Abstract
A large proportion of the recoding events mediated by RNA editing are in mRNAs that encode ion channels and transporters. The effects of these events on protein function have been characterized in only a few cases. In even fewer instances are the mechanistic underpinnings of these effects understood. This review focuses on how RNA editing affects protein function and higher order physiology. In mammals, particular attention is given to the GluA2, an ionotropic glutamate receptor subunit, and Kv 1.1, a voltage-dependent K+ channel, because they are particularly well understood. In Kv addition, work on cephalopod K+ channels and Na+/K+-ATPases has also provided important clues on the rules used by RNA editing to regulate excitability. Finally, we discuss some of the emerging targets for editing and how this process may be used to regulate nervous function in response to a variable environment.
Introduction
The recent improvements in DNA sequencing technologies have led to an explosion in the discovery of new RNA editing sites arising from adenosine deamination. Recent reports suggest that there are thousands of RNA editing sites in the human brain transcriptome, although only a small fraction of these are in open reading frames and recode amino acids (Li et al., 2009, 2011; Peng et al., 2012). Editing in invertebrates appears to be even more extensive. A recent analysis of transcriptomes from Drosophila melanogaster at different developmental stages uncovered over 600 recoding events (Graveley et al., 2011), and studies on a handful of individual mRNAs from cephalopods have uncovered close to a hundred such sites (Patton et al., 1997; Rosenthal and Bezanilla, 2002b; Palavicini et al., 2009; Colina et al., 2010; Garrett and Rosenthal, 2012). A disproportionately large number of these edits occur within mRNAs that encode proteins directly involved in excitability. Although it is now clear that RNA editing is modifying the primary sequences of many target proteins, surprisingly few of the editing sites have been characterized on a functional level. Of those that have been studied, the majority lie within mRNAs encoding ion channels and transporters.
This review aims to give the reader an up-to-date accounting of the best studied examples of functional changes caused by editing. First we would like to stress that a full functional characterization of an editing site is a quite difficult undertaking because, to be complete, it must be carried on many different organizational levels. For example, an edit in an ion channel could affect a specific biophysical property, which would then affect the action potential, which could affect a neural circuit, which could then affect a specific behaviour. For the most part, studies to date have focused on the beginning of this sequence, and to greatly different depths. In most cases, studies have been descriptive, defining a physiological property that is altered by RNA editing. In some cases the descriptions have been in exceptional detail, but they are descriptions nonetheless. In two cases the mechanistic underpinnings of specific changes are well understood. Only a few studies have explored the relevance of an editing event on higher order function.
This review focuses most heavily on the functional consequences of editing ion channel transcripts. Ion channels are exceptionally diverse and, for non-aficionados, the jargon associated with their physiology can be arcane. However, all ion channel function shares certain common features, and we will try to focus on these in our discussions. For example, channels open and close, or gate, in response to an extrinsic factor, such as changes in the transmembrane voltage or the concentration of a specific ligand. Voltage-dependent K+ channels are a good example of the former, and ionotropic glutamate receptors of the latter. The kinetics associated with opening and closing, in addition to a channel’s sensitivity to an extrinsic factor, are physiologically relevant characteristics. Once open, all channels permit ions to pass through them in response to an electrochemical gradient. The degree of selectivity for one ion species over another varies greatly between channels and is an important determinant for nerve physiology. Finally, open channels often spontaneously close, even when the extrinsic factor that commands them to stay open is still present. For voltage-dependent channels this process is known as inactivation, and for ligand-gated channels it is known as desensitization. Here too the kinetics of the entry and exit from these processes can be important. Depending on the target, almost all of these physiological characteristics can be modified by RNA editing.
Regulation of excitatory neurotransmission by recoding glutamate receptors
In the mammalian brain most fast excitatory neurotransmission is generated by ionotropic glutamate receptors located in the postsynaptic membrane. When glutamate is released into the synaptic cleft, these receptors open a cation selective pore, allowing extracellular Na+, and sometimes Ca2+, to enter the cell, causing a transient depolarization. Repeated stimulation by their agonist often makes the receptor less sensitive to further stimulation, a process known as desensitization. Because they play such a critical role in generating and shaping synaptic potentials, glutamate receptors are an exceptionally diverse gene family. Early studies identified three basic groups of ionotropic glutamate receptors based on pharmacological differences: α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor (AMPA) receptors, kainate receptors, and N-methyl-d-aspartate (NMDA) receptors. Molecular sequencing has supported this classification. For the purpose of this review we will focus only on AMPA and kainate receptors because their mRNAs are edited, and those encoding NMDA receptors are not as far as we know. In the mammalian brain, AMPA receptors generate the bulk of fast, excitatory neurotransmission. Kainate receptors are thought to play a more subtle, modulatory, role.
Glutamate receptors are tetramers and the individual subunits share a common design (Laube et al., 1998; Rosenmund et al., 1998; Nakagawa et al., 2005; Sobolevsky et al., 2009). The monomer consists of four membrane spans (M1–4). M2, which lines the ion conduction pathway, is atypical in that it does not fully span the membrane, both entering from – and exiting to – the cytoplasm. In mammals, both AMPA and kainate receptors are diverse. There are four AMPA receptors subunits named GluA1–4 using the most up-to-date nomenclature, although they have also been termed GluR-A to D or GluR1–4 in past studies, and their genes are referred to as Gria1–4. These subunits form both homo – and heterotetramers, with different combinations in different brain regions (Keinanen et al., 1990; Sommer et al., 1990). Alternative exon splicing further increases AMPA receptor diversity, generating two forms, known as ‘flip’ and ‘flop’, which have altered sequences in the large extracellular domain immediately before TM4 (Sommer et al., 1990), and have different kinetic properties (Monyer et al., 1991; Lambolez et al., 1996). There are five kainate receptors (GluK1–5), and GluK1 and GluK3 form both homo – and heteromultimers. Thus, for glutamate receptors, there is substantial molecular diversity even before RNA editing enters the equation.
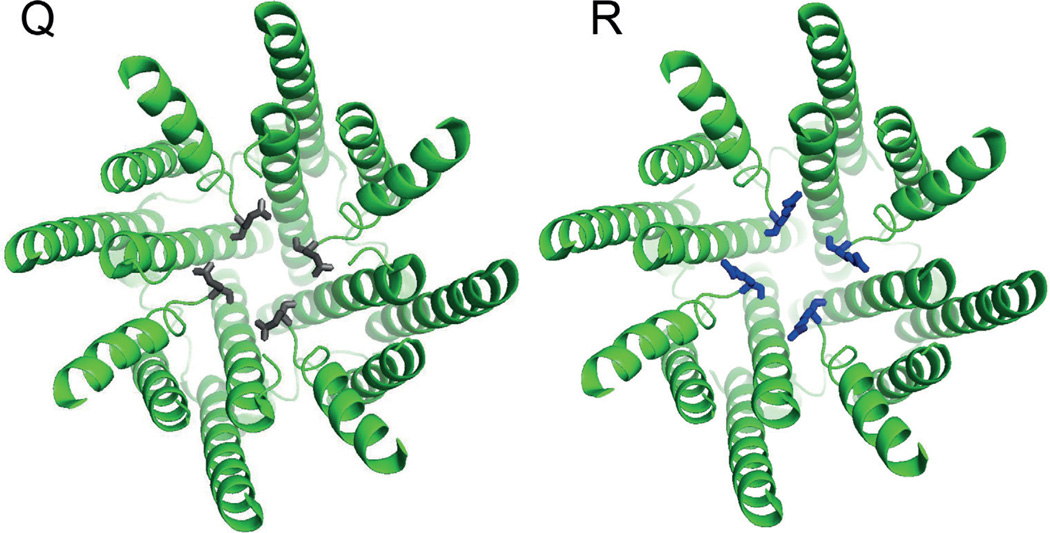
There are numerous editing sites scattered throughout the mRNAs encoding both AMPA and kainate receptors, however, a single site stands above the rest in terms of the thoroughness with which it has been studied, and its importance to the host’s physiology. Editing at codon 586 of GluA2, better known as the Q/R site, recodes a conserved glutamine within M2 to Arginine. This position lies at a critical juncture in the ion conduction pathway (Sommer et al., 1991). In the adult mammalian brain, this site is edited with near-perfect efficiency in GluA2, whereas in GluA1, 3 and 4 the codon remains a Glutamine. An Arginine at this site renders the receptors impermeable to Ca2+ (Figure 1) and decreases their conductance by an order of magnitude (Hollmann et al., 1991; Burnashev et al., 1992). Further mutagenesis of codon 586 demonstrated that the charge of the side chain was the main determinant in abolishing divalent permeability, thus the effect is likely electrostatic, however changes in side chain volume also influenced the relative permeability of Ca²+ and Mg²+. Another interesting feature of the Q/R site in heteromultimers is that arginine’s effect is dominant. Thus, because essentially 100% of GluA2 subunits are edited, we can conclude that this site is a major determinant for keeping Ca²+ from crossing the postsynaptic membrane into the cell during excitatory postsynaptic potentials.
Figure 1.
Two views of the ion conduction pathway of GluA2 with either a glutamine (grey) or a arginine (blue) at codon 586. Structures are from pdb 3KG2 (Sobolevsky et al., 2009). The view shows only the transmembrane spans and is from the inside looking out. As can be seen, the Q/R site sits directly in the ion permeation pathway. The Q/R substitution was generated with Pymol software. The position of the R side chain was not calculated via energy minimization, but merely reflects the most common rotamer for R.
The importance of editing the Q/R site of GluA2 was demonstrated in several elegant studies by Peter Seeburg and colleagues using transgenic mice. First, ADAR2 knockout mice suffered seizures and died soon after birth and Q/R site editing was very low in these animals (Higuchi et al., 2000). Interestingly, in wild-type background, abolishing only editing at the Q/R site by disrupting the ECS structure was also lethal, even when just applied to a single allele (Brusa et al., 1995). This phenotype could be rescued by hardwiring an arginine in codon 586 in the gene, thus obviating the need for editing the Q/R site (Higuchi et al., 2000). The lethality associated with the inability to edit the Q/R site of GluA2 was not a developmental problem. A conditional knock-in of an uneditable GluA2 throughout the adult forebrain also led to severe epileptic seizures and lethality (Krestel et al., 2004). Additionally, expressing different levels of the uneditable GluA2 led to a variety of neurological symptoms, from extreme lethargy to hyperactivity (Feldmeyer et al., 1999). Taken together, these results underscore the importance of editing this site completely.
This idea is reinforced by more recent data relating Q/R editing of GluA2 with disease. In post-mortem studies, under-editing of the Q/R site has been associated with sporadic amyotrophic lateral sclerosis (ALS) (Kawahara et al., 2004). Further studies indicated that the under editing could be attributed to a down-regulation of ADAR2 in motoneurons (Hideyama et al., 2011). Significantly, the reduction in Q/R site editing was modest. Studies on mice showed that ALS-like symptoms, including motoneuron death, could be reproduced by conditional knockouts of ADAR2 in wild-type animals, but not in mice carrying pre-edited versions of GluA2 at the Q/R site (Hideyama et al., 2010). These studies on humans and mice, in conjunction with a recent study that found few deficits in ADAR2-less mice hardwired with an arginine at the Q/R site of GluA2 (Horsch et al., 2011), led to the conclusion that this site is far and away the most important for mammalian physiology. Remarkably, other glutamate receptor mRNAs are also edited at the Q/R site, apparently with less significant consequences.
As with GluA2, the kainate receptors GluK1 and GluK2 are also edited at the Q/R site (Herb et al., 1996; Maas et al., 1996), although far less efficiently (Belcher and Howe, 1997; Paschen et al., 1997; Bernard et al., 1999). In GluK1 and GluK2, the Q/R site causes very similar functional changes, namely a drastic reduction in Ca²+ permeability and a significant reduction in single channel conductance (Egebjerg and Heinemann, 1993; Kohler et al., 1993; Swanson et al., 1996). In addition, editing the Q/R site renders channels insensitive to block by polyamines, but makes them sensitive to block by specific fatty acids such as arachidonic acid (Wilding et al., 2005, 2008, 2010). Thus, Q/R site editing can be used to regulate kainate receptors in a manner that is dependent on other cellular components. GluK2 mRNAs are also edited at two additional sites: codon 567, where an isoleucine is converted to a valine, and codon 571, where a tyrosine is converted to a cysteine (Kohler et al., 1993). Little is known about the physiological effects of these edits, but it was suggested that they fine-tune Ca2+ permeability.
AMPA receptor mRNAs in rats, mice and humans also undergo editing at codon 764, recoding an exonically encoded arginine to a glycine (Lomeli et al., 1994). This edit, at the end of the M3–M4 extracellular loop, occurs in GluA2–4, but not in GluA1. It has a marked and specific effect on receptor physiology; with a glycine at position 764, receptors recover from desensitization twice as quickly. This effect was apparent when receptors were activated by physiologically relevant applications of glutamate in terms of concentration and duration, suggesting that this edit could be important in shaping the magnitudes of excitatory postsynaptic potentials during repetitive firing. The overall physiological importance of editing glutamate receptor mRNAs cannot be understated. However, only in the case of the Q/R site in GluA2 are the consequences of not editing well understood. How the Q/R site in the kainate receptors GluK1–2, and the R/G site in GluA2–4 relate to higher-order physiological processes should prove to be fertile ground for further investigations.
Voltage-activated potassium channels
In neurons, voltage-activated potassium (Kv) channels play important roles in excitability. For example, once an action potential has been initiated and the membrane potential has depolarized, Kv channels are called into action to repolarize the membrane potential back towards the resting potential; therefore, their activity is a critical determinant of the action potential’s shape (Aldrich et al., 1979). More importantly, Kv channels play key roles in setting the firing properties of neurons (Connor and Stevens, 1971). Therefore, changes in Kv channel function can have significant consequences on a neuron’s excitability status. Not surprisingly, Kv channels from both invertebrates and vertebrates have been extensively targeted by the process of RNA editing (Patton et al., 1997; Rosenthal and Bezanilla, 2002b; Hoopengardner et al., 2003; Ryan et al., 2008; Ingleby et al., 2009).
Mammalian Kv1.1 channel inactivation is regulated by editing
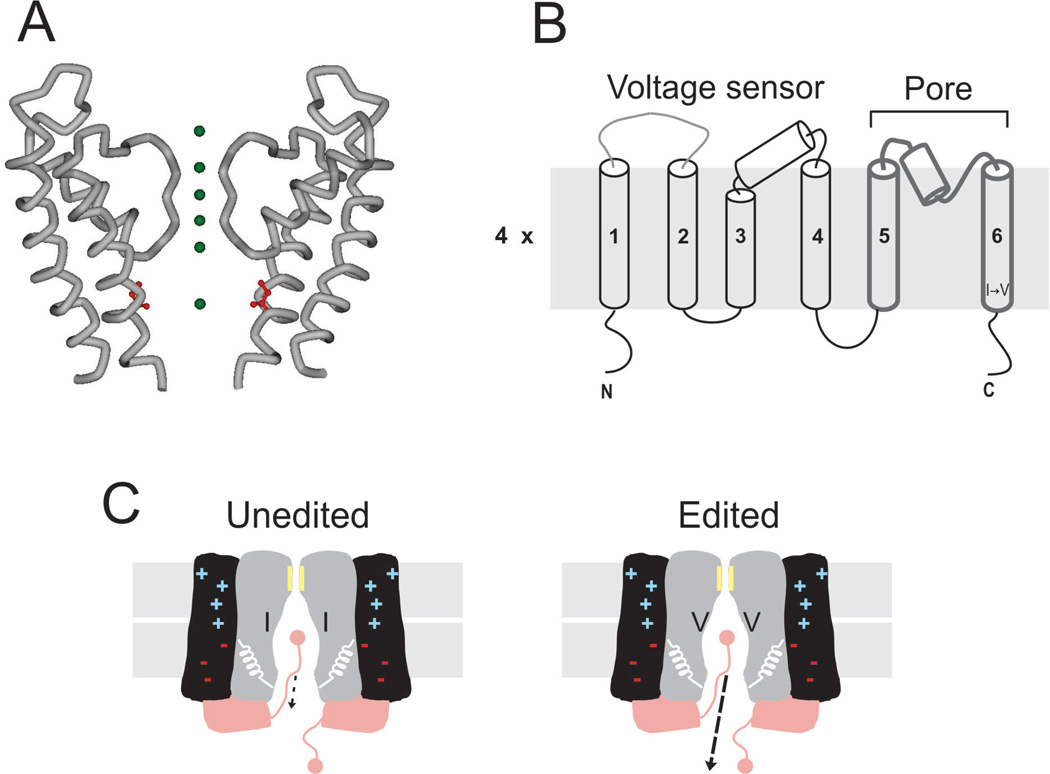
In mammals, RNA editing targets a highly conserved isoleucine located in the lining of the Kv1.1 channel’s permeation pathway and recodes it to valine (I400V). From functional (Liu et al., 1997) and structural work (Long et al., 2005), it is known that the side chain of this position faces the lumen of the ion pathway in a region called the intracellular cavity (Doyle et al., 1998) (Figure 2A). The conversion of I → V at this site is broadly understood. Editing is mediated by ADAR2, which recognizes a 114-nucleotide imperfect inverted repeat hairpin derived entirely from an exonic region (Bhalla et al., 2004). Levels of editing vary between different regions of the nervous system of humans, with high incidence in the spinal cord, medulla and thalamus and comparatively low incidence in cortex, cerebellum or hippocampus (Hoopengardner et al., 2003). Interestingly, a similar pattern was observed in rodents, suggesting that regulation of RNA editing at this site is evolutionarily conserved (Hoopengardner et al., 2003). Another relevant biological feature of this site is that it has also evolved in mRNAs encoding Kv2 channels in squid and fruit fly (Patton et al., 1997; Bhalla et al., 2004).
Figure 2.
RNA editing of mammalian Kv channels. (A) Crystal structure of the last two transmembrane segments from rat Kv1.2 channels (accession ID 2A79). Green spheres represent K+ along the ion permeation pathway. In red is shown the isoleucine edited by ADAR2. (B) Membrane topology of the Kv channel. A functional channel is a tetramer. In each subunit, the first four TM segments form the voltage-sensing domain, while the last two delimit the permeation pathway. The I → V edit is located at the intracellular end of the TM6. (C) Cartoon corresponding to the functional consequences of I → V substitution at the intracellular cavity of Kv channels. RNA editing targets exclusively the unbinding kinetics of the inactivation gate, increasing the off-rate by ∼ 20-fold.
Evolutionary convergence and the rather conspicuous location of this editing site were strong indicators that the I → V conversion would significantly alter functional properties. Generally, Kv channels assemble as homotetramers of α subunits (MacKinnon et al., 1993), each subunit containing a voltage-sensing domain (formed by the first four transmembrane segments) and a pore domain (formed by the last two transmembrane segments and a re-entrant loop between them which includes the selectivity filter; Figure 2B). In response to a depolarization, conformational changes within the voltage sensor drive the opening of an activation gate located at the intracellular end of the permeation pathway, about 7–10Å from the editing site. Mammalian Kv1.1 channels co-assemble with β-subunits to give rise to a macromolecular complex of eight subunits per channel (Gulbis et al., 2000). β-Subunits can confer fast inactivation to mammalian Kv1.1 channels (Rettig et al., 1994), a process in which permeation is shut by an intracellular inactivation gate that binds the ion permeation pathway once the activation gate has opened. Which gating mechanism is altered by the I → V conversion?
Expressing the human Kv1.1 α subunit alone renders voltage-activated K+ currents that do not inactivate, which allows the study of the activation gate mechanism in isolation. Edited and unedited versions of the hKv1.1 α-subunits showed little difference, opening and closing with similar kinetics and voltage dependence (Bhalla et al., 2004). Even though the editing site and the activation gate are just a few angstroms apart, these results are not surprising. For a long time it has been known that quaternary ammonium (QA) ions (∼ 8–10 Å in diameter) can block (but not permeate) at the intracellular end of Kv channels’ permeation pathway (Armstrong, 1966, 1969). Remarkably, if blocked channels are forced to close very quickly, the activation gate can shut the permeation pathway and trap the blocker inside (Armstrong, 1971). Activation gating in the absence and presence of the blocker are quite similar, suggesting that the architecture of the intracellular cavity remains relatively unchanged in open and closed channels (Holmgren et al., 1997). Similarly, the intracellular cavity in the crystal structure of KcsA (a bacterial K+ channel) with bound QA derivatives is almost identical to the cavity of an unbound channel (Zhou et al., 2001).
Co-expressing edited or unedited forms of hKv1.1 α subunits with the hKv1.1 β-subunit conferred, in both cases, fast inactivation to the ionic currents. The simplest kinetic scheme to describe inactivation is:
where C, O and I denote closed, open and inactivated states, respectively. At voltages where the relative probability of opening is maximal (>0mV), inactivation is determined by the on – and off-rate constants. The I → V substitution within the intracellular cavity of hKv1.1 channels specifically targeted the unbinding kinetics (koff), speeding it up by about 20-fold (Bhalla et al., 2004) (Figure 2C). The fact that the inactivation particle for hKv1.1 resides on a β-subunit presented significant complications for further analysing the effects of the I400V edit. For example, the α–β stoichiometry is difficult to control in heterologous expression systems and the hKv1.1 β-Subunit is known to be regulated by phosphorylation. However, the functional consequence of RNA editing could be transferred to Shaker Kv1.1 channels, whose inactivation particle is part of the α subunit. In fact, mimicking the editing event in Shaker produced an identical effect on the unbinding kinetics without changing other properties (Bhalla et al., 2004). This observation turned out to be critical, allowing us to pursue the precise mechanism by which inactivation is altered by RNA editing.
Since isoleucine and valine are both aliphatic amino acids, and the side chain of the edited position is facing the lumen of the pore, it was hypothesized that a reduction in hydrophobicity accompanying a valine substitution was responsible for speeding up the unbinding kinetics. To test this hypothesis, we devised a strategy to change the chemistry at the edited position during an experiment. The Shaker position corresponding to I400 was mutated to cysteine in order to serve as a target for chemical modification with methanethiosulphonate (MTS) reagents. As expected, the more hydrophobic a moiety attached to the edited codon, the slower the unbinding kinetics became (Gonzalez et al., 2011). In other words, once the inactivation particle is bound, hydrophobic interactions determine its off-rate. Such interactions demand close proximity between binding partners. Which specific amino acids of Kv 1.1 interacts with the edited position at the intracellular cavity of the protein?
Fast inactivation in Shaker Kv1.1 channels is a well-understood process. In their pioneering studies, Aldrich and coworkers (Hoshi et al., 1990; Zagotta et al., 1990) discovered that a mild intracellular treatment with trypsin removed the channels’ ability to inactivate. By studying the inactivation properties of a series of deletion mutations at the N-terminus of the protein, they concluded that the inactivation gate is formed by the first ∼ 20–25 amino acids of the protein. They also showed that inactivation is restored in the non-inactivating mutants by exposing the intracellular side of the protein to a synthetic peptide formed by the first 20 amino acids. It was also known that the inactivation gate (N-terminus) is an open channel blocker (Demo and Yellen, 1991) that can compete with intracellular, but not extracellular, QA ions, like tetraethylammonium (Choi et al., 1991). Further, crystallography data show that QA ions bind at the intracellular cavity (Zhou et al., 2001). All these results combined strongly suggest that to inactivate the current, the N-terminus enters the intracellular cavity once the channel has opened.
Is the very tip of the N-terminus entering deeply into the cavity of the channel? To approach this question, we extended our chemical approach by developing channel constructs in which an additional cysteine was substituted at positions near the N-terminus. The expectation was that if the two cysteines were close together in an inactivated channel, a disulfide bond would form, creating a stably blocked channel that required a reducing agent to recover. We made six double cysteine channels with one cysteine at the edited position and the second between positions 2 and 7 at the N-terminus. Only with the construct 2C–470C did we observed an irreversible current reduction when the channels were exposed to an oxidizing environment, indicating that position 2 is the binding partner for the edited codon (Gonzalez et al., 2011). In summary, once valine substitutes for isoleucine, the intracellular cavity of Kv1.1 channels loses an important hydrophobic component to its association with the very tip of the N-terminus, an interaction that determines the off kinetics of the inactivation gate.
For a neuron, what are the implications of editing a Kv1.1 channels’ intracellular cavity? Although as yet untested, we predict it to have a profound impact on excitability. Because this editing site accelerates the inactivation gate’s off rate, a neuron possessing a large proportion of edited channels is poised to have a larger pool of available channels, particularly during periods or repetitive firing. Consequently, RNA editing could play an important role in regulating action potential shape during repetitive firing (Aldrich et al., 1979), as well as helping to set the actual firing frequency of a neuron (Connor and Stevens, 1971). In fact, fast inactivating K+ currents have been shown to play key roles in determining the firing properties in axons and dendrites (Debanne et al., 1997; Hoffman et al., 1997). Additionally, tissue-specific regulation of the RNA editing levels (Hoopengardner et al., 2003; Decher et al., 2010) can have important pharmacological implications when a drug interacts near an editing site (Decher et al., 2010). It turns out that the apparent affinity of several open channel blockers which can be used as therapeutics is significantly reduced by RNA editing (Decher et al., 2010). Similarly, highly unsaturated fatty acids can also bind to the intracellular cavity of Kv1.1 channels and block their currents in a process similar to inactivation. Their apparent affinities are also influenced by RNA editing (Decher et al., 2010).
Cephalopod K+ channels are recoded at unprecedented levels
In mammals, recoding events caused by RNA editing are exceptionally rare. The same cannot be said for cephalopods, where more recoding events have been discovered in mRNAs encoding K+ channels alone than in the entire human brain transcriptome. The first K+ channel mRNA to be analysed in squid encoded a Kv2 subfamily member expressed in the optic lobe, a large ganglion within the central nervous system (Patton et al., 1997). In this mRNA only a short 360 nt region was examined for RNA editing and 12 editing sites were uncovered. These sites changed codons in the voltage-sensing fourth transmembrane span, and also in the helices encoding the pore. Two sites were selected for electrophysiological characterization: Y576C in the pore and I597V in S6. Y576C slowed both the rate of channel closing and slow inactivation. I597V had the opposite effect, increasing both. A subsequent study examined the entire mRNA encoding the squid delayed rectifier of the giant axon, a Kv1 family member (Rosenthal et al., 1996; Rosenthal and Bezanilla, 2002b). Here 14 recoding events were identified, mostly in the channel’s tetramerization domain, and S1 and S3 transmembrane spans. As with squid Kv2, many sites affected closing kinetics, and some also shifted the voltage-dependence of activation. One site in the tetramerization domain (R87G) reduced the α-subunits’ ability to oligomerize into tetramers by drastically reducing their affinity for each other. This effect could be expected to influence overall K+ conductance in the giant axon, a physiological property that is actively regulated between species of squid which inhabit different thermal environments (Rosenthal and Bezanilla, 2002a).
The bewildering array of RNA editing events in squid K+ channels makes us speculate on why these organisms use this process so extensively used to regulate excitability. RNA editing provides an organism with physiological options. In theory, they can choose to edit a specific position when conditions are favourable. RNA editing of the octopus orthologue of squid Kv1 gives some substance to this speculation (Garrett and Rosenthal, 2012). Different octopus species inhabit a tremendous range of temperature environments, from the poles to the equator. As with squid, mRNAs encoding octopus Kv1 are extensively edited. For seven species studied, the gene sequences for the same Kv1 orthologue were virtually identical, however the mRNAs were edited at 19 sites, some shared, some species specific. Although many of these sites affected Kv channel function, one site, which recoded an isoleucine to a valine in the fifth transmembrane span (I321V), doubled the speed of channel closing and the extent to which it was edited correlated closely with the host species’ thermal environment. I321V was edited almost to completion in Arctic and Antarctic species, yet scarcely edited in tropical species, and temperate species edited it to intermediate extents. Single-channel analysis revealed that the edit destabilizes the open ion conducting state, poising the channel on the edge of closing. These results were significant because they were the first to link RNA editing with an environmental factor. Temperature was chosen because it is easy to manipulate and has a direct, predictable effect on channel function. Future studies will help determine whether organisms can rapidly change the extent of RNA editing at specific, functionally relevant sites in acclimation to environmental pressures, or instead whether these sites evolve over generations by evolutionary adaptation. The relevant environmental pressure, and the molecular mechanisms underlying adaption or acclimation, should prove interesting for further studies.
Drosophila Kv channels are also edited extensively
mRNAs for Kv channels, including shaker, the most intensively studied channel in history, are edited in Drosophila (Ryan et al., 2008; Ingleby et al., 2009). These sites have a variety of functional effects, including changes to activation, deactivation and inactivation kinetics, and some small shifts to the channels’ voltage sensitivity. In shaker, there are four editing sites, giving rise to 16 different combinations of edited isoforms (Hoopengardner et al., 2003). Interestingly, the frequency of occurrence of these isoforms varied between different parts of the adult’s anatomy (Ingleby et al., 2009). It was also found that editing at different sites was linked, and the linkage varied between different regions. These editing patterns made a difference on a functional level. An interesting feature of editing in Shaker is that the physiological properties of different editing isoforms could not be predicted by the effects of the individual editing sites. This phenomenon, known as functional epistasis, means that the effect of editing a specific site is context dependent, greatly expanding the possibilities for regulation. On top of this, the shaker locus encodes numerous functionally different splice variants (Papazian et al., 1987; Tempel et al., 1987; Schwarz et al., 1988; Timpe et al., 1988), and the functional effects of editing in most of these has yet to be explored.
Voltage-activated calcium channels
Voltage-activated calcium (Cav) channels are broadly expressed in excitable cells, where they play diverse functions. In some muscle cells, like the pacemaker myocytes in the mammalian sinoatrial node, Cav channels are essential to the repetitive electrical activity of these cells (Mangoni et al., 2006). However, because intracellular Ca2+ is a second messenger, Cav channels are also used as an entryway for Ca2+ to regulate diverse cellular processes, from gene expression to motion (Hille, 2001). The primary sequence of the Cav channel pore-forming α-subunit contains four repeated domains (Tanabe et al., 1987; Catterall, 2000), each containing a voltage-sensing domain and a pore domain. Presumably, these repeats form a functional unit that should be similar to the assembly of the four individual α subunits of Kv channels (Long et al., 2005). Using genomic approaches in Drosophila, several editing sites have been identified in Cav channels (Hoopengardner et al., 2003; Graveley et al., 2011). At present we have no clues on the biological or physiological significance of these sites.
Recently, three RNA editing sites were discovered in mRNAs encoding rodent Cav1.3 channels, clustered in four contiguous amino acids located in the cytoplasmic C-terminus (i.e. after the fourth transmembrane repeat) (Huang et al., 2012). These amino acids (IQDY; in bold are represented the amino acids recoded by RNA editing) are part of a calmodulin (CaM)-binding domain. The Cav1.3-CaM–Ca2+ complex influences the gating properties of the channel, particularly the Ca2+-dependent inactivation kinetics (Shen et al., 2006; Yang et al., 2006; Huang et al., 2012).
I → M, Q → R and Y → C are edited to different extents, but none by more than ∼50%. Editing of this region is restricted to the central nervous system, with high levels of activity in the frontal cortex, hippocampus, medulla oblongata and cerebellum. Using a proteomic approach, the authors were able to identify peptides containing amino acid substitutions, demonstrating that all three possible conversions exist at the protein level. Further support for multiple variants was provided at the mRNA level, where seven out of the eight possible combinations of editing sites were identified. The most abundant variants were MQDY and MQDC. In ADAR2−/−/GluR-BR/R knockout mice (Higuchi et al., 2000) all three editing sites were absent. What are the functional consequences of these editing events?
Upon membrane depolarization, Cav1.3 channels open a gate that allows Ca2+ to flow into the cell, producing an inward current. Even if the depolarization is maintained, the Ca2+ current will inactivate. Kinetically, CDI is quite different than fast inactivation in Kv channels. On the one hand, the rate of inactivation depends on the levels of intracellular Ca2+ (Brehm and Eckert, 1978; Tillotson, 1979), a property which led this process to be named Ca2+-dependent inactivation (CDI). On the other hand, Cav channels enter inactivation from the closed state (Tadross et al., 2010). It has been previously shown that mutating the first two editing sites (IQ) to alanine almost completely abolishes CDI (Yang et al., 2006). Therefore, CDI was examined in all single RNA editing conversions (Huang et al., 2012). Only I → M and Q → R produced a substantial decrease in the levels of CDI, and the effect was additive when they both were edited (IQ → MR).
By transfecting primary hippocampal neurons with wild-type and edited channels, it was shown that all constructs reached the cell surface equally well (Huang et al., 2012), providing confidence that any potential consequence of editing on an animal’s behaviour would likely originate from the changes in channel function and not their density at the cell membrane. To explore the consequences of editing in neurons, the authors turned to a comparative study using wild-type and ADAR2−/−/GluR-BR/R knockout mice. As a first attempt, they selected neurons from the suprachiasmatic nucleus (SCN), which are known to contain Cav1.3 channels involved in the electrical rhythmic activity that governs circadian rhythms (Pennartz et al., 2002). SCN neurons do indeed edit the three sites, but to relatively low levels (<20%). Remarkably, Ca²+ currents recorded from neurons taken from knockout animals showed noticeably more inactivation, consistent with the expected lack of edited channel variants. SCN neurons spontaneously fire action potentials (Inouye and Kawamura, 1979; Reppert and Weaver, 2001). L-type Ca²+ channels (Cav1 family) play an essential role in setting the frequency of the spontaneous firing (Pennartz et al., 2002). Because neurons from knockout mice would have, at steady-state, fewer conducting Ca²+ channels available, they would be expected to fire at lower frequencies than neurons from wild-type animals, and this was found to be the case (Huang et al., 2012).
Ion pumps and transporters
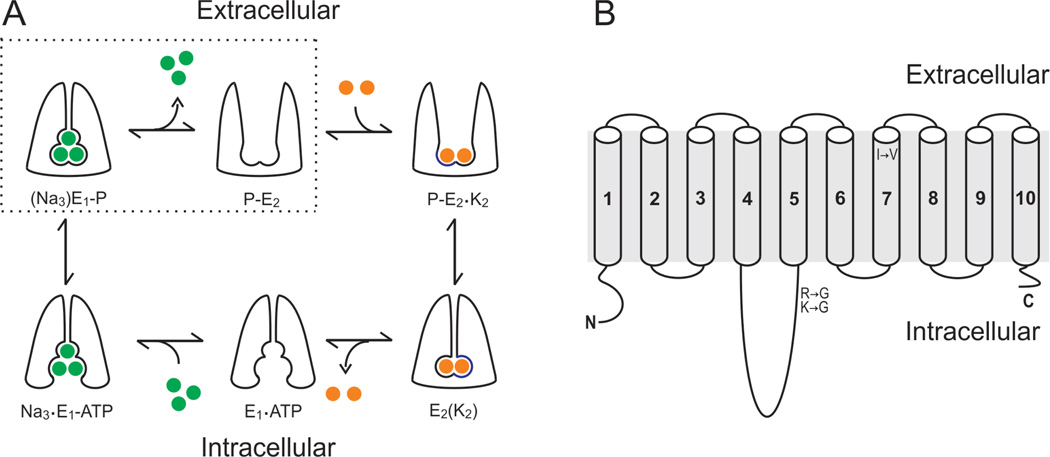
In contrast to ion channels, which allow ions to permeate at rates near diffusion, pumps and transporters do so at rates many orders of magnitude slower. This difference arises from the fundamental necessity of pumps and transporters to occlude, or trap, their substrates at some point during the transport process. For example, the Na+/K+-ATPase uses the energy derived from the hydrolysis of ATP to transport three Na+ out of the cell and import two K+, at rates of ∼100/s. Both, Na+ and K+ are transported against their electrochemical gradients, so pumps cannot afford the cost of having a continuous ion permeation pathway at any time. Figure 3A shows a schematic view of the transport mechanism of the Na+/K+-ATPase. The gates that control access of the ions to the inside or outside of the cell alternate opening. The extent to which the function of pumps and transporters are tuned by RNA editing appears to be substantially smaller than in ion channels. At present, editing sites have been identified in the Na+/K+-ATPase and the Na+/Ca²+–K+ exchanger in Drosophila (Stapleton et al., 2006; Graveley et al., 2011) and the Na+/ K+-ATPase of squid (Colina et al., 2010). Only for the squid Na+/K+-ATPase, however, do we have information on what these editing sites do. Interestingly, as discussed in detail below, RNA editing has targeted the kinetics of Na+ occlusion to speed up the pump’s transport velocity (Colina et al., 2010).
Figure 3.
RNA editing of squid Na+/K+-ATPase. (A) Albers–Post transport cycle. Green and orange spheres represent Na+ and K+, respectively. The alternate gating mechanism with occlusion states for Na+ (top-left corner) and K+ (bottom-right corner) are essentials for transporting these ions against their electrochemical gradients. Dotted lines enclose the states associated with the extracellular release of Na+, which were isolated by removing K+ from all solutions and the absence of intracellular ADR (B) Membrane topology of the α Na+/ K+-ATPase subunit. Afunctional Na+/K+-ATPase is formed by the co assembly of α – and β-subunits. The α-subunit, however, contains all requirements for transport: ATP binding domain, phosphorylation site and permeation pathway. Two amino acid substitutions by RNA editing were identified in the phosphorylation domain located within the large intracellular loop between TM segments 4 and 5. An additional site was identified at the extracellular end of TM segment 7.
Squid pump mRNAs are edited in four sites, targeting three different amino acids. Positions R663 and K666 are in the phosphorylation domain, located within the large intracellular loop between transmembrane (TM) segments 4 and 5 (Figure 3B). R663 had a single editing site, converting it to glycine. Both adenosines of the K666 codon (AAG) were found to be edited, giving rise to the possibility of three conversions: K → R, G or E. K → E, however, was never observed in any region of the squid nervous system. The fourth editing event is an I → V conversion at amino acid 877, which lies at the external end of the seventh TM segment (Figure 3B). R663G is consistently edited at percentages >65% throughout the squid’s nervous system. The other sites, however, showed high spatial regulation, suggesting that they might be edited differently depending on the demands of different types of neurons. What are the functional consequences of these editing events on the pump’s function?
The function of the Na+/K+-ATPase is to maintain the Na+ and K+ gradients between the inside and outside of the cell. How effectively the pump can do its job depends, ultimately, on its turnover rate (i.e. the number of transport cycles the pump can undergo per unit of time). Because the pump moves an unequal number of ions per cycle, it generates an electrical current (IP) that can be measured and used as a faithful metre of its turnover rate. We showed that the maximal turnover rates of the exonic and all edited versions of the pump were similar, suggesting that RNA editing does not target the rate limiting step when pumps are running at full speed. The pump’s transport velocity, however, is voltage dependent, reaching a maximum at voltages >∼0mV and decreasing monotonically at negative potentials. Therefore, from a cell’s perspective, the relevant issue is the pump’s speed at the resting membrane potential (∼−80mV), where it spends most of its time. We studied the voltage dependence of IP in all constructs and observed that I877V pumps were able to pump faster at negative potentials, i.e. the IP versus voltage curve was shifted towards more negative potentials. What is the mechanism of this change?
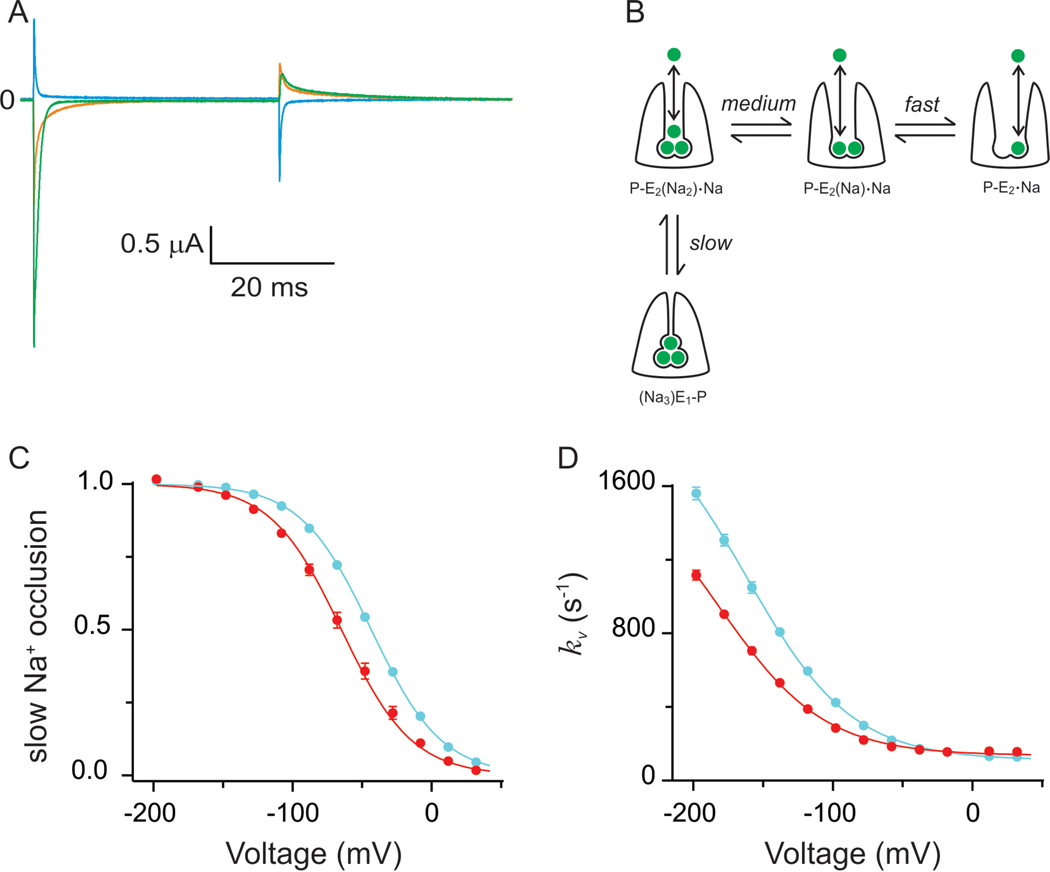
Before Na+ ions are released to the external bulk solution, they must travel through a narrow access channel (Gadsby et al., 1993; Hilgemann, 1994; Holmgren et al., 2000), along which there is a voltage drop. Therefore, negative potentials will drive external Na+ back to their binding sites, slowing down the turnover rate, as observed experimentally (Gadsby et al., 1985; Gadsby and Nakao, 1989; Nakao and Gadsby, 1989; Sagar and Rakowski, 1994). In order to acquire precise mechanistic information about the transitions involving the binding/release of external Na+ it is necessary to isolate them (Figure 3A, enclosed by dashed lines). We achieved these conditions by removing K+ from both the intracellular and extracellular solutions, while maintaining internal Na+ and ATP, but not ADP. We also used the cut-open oocyte voltage-clamp technique, which allowed us access to both sides of the membrane (Taglialatela et al., 1992). Figure 4A shows examples of pump-mediated currents obtained in response to voltage steps from 0mV to −198mV (green), −78 mV (orange) and 42 mV (brown). As expected, the pumpmediated currents have only transient components, which represent the redistribution of the Na+-bound states upon the fast voltage change. Clearly, these transient currents have multiple components. A careful dissection shows that the transient currents contain a fast (τ<100µs), medium (τ∼0.2–0.5ms) and a slow (τT∼1–10ms) component, which led us to propose that they represent the sequential and distinct interactions of each Na+ with the pump (Holmgren et al., 2000; Gadsby et al., 2012), as shown in Figure 4B. By estimating the amount of charge moved by the slow component (Qs) at many voltages, we can infer the voltage dependence of the steady-state occupancy of the deeply occluded state (Figure 4C). For genomic (filled cyan circles) and edited pumps (filled red circles), the distributions followed a Boltzmann function with the same steepness. However, RNA editing shifted the voltage dependence towards more negative potentials (Figure 4C). This means that at resting membrane potentials, an edited pump will less likely be in the deep occluded state, therefore favouring forward pumping. Does RNA editing directly influence the kinetics of the slow component? To address this question we studied the voltage dependence of the relaxation rates of the slow component (Figure 4D). The solid lines through the data represent the best fit to:
which describes the occlusion (kb) and deocclusion (Kb) rates as voltage independent, and the voltage dependence arising from the binding/ unbinding reaction with an apparent affinity (K0.5(0)) value at 0 mV. F, R and T are constants with their usual meaning and values. The parameter λ denotes the fractional ‘electrical depth’ of the access channel. In this model, the asymptote at positive potentials corresponds to kf. As observed in Figure 4D, RNA editing does not influence kf; in both edited and unedited pumps it is ∼ 100/s. The asymptote at negative potentials is the sum of kf and kb. Even though we cannot reach the voltages needed to observe the asymptotic value, we can confidently estimate it because λ was independently determined to be 0.67 for edited and unedited pumps. From the fits, kb showed a small change from ∼2000/s to ∼1600/s for unedited and edited pumps, respectively. These results suggest that the kinetics of the occlusion/deocclusion transition remain largely unaltered by RNA editing. The most significant change predicted by the model was a substantial reduction of the apparent affinity in edited pumps. K0.5(0) was ∼7.5M for genomic pumps, as compared with ∼13M for the I877V pumps. What could be the underlying reason for this effect?
Figure 4.
RNA editing of squid Na+/K+-ATPase: functional consequences. (A) Transient currents mediated by the Na+/K+-ATPase trapped in states linked to release of external Na+. Green, orange and cyan current traces represent pump currents in response to fast voltage steps from 0mV to −198, −78 and +42mV, respectively. It is noticeable the multi component nature of these pump-mediated transient currents. (B) Cartoon representing the kinetic scheme that best describes the transient currents shown in (A). In this model, each of the Na+ binding/release and occlusion/deocclusion transitions are kinetically distinct. The slow component symbolizes the deep occlusion transition, which is the rate limiting step of the transient pump currents. (C) Voltage distribution of the deep occlusion transition. Cyan and red circles represent deep occlusion of unedited (I877) and edited (V877) pumps. Solid lines represent Boltzmann fits. RNA editing shifted the distribution towards more negative potentials which translate in faster pump velocities at potentials near the resting state of a neuron. (D) Voltage dependence of the relaxation rate of the deep occlusion transition. Same colour scheme as C. Solid lines correspond to ‘access channel model’ fits. Edited pumps reduced their apparent affinity for Na+ binding to ∼13M from a value of ∼7M in unedited pumps.
From Na+/K+-ATPase models based on the Ca2+-ATPase crystal structure (Ogawa and Toyoshima, 2002) and Na+/K+-ATPase crystal structures (Morth et al., 2007; Shinoda et al., 2009) we know that the cation binding sites are far away from position I877, so it is unlikely that a valine substitution would directly alter the way Na+ binds. Another possibility would be that electrostatics influence the apparent affinity (K0.5(0)) for external Na+. In fact, this mechanism has been used by nature to help Na+/K+-ATPases from marine osmoconformers to adapt to the high concentrations of Na+ found in sea water (Colina et al., 2007). But I and V are both aliphatic amino acids, so it is also unlikely that they affect electrostatics. A more likely explanation is that the relative contribution of each component at different voltages is different between genomic and edited pumps. By quantifying the relative amounts of the slow, medium and fast charge components at many voltages, we discovered a clear shift in their distributions caused by the I877V edit: the fast component showed a greater fractional representation at the expense of the slow component.
Our studies clearly demonstrate that RNA editing tunes the Na+/K+-ATPase’s transport velocity. Interestingly, the I877V editing site is spatially regulated through the squid’s nervous system, suggesting that RNA editing regulates the specific Na+ homeostasis demands of different types of neurons. As we observed with both human and octopus K+ channels, the removal of a single methyl group by recoding I→V can produce a large effect, presumably because nature has selected key positions to edit. Many interesting questions remain concerning editing of the squid Na+/K+-ATPase. Mechanistically, how does this small change in primary structure at position 877 affect the larger-scale movements associated with Na+ release? What are the functional consequences of editing codons R663 and K666? Finally, how do these edits relate to the physiological demands of different neurons?
Emerging targets for RNA editing
Besides the examples listed thus far, the functional effects of a handful of editing sites are just beginning to be explored. For example, an isoleucine can be recoded to a methionine in the third transmembrane span of the human GABA-gated Cl− channel α3 subunit (Ohlson et al., 2007). A methionine at this site causes the multimeric channel to activate more slowly and deactivate more rapidly. It also largely abolished outward rectification of the current-voltage relationship, suggesting that the site also affects ion selectivity (Rula et al., 2008). The mechanisms underlying these effects are largely unknown. In Drosophila, a GABA-gated CI− channel is also edited, increasing the channel’s sensitivity to its agonist (Jones et al., 2009). A recent study on the Drosophila developmental transcriptome has predicted more than 600 recoding events due to adenosine deamination, and more than 100 of these occur in ion channels, transporters or the machinery for synaptic vesicle release (Graveley et al., 2011). Recent efforts on the human transcriptome have uncovered many new sites that have yet to be characterized. Clearly, much work remains before we get a clear understanding of how RNA editing is used to regulate animal physiology in general, and excitability in particular.
References
- Aldrich RW, Jr, Getting PA, Thompson SH. Mechanism of frequency-dependent broadening of molluscan neurone soma spikes. J. Physiol. 1979;297:531–544. doi: 10.1113/jphysiol.1979.sp012829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong CM. Time course of TEA -induced anomalous rectification in squid giant axons. J. Gen. Physiol. 1966;50:491–503. doi: 10.1085/jgp.50.2.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong CM. Inactivation of the potassium conductance and related phenomena caused by quaternary ammonium ion injection in squid axons. J. Gen. Physiol. 1969;54:553–575. doi: 10.1085/jgp.54.5.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong CM. Interaction of tetraethylammonium ion derivatives with the potassium channels of giant axons. J. Gen. Physiol. 1971;58:413–437. doi: 10.1085/jgp.58.4.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belcher SM, Howe JR. Characterization of RNA editing of the glutamate-receptor subunits GluR5 and GluR6 in granule cells during cerebellar development. Brain Res. Mol. Brain Res. 1997;52:130–138. doi: 10.1016/s0169-328x(97)00252-0. [DOI] [PubMed] [Google Scholar]
- Bernard A, Ferhat L, Dessi F, Charton G, Represa A, Ben-Ari Y, Khrestchatisky M. Q/R editing of the rat GluR5 and GluRδ kainate receptors in vivo and in vitro: evidence for independent developmental, pathological and cellular regulation. Eur. J. Neurosci. 1999;11:604–616. doi: 10.1046/j.1460-9568.1999.00479.x. [DOI] [PubMed] [Google Scholar]
- Bhalla T, Rosenthal JJ, Holmgren M, Reenan R. Control of human potassium channel inactivation by editing of a small mRNA hairpin. Nat. Struct. Mol. Biol. 2004;11:950–956. doi: 10.1038/nsmb825. [DOI] [PubMed] [Google Scholar]
- Brehm P, Eckert R. Calcium entry leads to inactivation of calcium channel in Paramecium. Science. 1978;202:1203–1206. doi: 10.1126/science.103199. [DOI] [PubMed] [Google Scholar]
- Brusa R, Zimmermann F, Koh DS, Feldmeyer D, Gass P, Seeburg PH, Sprengel R. Earlyonset epilepsy and postnatal lethality associated with an editing-deficient GluR-B allele in mice. Science. 1995;270:1677–1680. doi: 10.1126/science.270.5242.1677. [DOI] [PubMed] [Google Scholar]
- Burnashev N, Monyer H, Seeburg PH, Sakmann B. Divalent ion permeability of AMPA receptor channels is dominated by the edited form of a single subunit. Neuron. 1992;8:189–198. doi: 10.1016/0896-6273(92)90120-3. [DOI] [PubMed] [Google Scholar]
- Catterall WA. Structure and regulation of voltagegated Ca²+ channels. Annu. Rev. Cell. Dev. Biol. 2000;16:521–555. doi: 10.1146/annurev.cellbio.16.1.521. [DOI] [PubMed] [Google Scholar]
- Choi KL, Aldrich RW, Yellen G. Tetraethylammonium blockade distinguishes two inactivation mechanisms in voltage-activated K+ channels. Proc. Natl. Acad. Sci. U.S.A. 1991;88:5092–5095. doi: 10.1073/pnas.88.12.5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colina C, Rosenthal JJ, Degiorgis JA, Srikumar D, Iruku N, Holmgren M. Structural basis of Na+/K+-ATPase adaptation to marine environments. Nat. Struct. Mol. Biol. 2007;14:427–431. doi: 10.1038/nsmb1237. [DOI] [PubMed] [Google Scholar]
- Colina C, Palavicini JP, Srikumar D, Holmgren M, Rosenthal JJ. Regulation of Na+/K+ ATPase transport velocity by RNA editing. PLoS Biol. 2010;8:e1000540. doi: 10.1371/journal.pbio.1000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor JA, Stevens CF. Prediction of repetitive firing behaviour from voltage clamp data on an isolated neurone soma. J. Physiol. 1971;213:31–53. doi: 10.1113/jphysiol.1971.sp009366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debanne D, Guerineau NC, Gahwiler BH, Thompson SM. Action-potential propagation gated by an axonal I(A)-like K+ conductance in hippocampus. Nature. 1997;389:286–289. doi: 10.1038/38502. [DOI] [PubMed] [Google Scholar]
- Decher N, Streit AK, Rapedius M, Netter MF, Marzian S, Ehling P, Schlichthorl G, Craan T, Renigunta V, Kohler A, et al. RNA editing modulates the binding of drugs and highly unsaturated fatty acids to the open pore of Kv potassium channels. EMBO J. 2010;29:2101–2113. doi: 10.1038/emboj.2010.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demo SD, Yellen G. The inactivation gate of the Shaker K+ channel behaves like an open-channel blocker. Neuron. 1991;7:743–753. doi: 10.1016/0896-6273(91)90277-7. [DOI] [PubMed] [Google Scholar]
- Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- Egebjerg J, Heinemann SF. Ca²+ permeability of unedited and edited versions of the kainate selective glutamate receptor GluR6. Proc. Natl. Acad. Sci. U.S.A. 1993;90:755–759. doi: 10.1073/pnas.90.2.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmeyer D, Kask K, Brusa R, Kornau HC, Kolhekar R, Rozov A, Burnashev N, Jensen V, Hvalby O, Sprengel R, et al. Neurological dysfunctions in mice expressing different levels of the Q/R site-unedited AMPAR subunit GluR-B. Nat. Neurosci. 1999;2:57–64. doi: 10.1038/4561. [DOI] [PubMed] [Google Scholar]
- Gadsby DC, Nakao M. Steady-state current- voltage relationship of the Na/K pump in guinea pig ventricular myocytes. J. Gen. Physiol. 1989;94:511–537. doi: 10.1085/jgp.94.3.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadsby DC, Kimura J, Noma A. Voltage dependence of Na/K pump current in isolated heart cells. Nature. 1985;315:63–65. doi: 10.1038/315063a0. [DOI] [PubMed] [Google Scholar]
- Gadsby DC, Rakowski RF, De Weer P. Extracellular access to the Na,K pump: pathway similar to ion channel. Science. 1993;260:100–103. doi: 10.1126/science.7682009. [DOI] [PubMed] [Google Scholar]
- Gadsby DC, Bezanilla F, Rakowski RF, De Weer P, Holmgren M. The dynamic relationships between the three events that release individual Na+ ions from the Na+/K+-ATPase. Nat. Commun. 2012;3:669. doi: 10.1038/ncomms1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett S, Rosenthal JJ. RNA editing underlies temperature adaptation in K+ channels from polar octopuses. Science. 2012;335:848–851. doi: 10.1126/science.1212795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez C, Lopez-Rodriguez A, Srikumar D, Rosenthal JJ, Holmgren M. Editing of human K(V)1.1 channel mRNAs disrupts binding of the N-terminus tip at the intracellular cavity. Nat. Commun. 2011;2:436. doi: 10.1038/ncomms1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, et al. The developmental transcriptome of Drosophila melanogaster . Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulbis JM, Zhou M, Mann S, MacKinnon R. Structure of the cytoplasmic beta subunit-T1 assembly of voltage-dependent K+ channels. Science. 2000;289:123–127. doi: 10.1126/science.289.5476.123. [DOI] [PubMed] [Google Scholar]
- Herb A, Higuchi M, Sprengel R, Seeburg PH. Q/R site editing in kainate receptor GluR5 and GluR6 pre-mRNAs requires distant intronic sequences. Proc. Natl. Acad. Sci. U.S.A. 1996;93:1875–1880. doi: 10.1073/pnas.93.5.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hideyama T, Yamashita T, Suzuki T, Tsuji S, Higuchi M, Seeburg PH, Takahashi R, Misawa H, Kwak S. Induced loss of ADAR2 engenders slow death of motor neurons from Q/R site-unedited GluR2. J. Neurosci. 2010;30:11917–11925. doi: 10.1523/JNEUROSCI.2021-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hideyama T, Yamashita T, Aizawa H, Tsuji S, Kakita A, Takahashi H, Kwak S. Profound down-regulation of the RNA editing enzyme ADAR2 in ALS spinal motor neurons. Neurobiol. Dis. 2011;45:1121–1128. doi: 10.1016/j.nbd.2011.12.033. [DOI] [PubMed] [Google Scholar]
- Higuchi M, Maas S, Single FN, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R, Seeburg PH. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA e enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- Hilgemann DW. Channel-like function of the Na,K pump probed at microsecond resolution in giant membrane patches. Science. 1994;263:1429–1432. doi: 10.1126/science.8128223. [DOI] [PubMed] [Google Scholar]
- Hille B. Ion Channels of Excitable Membranes. 3rd. Suderland, MA: Sinauer Associates, Inc; 2001. [Google Scholar]
- Hoffman DA, Magee JC, Colbert CM, Johnston D. K+ channel regulation of signal propagation in dendrites of hippocampal pyramidal neurons. Nature. 1997;387:869–875. doi: 10.1038/43119. [DOI] [PubMed] [Google Scholar]
- Hollmann M, Hartley M, Heinemann S. Ca²+ permeability of KA-AMPA—gated glutamate receptor channels depends on subunit composition. Science. 1991;252:851–853. doi: 10.1126/science.1709304. [DOI] [PubMed] [Google Scholar]
- Holmgren M, Smith PL, Yellen G. Trapping of organic blockers by closing of voltage-dependent K+ channels: evidence for a trap door mechanism of activation gating. J. Gen. Physiol. 1997;109:527–535. doi: 10.1085/jgp.109.5.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren M, Wagg J, Bezanilla F, Rakowski RF, De Weer P, Gadsby DC. Three distinct and sequential steps in the release of sodium ions by the Na+/K+-ATPase. Nature. 2000;403:898–901. doi: 10.1038/35002599. [DOI] [PubMed] [Google Scholar]
- Hoopengardner B, Bhalla T, Staber C, Reenan R. Nervous system targets of RNA editing identified by comparative genomics. Science. 2003;301:832–836. doi: 10.1126/science.1086763. [DOI] [PubMed] [Google Scholar]
- Horsch M, Seeburg PH, Adler T, Aguilar-Pimentel JA, Becker L, Calzada-Wack J, Garrett L, Gotz A, Hans W, Higuchi M, et al. Requirement of the RNA editing enzyme ADAR2 for normal physiology in mice. J. Biol. Chem. 2011;286:18614–18622. doi: 10.1074/jbc.M110.200881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshi T, Zagotta WN, Aldrich RW. Biophysical and molecular mechanisms of Shaker potassium channel inactivation. Science. 1990;250:533–538. doi: 10.1126/science.2122519. [DOI] [PubMed] [Google Scholar]
- Huang H, Tan BZ, Shen Y, Tao J, Jiang F, Sung YY, Ng CK, Raida M, Kohr G, Higuchi M, et al. RNA editing of the IQ domain in Ca(v)1.3 channels modulates their Ca²+-dependent inactivation. Neuron. 2012;73:304–316. doi: 10.1016/j.neuron.2011.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingleby L, Maloney R, Jepson J, Horn R, Reenan R. Regulated RNA editing and functional epistasis in Shaker potassium channels. J. Gen. Physiol. 2009;133:17–27. doi: 10.1085/jgp.200810133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye ST, Kawamura H. Persistence of circadian rhythmicity in a mammalian hypothalamic ‘island’ containing the suprachiasmatic nucleus. Proc. Natl. Acad. Sci. U.S.A. 1979;76:5962–5966. doi: 10.1073/pnas.76.11.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AK, Buckingham SD, Papadaki M, Yokota M, Sattelle BM, Matsuda K, Sattelle DB. Splice-variant - and stage-specific RNA editing of the Drosophila GABA receptor modulates agonist potency. J. Neurosci. 2009;29:4287–4292. doi: 10.1523/JNEUROSCI.5251-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Ito K, Sun H, Aizawa H, Kanazawa I, Kwak S. Glutamate receptors: RNA editing and death of motor neurons. Nature. 2004;427:801. doi: 10.1038/427801a. [DOI] [PubMed] [Google Scholar]
- Keinanen K, Wisden W, Sommer B, Werner P, Herb A, Verdoorn TA, Sakmann B, Seeburg PH. A family of AMPA-selective glutamate receptors. Science. 1990;249:556–560. doi: 10.1126/science.2166337. [DOI] [PubMed] [Google Scholar]
- Kohler M, Burnashev N, Sakmann B, Seeburg PH. Determinants of Ca²+ permeability in both TM1 and TM2 of high affinity kainate receptor channels: diversity by RNA editing. Neuron. 1993;10:491–500. doi: 10.1016/0896-6273(93)90336-p. [DOI] [PubMed] [Google Scholar]
- Krestel HE, Shimshek DR, Jensen V, Nevian T, Kim J, Geng Y, Bast T, Depaulis A, Schonig K, Schwenk F, et al. A genetic switch for epilepsy in adult mice. J. Neurosci. 2004;24:10568–10578. doi: 10.1523/JNEUROSCI.4579-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambolez B, Ropert N, Perrais D, Rossier J, Hestrin S. Correlation between kinetics and RNA splicing of alpha-amino-3-hydroxy-5methylisoxazole-4-propionic acid receptors in neocortical neurons. Proc. Natl. Acad. Sci. U.S.A. 1996;93:1797–1802. doi: 10.1073/pnas.93.5.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laube B, Kuhse J, Betz H. Evidence for a tetrameric structure of recombinant NMDA receptors. J. Neurosci. 1998;18:2954–2961. doi: 10.1523/JNEUROSCI.18-08-02954.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JB, Levanon EY, Yoon JK, Aach J, Xie B, Leproust E, Zhang K, Gao Y, Church GM. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009;324:1210–1213. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- Li M, Wang IX, Li Y, Bruzel A, Richards AL, Toung JM, Cheung VG. Widespread RNA and DNA sequence differences in the human transcriptome. Science. 2011;333:53–58. doi: 10.1126/science.1207018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Holmgren M, Jurman ME, Yellen G. Gated access to the pore of a voltage-dependent K+ channel. Neuron. 1997;19:175–184. doi: 10.1016/s0896-6273(00)80357-8. [DOI] [PubMed] [Google Scholar]
- Lomeli H, Mosbacher J, Melcher T, Hoger T, Geiger JR, Kuner T, Monyer H, Higuchi M, Bach A, Seeburg PH. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science. 1994;266:1709–1713. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- Maas S, Melcher T, Herb A, Seeburg PH, Keller W, Krause S, Higuchi M, O’Connell MA. Structural requirements for RNA editing in glutamate receptor pre-mRNAs by recombinant doublestranded RNA adenosine deaminase. J. Biol. Chem. 1996;271:12221–12226. doi: 10.1074/jbc.271.21.12221. [DOI] [PubMed] [Google Scholar]
- MacKinnon R, Aldrich RW, Lee AW. Functional stoichiometry of Shaker potassium channel inactivation. Science. 1993;262:757–759. doi: 10.1126/science.7694359. [DOI] [PubMed] [Google Scholar]
- Mangoni ME, Couette B, Marger L, Bourinet E, Striessnig J, Nargeot J. Voltagedependent calcium channels and cardiac pacemaker activity: from ionic currents to genes. Prog. Biophys. Mol. Biol. 2006;90:38–63. doi: 10.1016/j.pbiomolbio.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Monyer H, Seeburg PH, Wisden W. Glutamate-operated channels: developmentally early and mature forms arise by alternative splicing. Neuron. 1991;6:799–810. doi: 10.1016/0896-6273(91)90176-z. [DOI] [PubMed] [Google Scholar]
- Morth JP, Pedersen BP, Toustrup-Jensen MS, Sorensen TL, Petersen J, Andersen JP, Vilsen B, Nissen P. Crystal structure of the sodium-potassium pump. Nature. 2007;450:1043–1049. doi: 10.1038/nature06419. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Cheng Y, Ramm E, Sheng M, Walz T. Structure and different conformational states of native AMPA receptor complexes. Nature. 2005;433:545–549. doi: 10.1038/nature03328. [DOI] [PubMed] [Google Scholar]
- Nakao M, Gadsby DC. [Na] and [K] dependence of the Na/K pump current-voltage relationship in guinea pig ventricular myocytes. J. Gen. Physiol. 1989;94:539–565. doi: 10.1085/jgp.94.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa H, Toyoshima C. Homology modeling of the cation binding sites of Na+K+-ATPase. Proc. Natl. Acad. Sci. U.S.A. 2002;99:15977–15982. doi: 10.1073/pnas.202622299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlson J, Pedersen JS, Haussler D, Ohman M. Editing modifies the GABA(A) receptor subunit alpha3. RNA. 2007;13:698–703. doi: 10.1261/rna.349107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palavicini JP, O’Connell MA, Rosenthal JJ. An extra double-stranded RNA binding domain confers high activity to a squid RNA editing enzyme. RNA. 2009;15:1208–1218. doi: 10.1261/rna.1471209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papazian DM, Schwarz TL, Tempel BL, Jan YN, Jan LY. Cloning of genomic and complementary DNA from Shaker, a putative potassium channel gene from Drosophila . Science. 1987;237:749–753. doi: 10.1126/science.2441470. [DOI] [PubMed] [Google Scholar]
- Paschen W, Schmitt J, Gissel C, Dux E. Developmental changes of RNA editing of glutamate receptor subunits GluR5 and GluR6: in vivo versus in vitro . Brain Res. Dev. Brain Res. 1997;98:271–280. doi: 10.1016/s0165-3806(96)00193-9. [DOI] [PubMed] [Google Scholar]
- Patton DE, Silva T, Bezanilla F. RNA editing generates a diverse array of transcripts encoding squid K+2 K+ channels with altered functional properties. Neuron. 1997;19:711–722. doi: 10.1016/s0896-6273(00)80383-9. [DOI] [PubMed] [Google Scholar]
- Peng Z, Cheng Y, Tan BC, Kang L, Tian Z, Zhu Y, Zhang W, Liang Y, Hu X, Tan X, et al. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat. Biotechnol. 2012;30:253–260. doi: 10.1038/nbt.2122. [DOI] [PubMed] [Google Scholar]
- Pennartz CM, de Jeu MT, Bos NP, Schaap J, Geurtsen AM. Diurnal modulation of pacemaker potentials and calcium current in the mammalian circadian clock. Nature. 2002;416:286–290. doi: 10.1038/nature728. [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. Molecular analysis of mammalian circadian rhythms. Annu. Rev. Physiol. 2001;63:647–676. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- Rettig J, Heinemann SH, Wunder F, Lorra C, Parcej DN, Dolly JO, Pongs O. Inactivation properties of voltage-gated K+ channels altered by presence of beta-subunit. Nature. 1994;369:289–294. doi: 10.1038/369289a0. [DOI] [PubMed] [Google Scholar]
- Rosenmund C, Stern-Bach Y, Stevens CF. The tetrameric structure of a glutamate receptor channel. Science. 1998;280:1596–1599. doi: 10.1126/science.280.5369.1596. [DOI] [PubMed] [Google Scholar]
- Rosenthal JJ, Bezanilla F. A comparison of propagated action potentials from tropical and temperate squid axons: different durations and conduction velocities correlate with ionic conductance levels. J. Experi. Biol. 2002a;205:1819–1830. doi: 10.1242/jeb.205.12.1819. [DOI] [PubMed] [Google Scholar]
- Rosenthal JJ, Bezanilla F. Extensive editing of mRNAs for the squid delayed rectifier K+ channel regulates subunit tetramerization. Neuron. 2002b;34:743–757. doi: 10.1016/s0896-6273(02)00701-8. [DOI] [PubMed] [Google Scholar]
- Rosenthal JJ, Vickery RG, Gilly WF. Molecular identification of SqKv1A. A candidate for the delayed rectifier K channel in squid giant axon. J. Gen. Physiol. 1996;108:207–219. doi: 10.1085/jgp.108.3.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rula EY, Lagrange AH, Jacobs MM, Hu N, Macdonald RL, Emeson RB. Developmental modulation of GABA(A) receptor function by RNA editing. J. Neurosci. 2008;28:6196–6201. doi: 10.1523/JNEUROSCI.0443-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan MY, Maloney R, Reenan R, Horn R. Characterization of five RNA editing sites in Shab potassium channels. Channels (Austin) 2008;2:202–209. doi: 10.4161/chan.2.3.6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagar A, Rakowski RF. Access channel model for the voltage dependence of the forwardrunning Na+/K+ pump. J. Gen. Physiol. 1994;103:869–893. doi: 10.1085/jgp.103.5.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz TL, Tempel BL, Papazian DM, Jan YN, Jan LY. Multiple potassium-channel components are produced by alternative splicing at the Shaker locus in Drosophila . Nature. 1988;331:137–142. doi: 10.1038/331137a0. [DOI] [PubMed] [Google Scholar]
- Shen Y, Yu D, Hiel H, Liao P, Yue DT, Fuchs PA, Soong TW. Alternative splicing of the Ca(v)1.3 channel IQ domain, a molecular switch for Ca²+-dependent inactivation within auditory hair cells. J. Neurosci. 2006;26:10690–10699. doi: 10.1523/JNEUROSCI.2093-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinoda T, Ogawa H, Cornelius F, Toyoshima C. Crystal structure of the sodium-potassium pump at 2.4 A resolution. Nature. 2009;459:446–450. doi: 10.1038/nature07939. [DOI] [PubMed] [Google Scholar]
- Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer B, Keinanen K, Verdoorn TA, Wisden W, Burnashev N, Herb A, Kohler M, Takagi T, Sakmann B, Seeburg PH. Flip and flop: a cell-specific functional switch in glutamate-operated channels of the CNS. Science. 1990;249:1580–1585. doi: 10.1126/science.1699275. [DOI] [PubMed] [Google Scholar]
- Sommer B, Kohler M, Sprengel R, Seeburg PH. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991;67:11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- Stapleton M, Carlson JW, Celniker SE. RNA editing in Drosophila melanogaster: New targets and functional consequences. RNA. 2006;12:1922–1932. doi: 10.1261/rna.254306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson GT, Feldmeyer D, Kaneda M, Cull-Candy SG. Effect of RNA editing and subunit co-assembly single-channel properties of recombinant kainate receptors. J. Physiol. 1996;492(Pt 1):129–142. doi: 10.1113/jphysiol.1996.sp021295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadross MR, Ben Johny M, Yue DT. Molecular endpoints of Ca²+/calmodulin - and voltage-dependent inactivation of Ca(v)1.3 channels. J. Gen. Physiol. 2010;135:197–215. doi: 10.1085/jgp.200910308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taglialatela M, Toro L, Stefani E. Novel voltage clamp to record small, fast currents from ion channels expressed in Xenopus oocytes. Biophys. J. 1992;61:78–82. doi: 10.1016/S0006-3495(92)81817-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe T, Takeshima H, Mikami A, Flockerzi V, Takahashi H, Kangawa K, Kojima M, Matsuo H, Hirose T, Numa S. Primary structure of the receptor for calcium channel blockers from skeletal muscle. Nature. 1987;328:313–318. doi: 10.1038/328313a0. [DOI] [PubMed] [Google Scholar]
- Tempel BL, Papazian DM, Schwarz TL, Jan YN, Jan LY. Sequence of a probable potassium channel component encoded at Shaker locus of Drosophila . Science. 1987;237:770–775. doi: 10.1126/science.2441471. [DOI] [PubMed] [Google Scholar]
- Tillotson D. Inactivation of Ca conductance dependent on entry of Ca ions in molluscan neurons. Proc. Natl. Acad. Sci. U.S.A. 1979;76:1497–1500. doi: 10.1073/pnas.76.3.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpe LC, Schwarz TL, Tempel BL, Papazian DM, Jan YN, Jan LY. Expression of functional potassium channels from Shaker cDNA in Xenopus oocytes. Nature. 1988;331:143–145. doi: 10.1038/331143a0. [DOI] [PubMed] [Google Scholar]
- Wilding TJ, Zhou Y, Huettner JE. Q/R site editing controls kainate receptor inhibition by membrane fatty acids. J. Neurosci. 2005;25:9470–9478. doi: 10.1523/JNEUROSCI.2826-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilding TJ, Fulling E, Zhou Y, Huettner JE. Amino acid substitutions in the pore helix of GluR6 control inhibition by membrane fatty acids. J. Gen. Physiol. 2008;132:85–99. doi: 10.1085/jgp.200810009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilding TJ, Chen K, Huettner JE. Fatty acid modulation and polyamine block of GluK2 kainate receptors analyzed by scanning mutagenesis. J. Gen. Physiol. 2010;136:339–352. doi: 10.1085/jgp.201010442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang PS, Alseikhan BA, Hiel H, Grant L, Mori MX, Yang W, Fuchs PA, Yue DT. Switching of Ca²+-dependent inactivation of Ca(v)1.3 channels by calcium binding proteins of auditory hair cells. J. Neurosci. 2006;26:10677–10689. doi: 10.1523/JNEUROSCI.3236-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagotta WN, Hoshi T, Aldrich RW. Restoration of inactivation in mutants of Shaker potassium channels by a peptide derived from ShB. Science. 1990;250:568–571. doi: 10.1126/science.2122520. [DOI] [PubMed] [Google Scholar]
- Zhou M, Morais-Cabral JH, Mann S, MacKinnon R. Potassium channel receptor site for the inactivation gate and quaternary amine inhibitors. Nature. 2001;411:657–661. doi: 10.1038/35079500. [DOI] [PubMed] [Google Scholar]