Abstract
Discovering small-molecule modulators of protein–protein interactions is a challenging task because of both the generally noncontiguous, large protein surfaces that form these interfaces and the shortage of high-throughput approaches capable of identifying such rare inhibitors. We describe here a robust and flexible methodology that couples disruption of protein–protein complexes to host cell survival. The feasibility of this approach was demonstrated through monitoring a small-molecule-mediated protein–protein association (FKBP12–rapamycin–FRAP) and two cases of dissociation (homodimeric HIV-1 protease and heterodimeric ribonucleotide reductase). For ribonucleotide reductase, we identified cyclic peptide inhibitors from genetically encoded libraries that dissociated the enzyme subunits. A solid-phase synthetic strategy and peptide ELISAs were developed to characterize these inhibitors, resulting in the discovery of cyclic peptides that operate in an unprecedented manner, thus highlighting the strengths of a functional approach. The ability of this method to process large libraries, coupled with the benefits of a genetic selection, allowed us to identify rare, uniquely active small-molecule modulators of protein–protein interactions at a frequency of less than one in 10 million.
Many regulatory processes in living organisms are often a consequence of specific protein–protein contacts, and interference with such interactions provides a means to exert control over cellular events. The de novo discovery of small molecules capable of disrupting such protein–protein complexes has been fraught with challenges, yielding very few inhibitors at a low success rate (1, 2, 3). These difficulties suggest that large, functionally diverse libraries might be essential for finding unique molecules that are capable of perturbing the intracellular levels of specific protein–protein interactions. The major challenge in sifting through such vast compound pools is the shortage of functional high-throughput assays for detection of the protein complex dissociation (4).
Genetic selection is uniquely capable of identifying individual molecules with desired properties from large libraries by using whole cells as reporters and correlating host growth to a desired functional property. Unlike recently popularized affinity-based selections (5), an intracellular genetic selection can directly assay for effects on enzymatic activity or the modulation of a protein–protein complex, thus bypassing the inherent limitations of in vitro approaches. Additionally, library members must function within the context of the entire host proteome, requiring positive candidates to have an enhanced level of selectivity for their target. This feature represents an important advantage over traditional screen-based methods in drug discovery by allowing both target affinity and selectivity to be simultaneously optimized. The application of a genetic selection to the identification of small-molecule modulators may yield both potent and selective activities as well as unique modes of action.
To develop such a selection, we integrated two existing technologies to pioneer a systematic method for discovering these small-molecule modulators. Protein complexation is monitored with two-hybrid technology, constructed originally for the discovery and characterization of protein–protein interactions in vivo (6). This method relies on linking protein complex formation to the expression of reporter genes, whose regulation can be monitored through chromogenic assays or host survival. The traditional forward design of various two-hybrid systems can be altered to couple cell growth to the disruption of protein complexes, an approach referred to as the reverse two-hybrid system (RTHS) (7, 8). As demonstrated previously with a small-molecule screen (9) and an aptamer-based selection (10), the RTHS presents a unique opportunity for functional discovery of inhibitors of protein–protein interactions. In our design, the RTHS is cocompartmentalized in host cells with genetically encoded small-molecule libraries, which allows the coupling of all system components to DNA encoding. The libraries are produced by using split intein-mediated circular ligation of peptides and proteins (SICLOPPS) technology, developed in our laboratory for intracellular synthesis of cyclic peptides (11, 12). The cyclization renders the peptides resistant to cellular catabolism and at the same time restricts conformational freedom, stabilizing the functional presentation of the peptide and potentially improving the binding affinity for target sites. We reasoned that interfacing SICLOPPS with the RHTS would create an innovative approach for the systematic identification of small molecules that modulate protein–protein interactions.
Materials and Methods
Culture Media and Growth Conditions. Antibiotics were provided at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 50 μg/ml; kanamycin, 50 μg/ml; spectinomycin, 50 μg/ml; and tetracycline, 20 μg/ml. For chromosomal markers, concentrations of antibiotics were reduced 2-fold. Minimal media A supplemented with 0.5% glycerol and 1 mM MgSO4 was used for all genetic selections (13).
Genetic Selections. SICLOPPS libraries were transformed into Escherichia coli strains containing integrated reporter and repressor constructs. Transformants were washed with minimal media A and plated on minimal media supplemented with 2 × 10-4% l-(+)-arabinose and 3-amino-1,2,4-triazole, kanamycin, and isopropyl β-d-thiogalactoside (IPTG), with respective concentrations determined for optimal stringency (see Supporting Materials and Methods, which is published as supporting information on the PNAS web site). After incubation at 37°C for 3–4 days, surviving colonies were restreaked onto the same media with and without arabinose. Plasmids from selected strains whose growth depended on the presence of arabinose were retransformed into the original selection strain and checked for phenotype retention. The variable insert regions on SICLOPPS plasmids were PCR-amplified, and their DNA sequence was determined.
Cyclic Peptide Synthesis. We reacted 69 mg (0.65 mmol) of 3-mercaptopropionic acid with 179 mg (0.81 mmol) of 2-Aldrithiol (Aldrich) in 500 μlof N,N-dimethylformamide (DMF), and the completion of the reaction was monitored by the release of 2-thiopyridone (λmax = 353 nm, ε = 8,080 M-1·cm-1). The reaction product was then coupled in situ with ≈0.325 mmol of amino polyethylene glycol acrylamide copolymer resin (PEGA, Nova Biochem) by using 125 mg (0.65 mmol) of 1-ethyl-3-(3′-dimethylaminopropyl)carbodiimide, 112 mg (0.98 mmol) of N-hydroxysuccinimide, and 210 mg (1.63 mmol) of N,N-diisopropylethylamine per 1 g of resin. Loading of the resulting disulfide resin (≈0.23 mmol/g) was established by displacing 2-thiopyridone with an excess of cysteine. A 0.006-mmol aliquot of the resin was incubated with cysteine-containing peptides (0.012 mmol) in 500 μl of DMF, and the progress of the peptide attachment was again monitored spectrophotometrically. Immobilized peptide was cyclized in the presence of 3.5 mg (0.018 mmol) of 1-ethyl-3-(3′-dimethylaminopropyl)carbodiimide and 4.9 mg (0.036 mmol) of 1-hydroxy-7-azabenzotriazole in 650 μl of DMF, and the progress of the cyclization was monitored by Kaiser assay (14). Finally, reductive cleavage with 17.2 mg (0.06 mmol) of tris-2-carboxyethylphosphine in 1 ml of 50% aqueous DMF released cyclic peptides from the resin. Crude peptide mixtures were subjected to reverse-phase chromatography [Partisil C-18 Magnum 9 (length, 50 cm; particle size, 10 μm) ODS-3 columns, Whatman] on a Waters HPLC system by using a water/acetonitrile gradient with 0.1% trifluoroacetic acid. Final peptide concentrations were determined with Ellman's reagent, and cyclic peptide yields ranged 20–71%. Mass analysis was performed on a Mariner mass spectrometer (PerSeptive Biosystems, Framingham, MA) (see Table 1, which is published as supporting information on the PNAS web site).
ELISA Methods. Two variations of solid-phase binding assays were used for analyzing the binding of peptide inhibitors to ribonucleotide reductase (RR) subunits: (i) protein competition ELISA in which peptides were competing with mR1 for binding to immobilized mR2 and (ii) binding and competition ELISA with covalently immobilized ligands. In general, the solid-phase assays were performed in microtiter plates (MaxiSorp, Nunc) or strip units (Reacti-Bind Maleimide Activated Clear Strip Plates, Pierce) involving continuous agitation by a Junior Orbit Shaker (Lab-Line Instruments) at a medium speed during all of the incubation steps. Sample volumes were 100 μl, unless specified otherwise. After coating was performed, a blocking step was conducted by incubating preloaded wells with 5% BSA in PBS for 1 h at room temperature. Wash procedures between any two successive incubations involved three washes with 200 μl of 0.5% Tween 20 in PBS, with the second wash involving a 5-min incubation. The detection of His-tagged proteins was performed with nickel·nitrilotriacetic acid-horseradish peroxidase (Ni·NTA-HRP) conjugate (Qiagen, Valencia, CA) and monitored by an absorbance change at 405 nm with 2,2′-azinobis[3-ethylbenzothiazoline-6-sulfonic acid] as a substrate.
Dissociative mR2·mR1 ELISA. Competition ELISA was performed with mR2 coated overnight (at 4°C) onto MaxiSorp 96-well microtiter plates at a concentration of 50 μg/ml in 50 mM carbonate-bicarbonate buffer (pH 9.6). After the blocking step, the wells were exposed to undersaturating amounts of His-tagged mR1 (typically 0.06 μM) with or without inhibitors. Retained mR1 was detected by the Ni·NTA-HRP conjugate.
Peptide Binding/Competition ELISA. Peptides (2 nmol per well) in 10% DMF/50 mM Tris·HCl (pH 7.5) with 1 mM tris-2-carboxyethylphosphine were reacted for 2 h at room temperature with maleimide-derivatized polystyrene wells. The unreacted sites were blocked by incubating wells with 5 μM cysteine in 50 mM Tris·HCl (pH 7.5) for 30 min. After the washing and blocking steps, the wells were incubated with His-tagged mR1 or mR2 in the presence or absence of inhibitors. The retained protein was detected with the Ni·NTA-HRP conjugate.
Results and Discussion
Design Strategy. Given our successful implementation of SICLOPPS in E. coli (11, 12) and our desire for high-throughput capacity, we designed a bacterial version of the RTHS that would function in parallel with SICLOPPS. As depicted in Fig. 1, our design was based on the bacteriophage regulatory circuit (15), linked to a positive genetic selection, which should be less likely to yield false positives resulting from RTHS-independent effects on growth rates. The RTHS design adapted elements from several reported bacterial systems to create a robust, flexible, and tunable genetic selection for molecules that modulate protein–protein interactions. The key features of this system are as follows: (i) chimeric repressors to monitor heterodimeric interactions (16); (ii) two conditionally selective reporters, HIS3 (17, 18) (imidazole glycerol phosphate dehydratase) and KanR (aminoglycoside 3′-phosphotransferase for kanamycin resistance), to amplify selection stringency and allow chemical tunability; and (iii) LacZ (β-galactosidase) for quantitative measurements of protein–protein interactions. Further details on plasmid designs (Fig. 5, which is published as supporting information on the PNAS web site), reporter constructions, and system implementation published are provided as Fig. 6 and Supporting Results, which are published as supporting information on the PNAS web site.
Fig. 1.
Schematic representation of the RTHS. (A) The expression of protein fusions containing DNA-binding domains is induced with IPTG, and they associate to repress a promoter that directs expression of three reporter genes: (i) HIS3, imidazole glycerol phosphate dehydratase; (ii) KanR (Kan), aminoglycoside 3′-phosphotransferase; (iii) lacZ, β-galactosidase. The formation of protein complexes inhibits growth on minimal media by blocking HIS3 expression, and residual background expression can be further adjusted with 3-amino-1,2,4-triazole (3-AT; competitive inhibitor of imidazole glycerol phosphate dehydratase) and kanamycin. The final reporter, β-galactosidase, quantitatively reports on the level of repression. (B) A small-molecule modulator capable of inhibiting the protein–protein interaction rescues growth by inducing HIS3 and KanR expression.
The ability of our RTHS to report on protein complex formation was investigated with a number of model systems. We used the wild-type 434 repressor protein and 434 repressor DNA-binding domain fusions with the Saccharomyces cerevisiae GCN4 leucine zipper and HIV-1 protease to monitor homodimeric interactions. We also used fusions with murine RR subunits to assay a heterodimeric complex. As shown in Fig. 7, which is published as supporting information on the PNAS web site, the fusion constructs repressed the lacZ reporter ≈4- to 9-fold over the unrepressed controls, a dynamic range typical of other repressor-based systems (16, 19). The chemical modulation of a protein–protein interaction in our RTHS was demonstrated with the FK506-binding protein (FKBP12) and FKBP12–rapamycin-associated protein (FRAP) pairing, whose dimerization depends on the presence of rapamycin (20), a naturally occurring chemical dimerizer. Cell growth and β-galactosidase assays demonstrated that rapamycin was taken up by E. coli, triggering the assembly of a functional repressor composed of the heterologously expressed FKBP12 and FRAP fusions (Fig. 7B). As expected, rapamycin functioned in a concentration-dependent manner (Fig. 2A), and similarly, varying the expression levels of FKBP12 and FRAP at fixed rapamycin concentrations correlated with the levels of β-galactosidase activity (data not shown). An IC50 of 209 ± 31 nM can be calculated from the repression of the RTHS (Fig. 2B), although this value is a composite of several events, including rapamycin membrane permeability. The ability to modulate our RTHS with small molecules and the satisfactory dynamic range suggest that this methodology could serve as a means to discover molecules interfering with protein–protein contacts.
Fig. 2.
The rapamycin-dependent association of the FKBP12–FRAP proteins. (A) Drops of rapamycin solution were applied at 1, 3, 10, 30, 100, and 300 μM concentrations on cell lawns containing integrated FKBP12–FRAP fusions to visualize the effect on cell growth on media containing kanamycin. Additional experiments demonstrated that the effect is due to the RTHS, indicating that rapamycin is not deleterious to E. coli at these concentrations (data not shown). (B) β-Galactosidase activity as a function of rapamycin concentration (IC50 = 209 ± 31 nM).
Control Peptide Inhibitors. Our initial RTHS efforts were focused on homodimeric HIV-1 protease and heterodimeric RR, whose enzymatic activities are known to depend on subunit association. These enzymes are attractive targets because of their importance in HIV infection (21) and cancer proliferation (22), respectively, as well as their long-term standing in multidisciplinary efforts to disrupt both homodimeric and heterodimeric protein–protein interfaces (1, 3). Perturbation of such complexes has been proposed as a superior alternative to chemotherapies targeting the active sites of enzymes, because of the intrinsically higher specificities and lower resistance frequencies associated with this approach (23). Toward this goal, both enzyme complexes were probed with known linear peptidic inhibitors. These peptides are a C-terminal-derived hexapeptide (TVSYEL) for HIV protease that inhibits the essential β-sheet interactions (24), and a heptapeptide (FTLDADF) for RR that competes with binding between subunits mR1 and mR2 (25). Gratifyingly, when coexpressed with target fusions, the inhibitor peptides and not scrambled controls relieved repression of the lacZ reporter (Fig. 8, which is published as supporting information on the PNAS web site). Furthermore, a mock selection with the RR heptapeptide control resulted in its exclusive retrieval from the inhibitor-expressing strains (see supporting information). The results of both preliminary tests confirm the advantages provided by our RTHS design, such as (i) positive selection format, (ii) amplified dynamic range, and (iii) chemical tunability, laying the groundwork for the identification of inhibitors from libraries.
Selection of Cyclic Peptide Modulators of RR. With the success of the mock selection, we exposed RR fusions to SICLOPPS libraries with the intent to discover cyclic peptides acting as dissociative inhibitors. Predictions from modeling studies (26) suggested that the reverse turn conformations of known complex disruptors should be well represented within libraries containing constrained scaffolds. A library, encoding hexapeptides with five random residues and an invariable cysteine as a cyclization nucleophile, was transformed into the RTHS E. coli strain expressing RR fusions. Approximately 108 transformants were plated on selective media (histidine-free minimal media supplemented with 3-amino-1,2,5-triazole and kanamycin) at a density of 107 per plate (100 × 15 mm), from libraries containing 108 individual plasmids. Plates were incubated until readily identifiable colonies (approximately one in 5 × 105 for RR) could be collected and processed further to confirm a relationship between the observed growth advantage and SICLOPPS expression. The low frequency of positives underscores the challenges inherent to the discovery of modulators of protein–protein interactions, an undertaking that has been aptly described as “genuinely difficult” (1).
To focus on the most promising library members and eliminate false positives, several filtering steps were developed that enabled rapid and convenient assessment of candidates. Aberrant selectants could arise because of the possibility of damage to the expression of SICLOPPS or the RR fusions, and they were detected by (i) the failure of IPTG, the inducer for the repressor hybrids, to inhibit growth (Fig. 9A, which is published as supporting information on the PNAS web site); (ii) the inability of arabinose, the inducer for SICLOPPS expression, to improve growth (Fig. 9B); and (iii) the inability to confirm the expected phenotype upon retransformation of the plasmids. Of the 108 total transformants, 262 potential candidates were processed, 24 of which passed each filtering test, resulting in ≈90% of the isolates being false positives (data not shown). Further ranking of these 24 candidates was achieved by spotting serial dilutions of cells expressing the peptides on selective plates (Fig. 10, which is published as supporting information on the PNAS web site), which allowed growth trends to be compared at each dilution level. Eight superior candidates were chosen and additionally compared against a control protein fusion (FKBP12 and FRAP) to assess target specificity (Fig. 11, which is published as supporting information on the PNAS web site), demonstrating that all selectants show a strong preference for RR. The final one in 10 million success rate highlights the advantages of having a high-throughput, readily implementable methodology capable of discovering these rare protein-complex modulators.
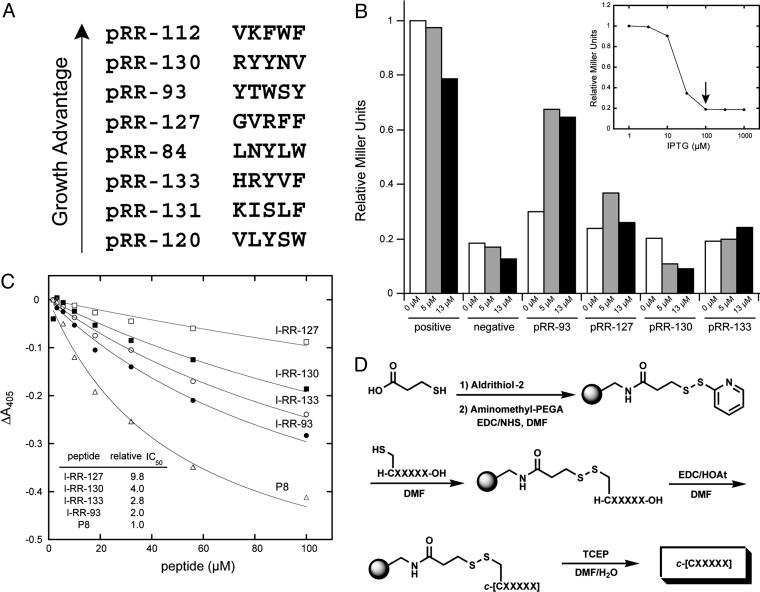
One of the principal benefits of genetically encoded combinatorial libraries is the ease of deciphering their chemical composition, in contrast to synthetically derived libraries. Thus, the amino acid sequence was readily determined for each candidate by DNA sequencing of the variable inserts present on the selected SICLOPPS plasmids. The sequences of the most potent in vivo selectants (Fig. 3A) can be tentatively grouped into neutral (e.g., RR84, RR93, and RR120) and charged (e.g., RR112, RR127, RR130, and RR133) classes. Remarkably, the neutral sequences resemble the Ar-X-F motif (where Ar is an aromatic amino acid and X is any amino acid) identified previously for several linear dissociative inhibitors of RR (27). Surprisingly, the C-terminal negative charge, deemed critical for the recognition of a large enzyme subunit, was noticeably absent in all identified sequences. From the eight candidates presented in Fig. 3A, the four with high target specificity, potent activity, and residues compatible with synthesis (RR93, RR127, RR130, and R133; Fig. 11) were then subjected to quantitative expression studies by using the lacZ reporter of the RTHS, with an intent to filter out nonspecific effects of selectants on host growth rates. All four peptides showed observable repression relief at background levels of expression, and, for three of the peptides (RR93, RR127, and RR133), this effect was further enhanced upon induction (Fig. 3B). Surprisingly, the fourth selectant, RR130, triggered arabinose-dependent repression of the lacZ reporter, suggesting a complex mode of action. These studies impelled an in vitro analysis to decipher the inhibition mechanism of these four selectants.
Fig. 3.
Processing of RR candidates. (A) Sequences of variable inserts listed in order of biological activity. (B) β-Galactosidase assays showing the in vivo activity of four expressed peptides as a function of the arabinose concentration. Positive (unrepressed strain) and negative (SICLOPPS control plasmid) controls are provided as reference points. Assays were performed at 100 μM IPTG to induce RR expression. (Inset) Titration that identified this optimal level of IPTG. (C) Competition ELISA comparing the binding affinity of the four linear peptides with P8 control. (Inset) Relative IC50 values. (D) Solid-phase synthesis of cyclic peptides (see Materials and Methods).
The absence of a dissociative assay for RR prompted the development of a screen based on a competitive ELISA. The screen involves immobilization of the small subunit (mR2) on a polystyrene surface, followed by its specific recognition with a His-tagged large subunit (mR1), whose presence is detected in turn by a Ni·NTA-HRP conjugate. The activity of inhibitors can be monitored by a concentration-dependent reduction in the horseradish peroxidase signal, because of the disruption of the complex. Gratifyingly, the synthetic linear peptides corresponding to the four genetically selected sequences promoted dissociation of the immobilized complex as shown in Fig. 3C and generally matched the in vivo observed trends, with l-RR93 showing the highest level of activity. The C-terminal octapeptide control (P8) can provide a Kd reference point for the competition ELISA, because its N-terminal truncate, P7, has a known Kd of 9 μM for the mR1 subunit (28). Although none of these peptides surpassed the potency of P8, the observation that all functioned as dissociative inhibitors in the in vitro assay demonstrates the power of genetic selection in identifying rare solutions to the problem of inhibiting protein complexation. Moreover, the cyclization of these peptides was expected to restrict the conformational flexibility of the active epitope and thus improve their potency.
Because of the challenges inherent to peptide head-to-tail backbone cyclization, a solid-phase strategy was devised that exploited immobilization of linear sequences through a cysteine side chain as a mixed disulfide (Fig. 3D). This approach was expected to favor monomolecular cyclization over bimolecular side reactions because of a solid-phase dilution effect. In addition to other advantages of solid-phase synthesis, such as improved yield and ease of purification, the disulfide immobilization strategy allows convenient isolation of the product by means of mild reductive cleavage with suitable thiol or phosphine reagents. By using this approach, all four linear peptides under investigation (l-RR93, l-RR127, l-RR130, and l-RR133) were cyclized, and their chemical nature was confirmed by a combination of Kaiser assay (14), reverse-phase HPLC, and electrospray ionization mass spectrometry (Table 1).
The cyclized peptides were tested against immobilized RR complex in the dissociative ELISA assay (data not shown). Compared with their linear forms, both cyclic RR93 and RR127 (c-RR93 and c-RR127) exhibited an ≈2-fold enhanced activity over the corresponding linear forms, confirming the entropic benefits of a constrained scaffold. The dissociative ELISA could not confirm the properties of c-RR130 and c-RR133, yielding a response pattern consistent with nonspecific peptide adherence to the plastic surface.
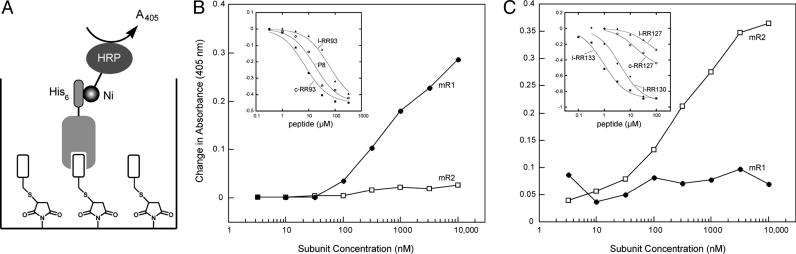
The functional nature of the genetic assay implies that peptides targeting either RR surface (mR1 or mR2) would be capable of perturbing the repressor complex. While confirming the dissociative properties of the four selected sequences, the functional format of the protein ELISA, relying merely on the complex disruption for read-out, is incapable of unambiguously identifying the receptor for the peptide ligands. To determine the mechanism of action of the four identified sequences, an alternative assay format was devised (Fig. 4A) whereby a peptide with a residual activity in its immobilized form can serve as a specific ligand for receptor capture in both binding and competition ELISA. The success of such an assay relies on both an efficient peptide immobilization strategy and a sufficient level of affinity that is uncompromised by this display strategy. The peptide immobilization becomes feasible through implementation of a cysteine, a nucleophile used in splicing, as a universal chemoselective handle allowing covalent attachment through a suitable electrophile. Chemoselective attachment of such peptides to appropriately derivatized surfaces should both display the small molecules for detection with a corresponding protein receptor and allow binding site competition analysis, not unlike in a traditional immunosorbent format. When immobilized on maleimide plates by means of its N-terminal cysteine, the P8 peptide maintained its specific affinity toward mR1 (Fig. 4B). Moreover, the resulting P8·mR1-immobilized complex was disrupted by both l-RR93 and c-RR93 in a concentration-dependent manner, with c-RR93 showing a 2-fold enhancement over a P8 reference and an 8-fold improvement over l-RR93 (Fig. 4B Inset). These results point to direct competition for the common binding site on mR1 by both the C terminus of mR2 (P8) and the selected RR93 sequence. The presence of the Ar-X-Ar motif in RR93 and other selectant sequences (e.g., RR84 and RR120) is consistent with previous observations (27).
Fig. 4.
Immoblized peptide ELISA. (A) An assay schematic showing an immobilized peptide being recognized by a protein receptor, which in turn is being detected by means of its His-6-tag by a Ni·NTA-HRP conjugate. (B) Immobilized P8 ELISA. Data demonstrates specific recognition of the P8 control peptide by the RR large subunit (mR1). (Inset) Comparison of the effectiveness of l-RR93 and c-RR93 versus P8 as a reference peptide by using an immobilized P8·mR1 complex. (C) Immobilized c-RR130 ELISA. Data demonstrate specific recognition of the c-RR130 control peptide by the RR small subunit (mR2). (Inset) Comparison of the activities of l-RR127, c-RR127, l-RR130, and 1-RR133 in disrupting the c-RR130·mR2 complex.
The fact that none of the positively charged sequences (RR127, RR130, and RR133) retained mR1 when immobilized or competed with P8·mR1 complex (data not shown) suggested an alternative and, perhaps, common mode of RR complex disruption. A systematic analysis of immobilized linear and cyclic forms yielded an unexpected observation that, unlike P8, a c-RR130-derivatized surface selectively captured the His-6-tagged mR2 subunit in a concentration-dependent manner (Fig. 4C). The target specificity was further verified by the failure of c-RR130 to complex with His-6-tagged mR1, even at high protein concentrations. Thus, binding partners for both small and large RR subunits were identified, demonstrating the capacity of genetic selection to discover not only inhibitors of protein–protein interaction but also alternative modes of action. Furthermore, confirming the original division of the selectants into charged and neutral categories, all three positively charged peptides (i.e., RR127, RR130, and RR133) competed with the c-RR130·mR2 complex (Fig. 4C Inset), with activities generally consistent with the protein ELISA observations. Thus, l-RR133 showed the highest capacity in dislodging mR2 from the cyclic peptide anchor, followed by l-RR130, and both forms of RR127. Although both c-RR130 and c-RR133 proved again to be incompatible with the ELISA, presumably because of nonspecific adsorption, their Kd values were determined by quenching of intrinsic mR2 tryptophan fluorescence to be 53 ± 5 μM and 133 ± 42 μM, respectively (data not shown). The activities of the corresponding linear counterparts were significantly lower in the fluorescence-quenching assay, precluding their thermodynamic characterization, because of the peptide fluorescence interference and solubility limits. These observations point again to the reduction of a conformational population by constraining flexible molecules as means of improving activity of protein modulators.
With the discovery of effective RR inhibitors, focused cyclic peptide libraries could be designed based on the best available candidates. The combination of a tailored library with the ability to chemically amplify the selection stringency should allow the identification of more potent inhibitors of RR subunit interactions. Further efforts will be required to kinetically characterize the effect of isolated peptide inhibitors on RR activity.
Conclusions
The challenges inherent in disrupting protein complexes demand an effective, robust methodology capable of high-throughput implementation. Herein, we have described such an approach that allows the discovery of cyclic peptide-based dissociative inhibitors through the combination of SICLOPPS technology with the RTHS. Affinity-based methods also provide these library-processing capabilities, but such approaches are indirect, handicapped by the problem of translating affinity into function (29). The ability to perform a functional assay within a genetic selection format, although retaining the throughput capacity, bypasses this issue, as highlighted by the RR case study. In this demonstration, a library of 108 members was rapidly focused to eight candidates through convenient filtering steps, resulting in cyclic peptides with comparable activities to known inhibitors and others with unprecedented binding modes. Furthermore, the selected epitopes are now presented from pharmacologically tractable, structurally better-defined scaffolds, amenable to further optimization as peptidomimetics (30). Also significant is the fact that the candidates passed the challenge of the host's proteome, without eliciting detectable toxic effects, suggesting that the final selectants should display a degree of target selectivity, a critical concern for drug development. Considering the key nature of protein–protein interactions for many physiological functions and the unique properties of these interfaces, the ability to systematically identify small-molecule modulators of these interactions could open new avenues in drug discovery.
Supplementary Material
Acknowledgments
We thank B. S. Cooperman (University of Pennsylvania, Philadelphia), G. B. Koudelka (University of Buffalo, Buffalo, NY), J. Tang (University of Oklahoma, Norman), J. C. Hu (Texas A&M University, College Station), J. D. Keasling (University of California, Berkley), B. L. Wanner (Purdue University, West Lafayette, Indiana), G. J. Phillips (Iowa State University, Ames), and the E. coli Genetic Stock Center for strains and plasmids. We thank GlaxoSmithKline (King of Prussia, PA) for the generous donation of rapamycin. This work was funded by National Institutes of Health Grant GM24129 (to S.J.B.). A.R.H. is a Damon Runyon Fellow supported by Damon Runyon Cancer Research Foundation Fellowship DRG-1729-02.
Author contributions: A.R.H., S.N.S., and S.J.B. designed research; A.R.H., S.N.S., and S.J.B. performed research; A.R.H. and S.N.S. contributed new reagents/analytical tools; A.R.H. and S.N.S. analyzed data; and A.R.H., S.N.S., and S.J.B. wrote the paper.
Abbreviations: RR, ribonucleotide reductase; RTHS, reverse two-hybrid system; IPTG, isopropyl β-d-thiogalactoside; DMF, N,N-dimethylformamide; nickel·NTA-HRP, nickel·nitrilotriacetic acid-horseradish peroxidase; SICLOPPS, split intein-mediated circular ligation of peptides and proteins.
References
- 1.Cochran, A. G. (2000) Chem. Biol. 7, R85-R94. [DOI] [PubMed] [Google Scholar]
- 2.Toogood, P. L. (2002) J. Med. Chem. 45, 1543-1558. [DOI] [PubMed] [Google Scholar]
- 3.Berg, T. (2003) Angew. Chem. Int. Ed. 42, 2462-2481. [DOI] [PubMed] [Google Scholar]
- 4.Troitskaya, L. A. & Kodadek, T. (2004) Methods 32, 406-415. [DOI] [PubMed] [Google Scholar]
- 5.Lin, H. & Cornish, V. W. (2002) Angew. Chem. Int. Ed. Engl. 41, 4402-4425. [DOI] [PubMed] [Google Scholar]
- 6.Fields, S. & Song, O. (1989) Nature 340, 245-246. [DOI] [PubMed] [Google Scholar]
- 7.Leanna, C. A. & Hannink, M. (1996) Nucleic Acids Res. 24, 3341-3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vidal, M., Brachmann, R. K., Fattaey, A., Harlow, E. & Boeke, J. D. (1996) Proc. Natl. Acad. Sci. USA 93, 10315-10320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang, J. & Schreiber, S. L. (1997) Proc. Natl. Acad. Sci. USA 94, 13396-13401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park, S. H. & Raines, R. T. (2000) Nat. Biotechnol. 18, 847-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scott, C. P., Abel-Santos, E., Jones, A. D. & Benkovic, S. J. (2001) Chem. Biol. 8, 801-815. [DOI] [PubMed] [Google Scholar]
- 12.Scott, C. P., Abel-Santos, E., Wall, M., Wahnon, D. C. & Benkovic, S. J. (1999) Proc. Natl. Acad. Sci. USA 96, 13638-13643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A. & Struhl, K. (2002) Short Protocols in Molecular Biology (Wiley, New York).
- 14.Kaiser, E., Colescott, R. L., Bossinger, C. D. & Cook, P. I. (1970) Anal. Biochem. 34, 595-598. [DOI] [PubMed] [Google Scholar]
- 15.Hu, J. C., O'Shea, E. K., Kim, P. S. & Sauer, R. T. (1990) Science 250, 1400-1403. [DOI] [PubMed] [Google Scholar]
- 16.Di Lallo, G., Castagnoli, L., Ghelardini, P. & Paolozzi, L. (2001) Microbiology 147, 1651-1656. [DOI] [PubMed] [Google Scholar]
- 17.Joung, J. K., Ramm, E. I. & Pabo, C. O. (2000) Proc. Natl. Acad. Sci. USA 97, 7382-7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brennan, M. B. & Struhl, K. (1980) J. Mol. Biol. 136, 333-338. [DOI] [PubMed] [Google Scholar]
- 19.Hays, L. B., Chen, Y. S. & Hu, J. C. (2000) BioTechniques 29, 288-294. [DOI] [PubMed] [Google Scholar]
- 20.Brown, E. J., Albers, M. W., Shin, T. B., Ichikawa, K., Keith, C. T., Lane, W. S. & Schreiber, S. L. (1994) Nature 369, 756-758. [DOI] [PubMed] [Google Scholar]
- 21.Kohl, N. E., Emini, E. A., Schleif, W. A., Davis, L. J., Heimbach, J. C., Dixon, R. A., Scolnick, E. M. & Sigal, I. S. (1988) Proc. Natl. Acad. Sci. USA 85, 4686-4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szekeres, T., Fritzer-Szekeres, M. & Elford, H. L. (1997) Crit. Rev. Clin. Lab. Sci. 34, 503-528. [DOI] [PubMed] [Google Scholar]
- 23.Zutshi, R., Brickner, M. & Chmielewski, J. (1998) Curr. Opin. Chem. Biol. 2, 62-66. [DOI] [PubMed] [Google Scholar]
- 24.Schramm, H. J., Boetzel, J., Buttner, J., Fritsche, E., Gohring, W., Jaeger, E., Konig, S., Thumfart, O., Wenger, T., Nagel, N. E. & Schramm, W. (1996) Antiviral Res. 30, 155-170. [DOI] [PubMed] [Google Scholar]
- 25.Yang, F. D., Spanevello, R. A., Celiker, I., Hirschmann, R., Rubin, H. & Cooperman, B. S. (1990) FEBS Lett. 272, 61-64. [DOI] [PubMed] [Google Scholar]
- 26.Pellegrini, M., Liehr, S., Fisher, A. L., Laub, P. B., Cooperman, B. S. & Mierke, D. F. (2000) Biochemistry 39, 12210-12215. [DOI] [PubMed] [Google Scholar]
- 27.Gao, Y., Liehr, S. & Cooperman, B. S. (2002) Bioorg. Med. Chem. Lett. 12, 513-515. [DOI] [PubMed] [Google Scholar]
- 28.Pender, B. A., Wu, X., Axelsen, P. H. & Cooperman, B. S. (2001) J. Med. Chem. 44, 36-46. [DOI] [PubMed] [Google Scholar]
- 29.Wirsching, P., Ashley, J. A., Lo, C. H., Janda, K. D. & Lerner, R. A. (1995) Science 270, 1775-1782. [DOI] [PubMed] [Google Scholar]
- 30.Hirschmann, R., Hynes, J., Jr., Cichy-Knight, M. A., van Rijn, R. D., Sprengeler, P. A., Spoors, P. G., Shakespeare, W. C., Pietranico-Cole, S., Barbosa, J., Liu, J., et al. (1998) J. Med. Chem. 41, 1382-1391. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.