stang E. Molecular and Cellular Biology. 2006;26(2):389–401. doi: 10.1128/MCB.26.2.389-401.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kazazic M, Bertelsen V, Pedersen KW, Vuong TT, Grandal MV, Rodland MS, Traub LM, Stang E, Madshus IH. Traffic. 2009;10(2):235–245. doi: 10.1111/j.1600-0854.2008.00858.x. [DOI] [PubMed] [Google Scholar]
- 43.Sorkin AD, Teslenko LV, Nikolsky NN. Experimental Cell Research. 1988;175(1):192–205. doi: 10.1016/0014-4827(88)90266-2. [DOI] [PubMed] [Google Scholar]
- 44.Sorkin A, Carpenter G. The Journal of Biological Chemistry. 1991;266(34):23453–23460. [PubMed] [Google Scholar]
- 45.Sorkin A, Kornilova E, Teslenko L, Sorokin A, Nikolsky N. Biochimica et Biophysica Acta. 1989;1011(1):88–96. doi: 10.1016/0167-4889(89)90083-9. [DOI] [PubMed] [Google Scholar]
- 46.Sorkin A, Krolenko S, Kudrjavtceva N, Lazebnik J, Teslenko L, Soderquist AM, Nikolsky N. The Journal of Cell Biology. 1991;112(1):55–63. doi: 10.1083/jcb.112.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stoscheck CM, Carpenter G. The Journal of Cell Biology. 1984;98(3):1048–1053. doi: 10.1083/jcb.98.3.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferrara N, Gerber HP, LeCouter J. Nature Medicine. 2003;9(6):669–676. doi: 10.1038/nm0603-669. [DOI] [PubMed] [Google Scholar]
- 49.Ruhrberg C. BioEssays. 2003;25(11):1052–1060. doi: 10.1002/bies.10351. [DOI] [PubMed] [Google Scholar]
- 50.Kobayashi S, Sawano A, Nojima Y, Shibuya M, Maru Y. The FASEB Journal. 2004;18(7):929–931. doi: 10.1096/fj.03-0767fje. [DOI] [PubMed] [Google Scholar]
- 51.Dougher M, Terman BI. Oncogene. 1999;18(8):1619–1627. doi: 10.1038/sj.onc.1202478. [DOI] [PubMed] [Google Scholar]
- 52.Ewan LC, Jopling HM, Jia H, Mittar S, Bagherzadeh A, Howell GJ, Walker JH, Zachary IC, Ponnambalam S. Traffic. 2006;7(9):1270–1282. doi: 10.1111/j.1600-0854.2006.00462.x. [DOI] [PubMed] [Google Scholar]
- 53.Gampel A, Moss L, Jones MC, Brunton V, Norman JC, Mellor H. Blood. 2006;108(8):2624–2631. doi: 10.1182/blood-2005-12-007484. [DOI] [PubMed] [Google Scholar]
- 54.Ballmer-Hofer K, Andersson AE, Ratcliffe LE, Berger P. Blood. 2011;118(3):816–826. doi: 10.1182/blood-2011-01-328773. [DOI] [PubMed] [Google Scholar]
- 55.Jopling HM, Howell GJ, Gamper N, Ponnambalam S. Biochemical and Biophysical Research Communications. 2011;410(2):170–176. doi: 10.1016/j.bbrc.2011.04.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kroll J, Waltenberger J. The Journal of Biological Chemistry. 1997;272(51):32521–32527. doi: 10.1074/jbc.272.51.32521. [DOI] [PubMed] [Google Scholar]
- 57.Laramee M, Chabot C, Cloutier M, Stenne R, Holgado-Madruga M, Wong AJ, Royal I. The Journal of Biological Chemistry. 2007;282(11):7758–7769. doi: 10.1074/jbc.M611327200. [DOI] [PubMed] [Google Scholar]
- 58.Wu C, Agrawal S, Vasanji A, Drazba J, Sarkaria S, Xie J, Welsch CM, Anand-Apte B, Horowitz A. The Journal of Biological Chemistry. 2011;286:23511–23520. doi: 10.1074/jbc.M111.245209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barbieri MA, Roberts RL, Gumusboga A, Highfield H, Alvarez-Dominguez C, Wells A, Stahl PD. The Journal of Cell Biology. 2000;151(3):539–550. doi: 10.1083/jcb.151.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jopling HM, Odell AF, Hooper NM, Zachary IC, Walker JH, Ponnambalam S. Arteriosclerosis, Thrombosis, and Vascular Biology. 2009;29(7):1119–1124. doi: 10.1161/ATVBAHA.109.186239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stenmark H. Nature Reviews. Molecular Cell Biology. 2009;10(8):513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 62.Cebe Suarez S, Pieren M, Cariolato L, Arn S, Hoffmann U, Bogucki A, Manlius C, Wood J, Ballmer-Hofer K. Cellular and Molecular Life Sciences. 2006;63(17):2067–2077. doi: 10.1007/s00018-006-6254-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cai H, Reed RR. The Journal of Neuroscience. 1999;19(15):6519–6527. doi: 10.1523/JNEUROSCI.19-15-06519.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Naccache SN, Hasson T, Horowitz A. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(34):12735–12740. doi: 10.1073/pnas.0605317103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aschenbrenner L, Lee T, Hasson T. Molecular Biology of the Cell. 2003;14(7):2728–2743. doi: 10.1091/mbc.E02-11-0767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Varsano T, Dong MQ, Niesman I, Gacula H, Lou X, Ma T, Testa JR, Yates III JR, Farquhar MG. Molecular and Cellular Biology. 2006;26(23):8942–8952. doi: 10.1128/MCB.00305-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Karpanen T, Heckman CA, Keskitalo S, Jeltsch M, Ollila H, Neufeld G, Tamagnone L, Alitalo K. The FASEB Journal. 2006;20(9):1462–1472. doi: 10.1096/fj.05-5646com. [DOI] [PubMed] [Google Scholar]
- 68.Koch S, Tugues S, Li X, Gualandi L, Claesson-Welsh L. The Biochemical Journal. 2011;437(2):169–183. doi: 10.1042/BJ20110301. [DOI] [PubMed] [Google Scholar]
- 69.Anderie I, Schulz I, Schmid A. Cellular Signalling. 2007;19(3):582–592. doi: 10.1016/j.cellsig.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 70.Haugsten EM, Malecki J, Bjorklund SM, Olsnes S, Wesche J. Molecular Biology of the Cell. 2008;19(8):3390–3403. doi: 10.1091/mbc.E07-12-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bruns AF, Herbert SP, Odell AF, Jopling HM, Hooper NM, Zachary IC, Walker JH, Ponnambalam S. Traffic. 2010;11(1):161–174. doi: 10.1111/j.1600-0854.2009.01001.x. [DOI] [PubMed] [Google Scholar]
- 72.Chen TT, Luque A, Lee S, Anderson SM, Segura T, Iruela-Arispe ML. The Journal of Cell Biology. 2010;188(4):595–609. doi: 10.1083/jcb.200906044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hasseine LK, Murdaca J, Suavet F, Longnus S, Giorgetti-Peraldi S, Van Obberghen E. Experimental Cell Research. 2007;313(9):1927–1942. doi: 10.1016/j.yexcr.2007.02.034. [DOI] [PubMed] [Google Scholar]
- 74.Rousseau S, Houle F, Landry J, Huot J. Oncogene. 1997;15(18):2169–2177. doi: 10.1038/sj.onc.1201380. [DOI] [PubMed] [Google Scholar]
- 75.He Y, Zhang H, Yu L, Gunel M, Boggon TJ, Chen H, Min W. Science Signaling. 2010;3(116):ra26. doi: 10.1126/scisignal.2000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Czubayko M, Knauth P, Schluter T, Florian V, Bohnensack R. Biochemical and Biophysical Research Communications. 2006;345(3):1264–1272. doi: 10.1016/j.bbrc.2006.04.129. [DOI] [PubMed] [Google Scholar]
- 77.van Kerkhof P, Lee J, McCormick L, Tetrault E, Lu W, Schoenfish M, Oorschot V, Strous GJ, Klumperman J, Bu G. The EMBO Journal. 2005;24(16):2851–2861. doi: 10.1038/sj.emboj.7600756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Manickam V, Tiwari A, Jung JJ, Bhattacharya R, Goel A, Mukhopadhyay D, Choudhury A. Blood. 2011;117(4):1425–1435. doi: 10.1182/blood-2010-06-291690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Choudhury A, Marks DL, Proctor KM, Gould GW, Pagano RE. Nature Cell Biology. 2006;8(4):317–328. doi: 10.1038/ncb1380. [DOI] [PubMed] [Google Scholar]
- 80.Lee S, Chen TT, Barber CL, Jordan MC, Murdock J, Desai S, Ferrara N, Nagy A, Roos KP, Iruela-Arispe ML. Cell. 2007;130(4):691–703. doi: 10.1016/j.cell.2007.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sigismund S, Confalonieri S, Ciliberto A, Polo S, Scita G, Di Fiore PP. Physiological Reviews. 2012;92(1):273–366. doi: 10.1152/physrev.00005.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Murdaca J, Treins C, Monthouel-Kartmann MN, Pontier-Bres R, Kumar S, Van Obberghen E, Giorgetti-Peraldi S. The Journal of Biological Chemistry. 2004;279(25):26754–26761. doi: 10.1074/jbc.M311802200. [DOI] [PubMed] [Google Scholar]
- 83.Giorgetti-Peraldi S, Murdaca J, Mas JC, Van Obberghen E. Oncogene. 2001;20(30):3959–3968. doi: 10.1038/sj.onc.1204520. [DOI] [PubMed] [Google Scholar]
- 84.Singh AJ, Meyer RD, Navruzbekov G, Shelke R, Duan L, Band H, Leeman SE, Rahimi N. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(13):5413–5418. doi: 10.1073/pnas.0700809104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Meyer RD, Srinivasan S, Singh AJ, Mahoney JE, Gharahassanlou KR, Rahimi N. Molecular and Cellular Biology. 2011;31(10):2010–2025. doi: 10.1128/MCB.01006-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Haglund K, Sigismund S, Polo S, Szymkiewicz I, Di Fiore PP, Dikic I. Nature Cell Biology. 2003;5(5):461–466. doi: 10.1038/ncb983. [DOI] [PubMed] [Google Scholar]
- 87.Pitulescu ME, Adams RH. Genes & Development. 2010;24(22):2480–2492. doi: 10.1101/gad.1973910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang HU, Chen ZF, Anderson DJ. Cell. 1998;93(5):741–753. doi: 10.1016/s0092-8674(00)81436-1. [DOI] [PubMed] [Google Scholar]
- 89.Gerety SS, Wang HU, Chen ZF, Anderson DJ. Molecular Cell. 1999;4(3):403–414. doi: 10.1016/s1097-2765(00)80342-1. [DOI] [PubMed] [Google Scholar]
- 90.Meyer G, Varoqueaux F, Neeb A, Oschlies M, Brose N. Neuropharmacology. 2004;47(5):724–733. doi: 10.1016/j.neuropharm.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 91.Pan Q, Chanthery Y, Liang WC, Stawicki S, Mak J, Rathore N, Tong RK, Kowalski J, Yee SF, Pacheco G, Ross S, Cheng Z, Le Couter J, Plowman G, Peale F, Koch AW, Wu Y, Bagri A, Tessier-Lavigne M, Watts RJ. Cancer Cell. 2007;11(1):53–67. doi: 10.1016/j.ccr.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 92.Soker S, Takashima S, Miao HQ, Neufeld G, Klagsbrun M. Cell. 1998;92(6):735–745. doi: 10.1016/s0092-8674(00)81402-6. [DOI] [PubMed] [Google Scholar]
- 93.Prahst C, Heroult M, Lanahan AA, Uziel N, Kessler O, Shraga-Heled N, Simons M, Neufeld G, Augustin HG. The Journal of Biological Chemistry. 2008;283(37):25110–25114. doi: 10.1074/jbc.C800137200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gao Y, Li M, Chen W, Simons M. Journal of Cellular Physiology. 2000;184(3):373–379. doi: 10.1002/1097-4652(200009)184:3<373::AID-JCP12>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 95.Jeanneteau F, Diaz J, Sokoloff P, Griffon N. Molecular Biology of the Cell. 2004;15(2):696–705. doi: 10.1091/mbc.E03-05-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang L, Zeng H, Wang P, Soker S, Mukhopadhyay D. The Journal of Biological Chemistry. 2003;278(49):48848–48860. doi: 10.1074/jbc.M310047200. [DOI] [PubMed] [Google Scholar]
- 97.Soares R, Guo S, Gartner F, Schmitt FC, Russo J. Angiogenesis. 2003;6(4):271–281. doi: 10.1023/B:AGEN.0000029413.32882.dd. [DOI] [PubMed] [Google Scholar]
- 98.Wang L, Mukhopadhyay D, Xu X. The FASEB Journal. 2006;20(9):1513–1515. doi: 10.1096/fj.05-5504fje. [DOI] [PubMed] [Google Scholar]
- 99.Bosanac I, Wertz IE, Pan B, Yu C, Kusam S, Lam C, Phu L, Phung Q, Maurer B, Arnott D, Kirkpatrick DS, Dixit VM, Hymowitz SG. Molecular Cell. 2010;40(4):548–557. doi: 10.1016/j.molcel.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 100.Bechara A, Nawabi H, Moret F, Yaron A, Weaver E, Bozon M, Abouzid K, Guan JL, Tessier-Lavigne M, Lemmon V, Castellani V. The EMBO Journal. 2008;27(11):1549–1562. doi: 10.1038/emboj.2008.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jiang SX, Whitehead S, Aylsworth A, Slinn J, Zurakowski B, Chan K, Li J, Hou ST. The Journal of Biological Chemistry. 2010;285(13):9908–9918. doi: 10.1074/jbc.M109.080689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Herzog B, Pellet-Many C, Britton G, Hartzoulakis B, Zachary IC. Molecular Biology of the Cell. 2011;22(15):2766–2776. doi: 10.1091/mbc.E09-12-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Avraham H, Park SY, Schinkmann K, Avraham S. Cellular Signalling. 2000;12(3):123–133. doi: 10.1016/s0898-6568(99)00076-5. [DOI] [PubMed] [Google Scholar]
- 104.Fantin A, Schwarz Q, Davidson K, Normando EM, Denti L, Ruhrberg C. Development (Cambridge, England) 2011;138(19):4185–4191. doi: 10.1242/dev.070037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Becker PM, Waltenberger J, Yachechko R, Mirzapoiazova T, Sham JS, Lee CG, Elias JA, Verin AD. Circulation Research. 2005;96(12):1257–1265. doi: 10.1161/01.RES.0000171756.13554.49. [DOI] [PubMed] [Google Scholar]
- 106.Murga M, Fernandez-Capetillo O, Tosato G. Blood. 2005;105(5):1992–1999. doi: 10.1182/blood-2004-07-2598. [DOI] [PubMed] [Google Scholar]
- 107.Goel HL, Pursell B, Standley C, Fogarty K, Mercurio AM. Journal of Cell Science. 2012;125(Pt 2):497–506. doi: 10.1242/jcs.094433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gavard J, Gutkind JS. Nature Cell Biology. 2006;8(11):1223–1234. doi: 10.1038/ncb1486. [DOI] [PubMed] [Google Scholar]
- 109.Murakami T, Felinski EA, Antonetti DA. The Journal of Biological Chemistry. 2009;284(31):21036–21046. doi: 10.1074/jbc.M109.016766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.di Blasio L, Droetto S, Norman J, Bussolino F, Primo L. Traffic. 2010;11(8):1107–1118. doi: 10.1111/j.1600-0854.2010.01077.x. [DOI] [PubMed] [Google Scholar]
- 111.Alexander RA, Prager GW, Mihaly-Bison J, Uhrin P, Sunzenauer S, Binder BR, Schutz GJ, Freissmuth M, Breuss JM. Cardiovascular Research. 2012;94(1):125–135. doi: 10.1093/cvr/cvs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Eden G, Archinti M, Furlan F, Murphy R, Degryse B. Current Pharmaceutical Design. 2011;17(19):1874–1889. doi: 10.2174/138161211796718215. [DOI] [PubMed] [Google Scholar]
- 113.Weis S, Shintani S, Weber A, Kirchmair R, Wood M, Cravens A, McSharry H, Iwakura A, Yoon YS, Himes N, Burstein D, Doukas J, Soll R, Losordo D, Cheresh D. The Journal of Clinical Investigation. 2004;113(6):885–894. doi: 10.1172/JCI20702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Carmeliet P, Lampugnani MG, Moons L, Breviario F, Compernolle V, Bono F, Balconi G, Spagnuolo R, Oosthuyse B, Dewerchin M, Zanetti A, Angellilo A, Mattot V, Nuyens D, Lutgens E, Clotman F, de Ruiter MC, Gittenberger-de Groot A, Poelmann R, Lupu F, Herbert JM, Collen D, Dejana E. Cell. 1999;98(2):147–157. doi: 10.1016/s0092-8674(00)81010-7. [DOI] [PubMed] [Google Scholar]
- 115.Grazia Lampugnani M, Zanetti A, Corada M, Takahashi T, Balconi G, Breviario F, Orsenigo F, Cattelino A, Kemler R, Daniel TO, Dejana E. The Journal of Cell Biology. 2003;161(4):793–804. doi: 10.1083/jcb.200209019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shukla AK, Xiao K, Lefkowitz RJ. Trends in Biochemical Sciences. 2011;36(9):457–469. doi: 10.1016/j.tibs.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Almagro S, Durmort C, Chervin-Petinot A, Heyraud S, Dubois M, Lambert O, Maillefaud C, Hewat E, Schaal JP, Huber P, Gulino-Debrac D. Molecular and Cellular Biology. 2010;30(7):1703–1717. doi: 10.1128/MCB.01226-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wallez Y, Cand F, Cruzalegui F, Wernstedt C, Souchelnytskyi S, Vilgrain I, Huber P. Oncogene. 2007;26(7):1067–1077. doi: 10.1038/sj.onc.1209855. [DOI] [PubMed] [Google Scholar]
- 119.Dodane V, Kachar B. The Journal of Membrane Biology. 1996;149(3):199–209. doi: 10.1007/s002329900020. [DOI] [PubMed] [Google Scholar]
- 120.Le Roy C, Wrana JL. Nature Reviews. Molecular Cell Biology. 2005;6(2):112–126. doi: 10.1038/nrm1571. [DOI] [PubMed] [Google Scholar]
- 121.Sundstrom JM, Tash BR, Murakami T, Flanagan JM, Bewley MC, Stanley BA, Gonsar KB, Antonetti DA. Journal of Proteome Research. 2009;8(2):808–817. doi: 10.1021/pr7007913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Furuse M. Cold Spring Harbor Perspectives in Biology. 2010;2(1):a002907. doi: 10.1101/cshperspect.a002907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Caswell PT, Vadrevu S, Norman JC. Nature Reviews. Molecular Cell Biology. 2009;10(12):843–853. doi: 10.1038/nrm2799. [DOI] [PubMed] [Google Scholar]
- 124.Humphries JD, Byron A, Humphries MJ. Journal of Cell Science. 2006;119(Pt 19):3901–3903. doi: 10.1242/jcs.03098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Soldi R, Mitola S, Strasly M, Defilippi P, Tarone G, Bussolino F. The EMBO Journal. 1999;18(4):882–892. doi: 10.1093/emboj/18.4.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wong C, Jin ZG. The Journal of Biological Chemistry. 2005;280(39):33262–33269. doi: 10.1074/jbc.M503198200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ivaska J, Heino J. Annual Review of Cell and Developmental Biology. 2011;27:291–320. doi: 10.1146/annurev-cellbio-092910-154017. [DOI] [PubMed] [Google Scholar]
- 128.Blasi F, Sidenius N. FEBS Letters. 2010;584(9):1923–1930. doi: 10.1016/j.febslet.2009.12.039. [DOI] [PubMed] [Google Scholar]
- 129.Kugler MC, Wei Y, Chapman HA. Current Pharmaceutical Design. 2003;9(19):1565–1574. doi: 10.2174/1381612033454658. [DOI] [PubMed] [Google Scholar]
- 130.Czekay RP, Kuemmel TA, Orlando RA, Farquhar MG. Molecular Biology of the Cell. 2001;12(5):1467–1479. doi: 10.1091/mbc.12.5.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Byzova TV, Goldman CK, Pampori N, Thomas KA, Bett A, Shattil SJ, Plow EF. Molecular Cell. 2000;6(4):851–860. [PubMed] [Google Scholar]
- 132.Mahabeleshwar GH, Feng W, Reddy K, Plow EF, Byzova TV. Circulation Research. 2007;101(6):570–580. doi: 10.1161/CIRCRESAHA.107.155655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.West XZ, Meller N, Malinin NL, Deshmukh L, Meller J, Mahabeleshwar GH, Weber ME, Kerr BA, Vinogradova O, Byzova TV. PloS One. 2012;7(2):e31071. doi: 10.1371/journal.pone.0031071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Palamidessi A, Frittoli E, Garre M, Faretta M, Mione M, Testa I, Diaspro A, Lanzetti L, Scita G, Di Fiore PP. Cell. 2008;134(1):135–147. doi: 10.1016/j.cell.2008.05.034. [DOI] [PubMed] [Google Scholar]
- 135.Morimoto S, Nishimura N, Terai T, Manabe S, Yamamoto Y, Shinahara W, Miyake H, Tashiro S, Shimada M, Sasaki T. The Journal of Biological Chemistry. 2005;280(3):2220–2228. doi: 10.1074/jbc.M406906200. [DOI] [PubMed] [Google Scholar]
- 136.Terai T, Nishimura N, Kanda I, Yasui N, Sasaki T. Molecular Biology of the Cell. 2006;17(5):2465–2475. doi: 10.1091/mbc.E05-09-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mili S, Moissoglu K, Macara IG. Nature. 2008;453(7191):115–119. doi: 10.1038/nature06888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Jeansonne B, Lu Q, Goodenough DA, Chen YH. Cellular and Molecular Biology (Noisy-le-Grand, France) 2003;49(1):13–21. [PubMed] [Google Scholar]
- 139.Hamazaki Y, Itoh M, Sasaki H, Furuse M, Tsukita S. The Journal of Biological Chemistry. 2002;277(1):455–461. doi: 10.1074/jbc.M109005200. [DOI] [PubMed] [Google Scholar]
- 140.Estevez MA, Henderson JA, Ahn D, Zhu XR, Poschmann G, Lubbert H, Marx R, Baraban JM. Journal of Neurochemistry. 2008;106(3):1287–1297. doi: 10.1111/j.1471-4159.2008.05472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Heller B, Adu-Gyamfi E, Smith-Kinnaman W, Babbey C, Vora M, Xue Y, Bittman R, Stahelin RV, Wells CD. The Journal of Biological Chemistry. 2010;285(16):12308–12320. doi: 10.1074/jbc.M109.096230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lindmo K, Stenmark H. Journal of Cell Science. 2006;119(Pt 4):605–614. doi: 10.1242/jcs.02855. [DOI] [PubMed] [Google Scholar]
- 143.Meyer RD, Husain D, Rahimi N. Oncogene. 2011;30(19):2198–2206. doi: 10.1038/onc.2010.597. [DOI] [PMC free article] [PubMed] [Google Scholar]