Abstract
Macrophage migration inhibitory factor (MIF) is a key cytokine in autoimmune and inflammatory diseases that attracts and then retains activated immune cells from the periphery to the tissues. MIF exists as a homotrimer and its effects are mediated through its primary receptor, CD74 (the class II invariant chain that exhibits a highly structured trimerization domain), present on class II expressing cells. Although a number of binding residues have been identified between MIF and CD74 trimers, their spatial orientation has not been established. Using a docking program in silico, we have modeled binding interactions between CD74 and MIF as well as CD74 and a competitive MIF inhibitor, RTL1000, a partial MHC class II construct that is currently in clinical trials for multiple sclerosis. These analyses revealed 3 binding sites on the MIF trimer that each were predicted to bind one CD74 trimer through interactions with two distinct 5 amino acid determinants. Surprisingly, predicted binding of one CD74 trimer to a single RTL1000 antagonist utilized the same two 5 residue determinants, providing strong suggestive evidence in support of the MIF binding regions on CD74. Taken together, our structural modeling predicts a new MIF(CD74)3 dodecamer that may provide the basis for increased MIF potency and the requirement for ~3-fold excess RTL1000 to achieve full antagonism.
Keywords: Inflammation, Molecular modeling, Protein interactions, Binding residues
Introduction
Multiple Sclerosis (MS) is a CNS chronic neuroinflammatory and demyelinating disorder characterized by axon degeneration and tissue destruction (Morandi et al. 2015; Steinman 2014). These and many other autoimmune diseases are characterized by a severe inflammatory cell mediated response that eventually leads to the destruction of the local tissue. Macrophage migration inhibitory factor (MIF) is a pleiotropic cytokine that plays a pivotal role in the development and pathogenesis of several inflammatory diseases, like MS, (Cox et al. 2013) and is necessary for the progression of experimental autoimmune encephalomyelitis (EAE) in experimental animals (Powell et al. 2005). Recent work described a positive correlation between MS and a high level of expression of CD74, the cognate receptor for MIF (Benedek et al. 2013; Leng et al. 2003). Moreover, we have shown that the class II derived recombinant proteins, RTL1000 and DRα1-MOG-35-55, antagonize MIF binding to CD74 on the cell surface of macrophages (Meza-Romero et al. 2014; Vandenbark et al. 2013).
The cytokine MIF is encoded in a functionally polymorphic locus that has been linked to several autoimmune inflammatory diseases and MIF directed therapies are in advanced clinical testing. Although it is well accepted that many of MIF’s biological activities are initiated by the binding and activation of CD74 (Leng et al. 2003; Shi et al. 2006), their precise molecular interactions remain a mystery. Structurally, MIF has been crystallized and characterized extensively from the biochemical standpoint for the role of individual amino acid residues in its biological function (El-Turk et al. 2008; Pantouris et al. 2015; Suzuki et al. 1996). The mode of interaction between MIF and CD74 and the role of specific amino acid residues on the binding energy of this interaction have not been studied in detail due to the lack of a suitable source of CD74 and its highly flexible structural conformation.
CD74 is a type II transmembrane protein with a dormant transcription factor in the intracytoplasmic domain, a transmembrane region and an extracellular domain thought to be the target for MIF binding. CD74, also known as the class II invariant chain, is associated with multiple biological functions as an intracellular chaperone for class II molecules (Landsverk et al. 2009), a MIF co-receptor (Leng et al. 2003) that promotes cell survival (Starlets et al. 2006) and a potential biomarker for several inflammatory diseases (Grieb et al. 2014). CD74 has been difficult to study structurally and to date information has been limited to an NMR analysis of a 74 amino acid portion of the protein’s ectodomain (Jasanoff et al. 1998). Given recent promise in the design of inhibitors of the MIF:CD74 interaction, there is significant interest in defining the molecular basis for MIF activation of CD74.
For this study, the application of computer assisted molecular docking algorithms enabled a detailed in silico analysis of the putative contact residues between the MIF trimer and the CD74 trimer, as well as between antagonistic MHC class II DRα1 and β1 domains and the CD74 trimer. Using the spatial coordinates of the paired polypeptides deposited in the Protein Data Bank and the protein/protein docking algorithm ZDOCK (Pierce et al. 2011; Pierce et al. 2014), we developed a working model consistent with experimental results described in the literature (El-Turk et al. 2008; Pantouris et al. 2015) that predicts some rather surprising but informative structural interactions.
Materials and Methods
Our structural modeling utilized the Molecular Docking Software ZDOCK (Pierce et al. 2014) and included published spatial coordinates available in the Protein Data Bank for the 36kD human (h)MIF homotrimer (Sun et al. 1996) (PDB ID: 1MIF), the extracellular coordinates obtained by NMR structure for the CD74 trimerization domain of the 33kD isoform of human CD74, residues 118–192 with N and C terminal unstructured residues (Jasanoff et al. 1998) (PDB ID: 1IIE) and a 25kD HLA-DRα1β1/hMOG-35-55 construct (also known as Recombinant T cell receptor Ligand, RTL1000) that demonstrated antagonistic inhibition of MIF binding to CD74, mainly through the DRα1 moiety (Meza-Romero et al. 2014). The structure of the HLADRα1β1/ hMOG-35-55 construct was homology-modeled using HLA-DR3 bound to CLIP (PDB ID: 1A6A) and HLA-DR2 complexed to human MBP (PDB ID: 1BX2) as templates with the assistance of Pymol to generate the theoretical structure of the DRα1β1/hMOG-35-55 construct.
The coordinates were entered as whole molecules without selecting or highlighting any particular amino acid residues along with the ZDOCK protein:protein docking algorithm for the body-rigid search of docking orientations between the two polypeptides. This docking approach generated 10 different energy-minimized conformational predictions ranking from prediction 1 (with the minimal energy and most stable molecular complex) to prediction 10 (the least stable) of the MIF/CD74 complex. All predictions showed a binding mode in which the two components interacted through the most flexible unstructured regions. All the predictions indicated (with small variations in the orientation) that there are 3 CD74 trimers per MIF trimer in the complex and that each of the CD74 trimers bind to the interface of two MIF subunits. We chose Prediction 03 for further description given the experimental support that mutational data of MIF amino acid residues lend to this mode of interaction. The docking of the CD74 with the RTL1000 followed a similar strategy. In this case we chose the most energy-minimized model (Prediction 1) predicted by the ZDOCK algorithm. This model showed that there is one RTL1000 complex per CD74 trimer and that mode of binding is, as in the MIF model, through the most flexible regions of the CD74 trimer contacting residues located mostly on the DRα1 domain of the RTL1000 and, to a lesser extent, amino acid residues on the DRα1 domain.
Results and Discussion
MIF/CD74 interactions
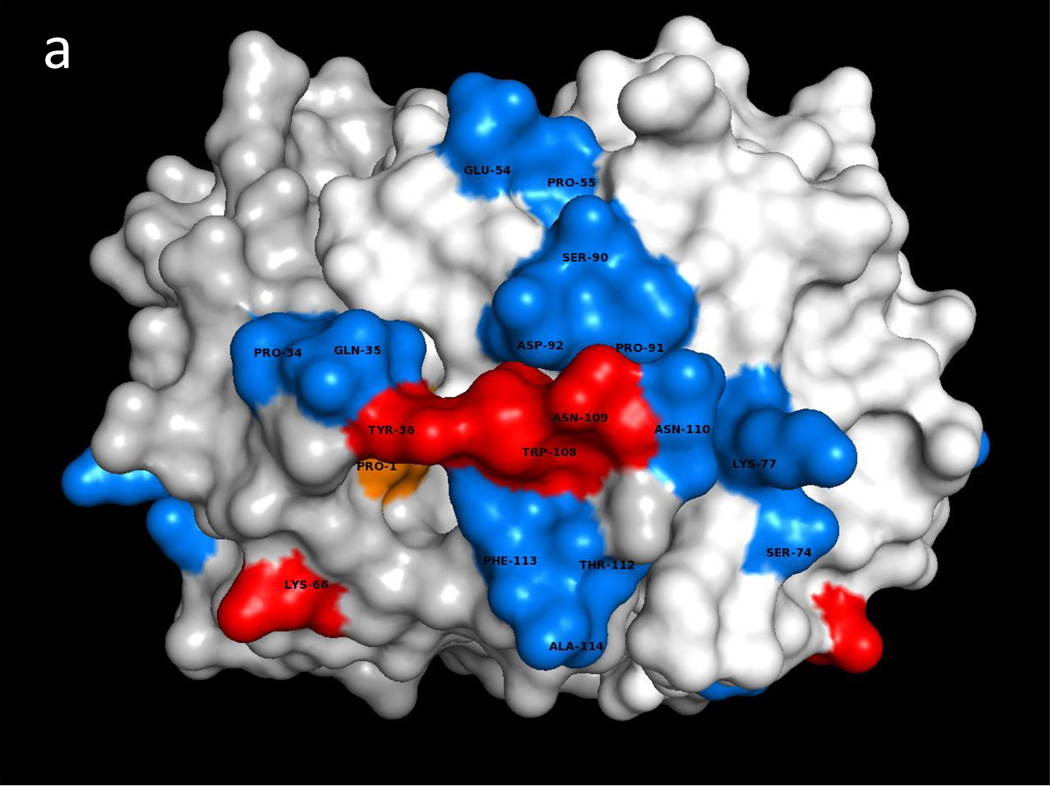
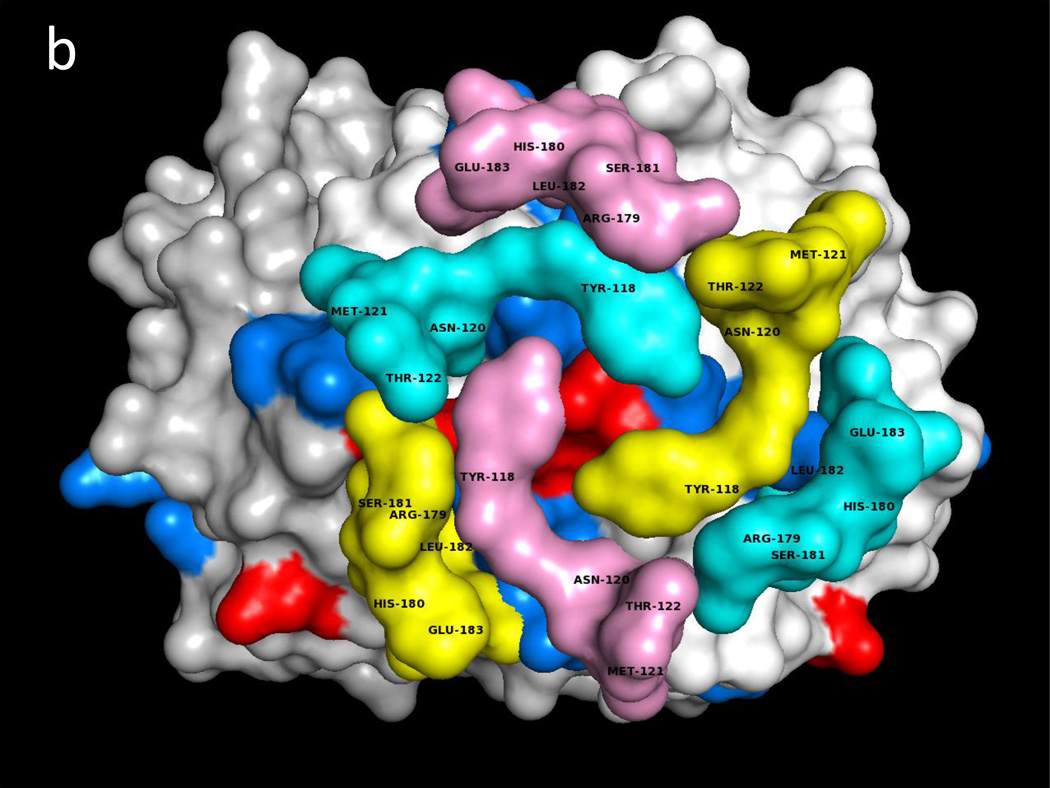
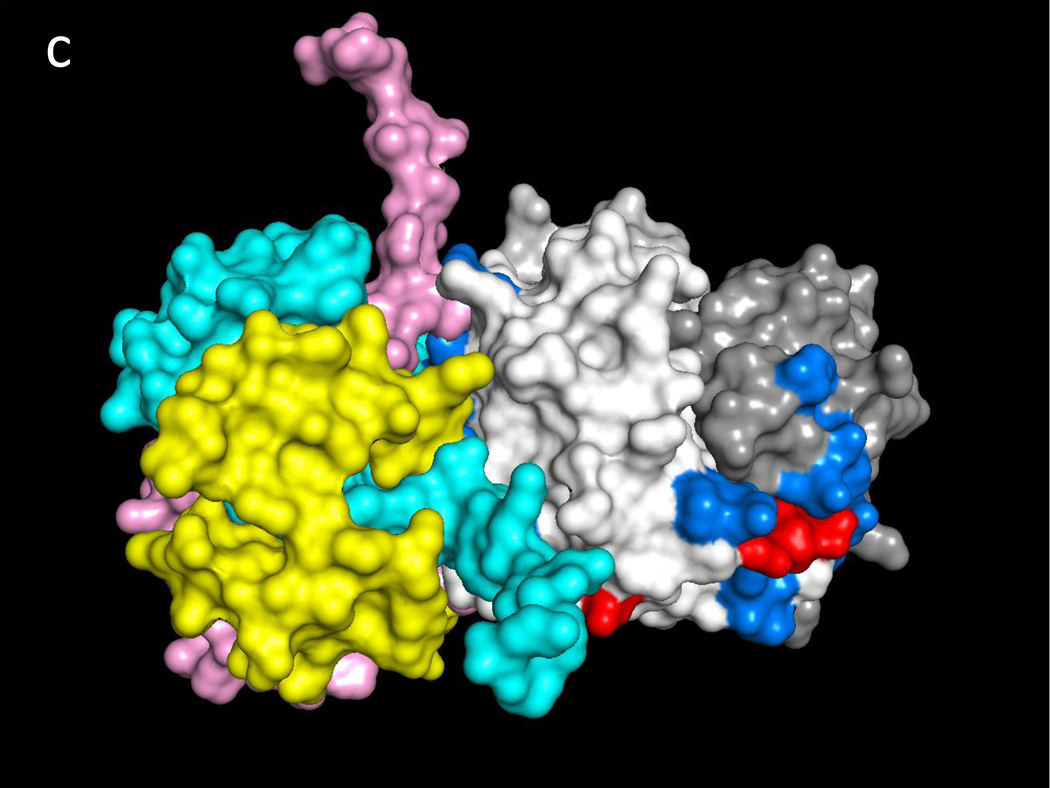
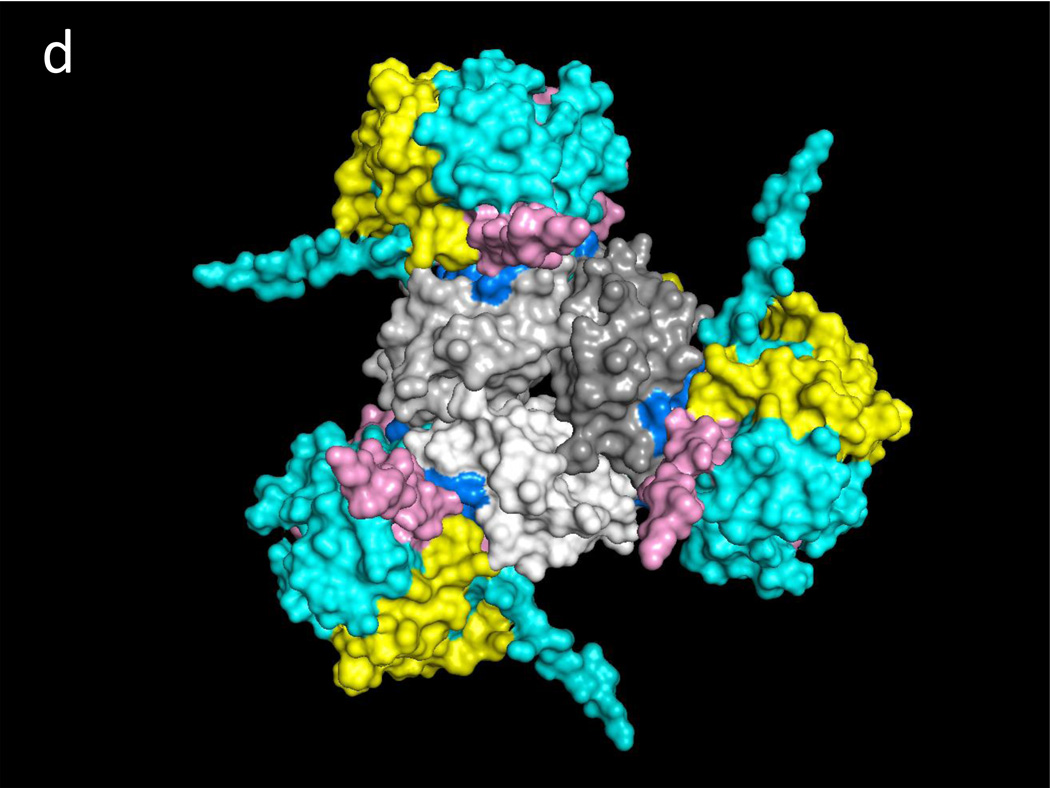
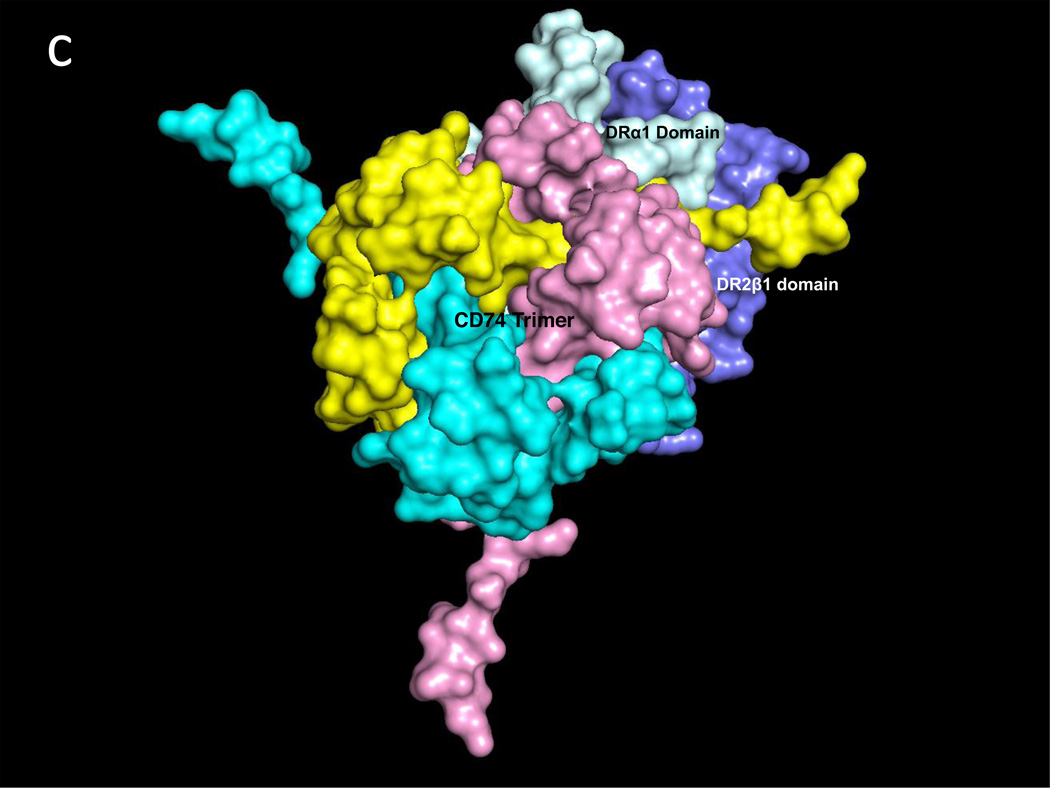
Fig. 1a illustrates one CD74 binding face of hMIF at the junction between monomers A and B of the hMIF trimer that includes four key CD74 activation residues, Trp108-Asn109, Tyr36 and Lys66 (Red) surrounded by additional predicted CD74-TD binding residues (Blue). Fig. 1b shows the same view of the hMIF trimer with overlaid residues 118YGNMT122 and 179RHSLE183 from each CD74 monomer that interface with hMIF Chain A residues 50FGGSEP55, K76, 90SPDR93 and 109NNS111; and hMIF Chain B residues 34PQ35, 108WNN110 & 111STFA114. It is noteworthy that 8 out of the top 10 docking solutions showed insertion of the CD74 residue Leu182 into the hMIF catalytic pocket with possible interaction with the key Pro1 residue (shown in Fig. 1a in Orange). The interactions between residues on individual CD74 monomers and their binding partners on a single hMIF binding face is shown in Table 1. Based on these interactions, Fig. 1c shows optimized docking from the lateral view between the CD74 trimer and one binding face of the hMIF trimer as well as one of two additional unoccupied CD74 binding faces on hMIF. Because our model predicts that each hMIF trimer has three CD74 binding faces, we show in Fig. 1d (top view) a dodecamer comprised of a single hMIF trimer with all three binding sites filled with CD74 trimers.
Fig. 1. Predicted binding interactions between the MIF trimer and the CD74 trimerization domain (TD).
a. Human (h)MIF Chains A, B, and C shown in white, light grey and grey (out of plane), respectively, illustrating one binding face at the junction between Chains A and B for the CD74-TD. Shown in Red are labeled residues Trp108-Asn109 and Tyr36 (Chain B, center) and Lys66 (Chain B, bottom left) that have been described previously as important for CD74 activation; in Blue are labeled residues located on both Chains A and B that were identified in these docking analyses as contacting CD74-TD; in Orange is the Pro1 known to be part of the active site of MIF. b. One binding face of the hMIF trimer overlaid with N-terminal 118YGNMT122 and C-terminal 179RHSLE183 binding residues from CD74-TD Chains A (Yellow), B (Pink) and C (Teal). c. One binding face of the hMIF trimer overlaid with one CD74-TD trimer (side view). Spikes protruding from the complex represent unstructured C-terminal extensions of each CD74-TD Chain. d. Each of the three binding faces of the hMIF trimer bound to a different CD74-TD trimer, producing a novel MIF(CD74-TD)3 dodecamer (top view)
Table 1.
CD74 binding residues and their binding partner residues on one binding face of MIF vs. one RTL1000 construct.
| CD74-TD Chain | MIF/RTL1000 |
|---|---|
| CD74-TD Chain A residues 118YGNMT122 (Yellow) | MIF Chain A (White) residue K77; and MIF Chain B (Light Grey) residues 108WNN110 |
| CD74-TD Chain A residues 179RHSLE183 (Yellow) | MIF Chain B (Light Grey) residues 111STFA114 |
| CD74-TD Chain C 118YGNMT122 residues (Cyan) | MIF Chain A (White) residues 50FGGS52 and 109NNS111; and Chain B (Light Grey) residues 34PQ35 |
| CD74-TD Chain C residues 179RHSLE183 (Cyan) | MIF Chain A (White) residues 49FGG51, S74, K77 & 90SPDR93 |
| CD74-TD Chain B residues 118YGNMT122 (Pink) | MIF Chain B (Light Grey) residues 108WNN110 |
| CD74-TD Chain B residues 179RHSLE183 (Pink) | MIF Chain A (White) residues 53SEP55 |
| CD74-TD Chain A (Yellow) & Chain B (Pink) residues 118YGNMT122 and Chain A (Yellow) & Chain C (Cyan) residues 179RHSLE183 |
DRα1 (Light Blue) residues F12, L14, C16, D17 & S19 |
| CD74 Chain C residues 118YGNMT122 (Cyan) and Chain B residues 179RHSLE183 (Pink) |
DRα1 (Light Blue) residues Y79 & T80 |
| CD74-TD Chain C residues 118YGNMT122 (Cyan) | DR2β1 (Dark Blue) residue Q34 |
RTL1000/CD74 interactions
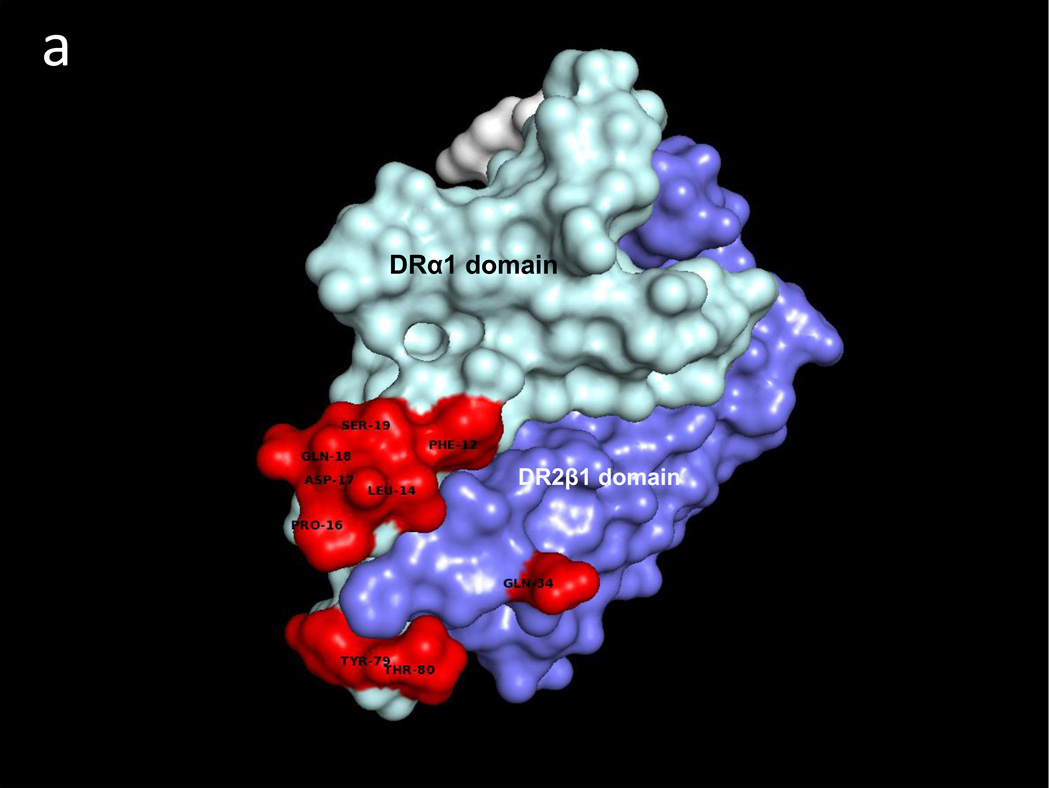
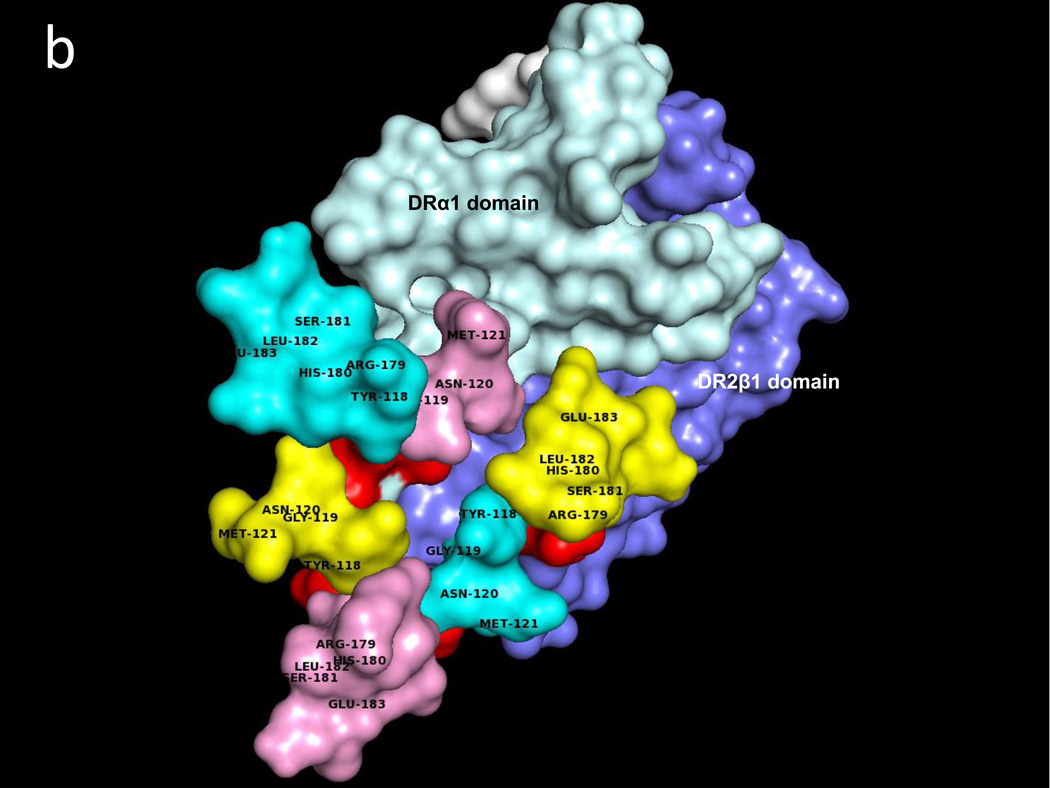
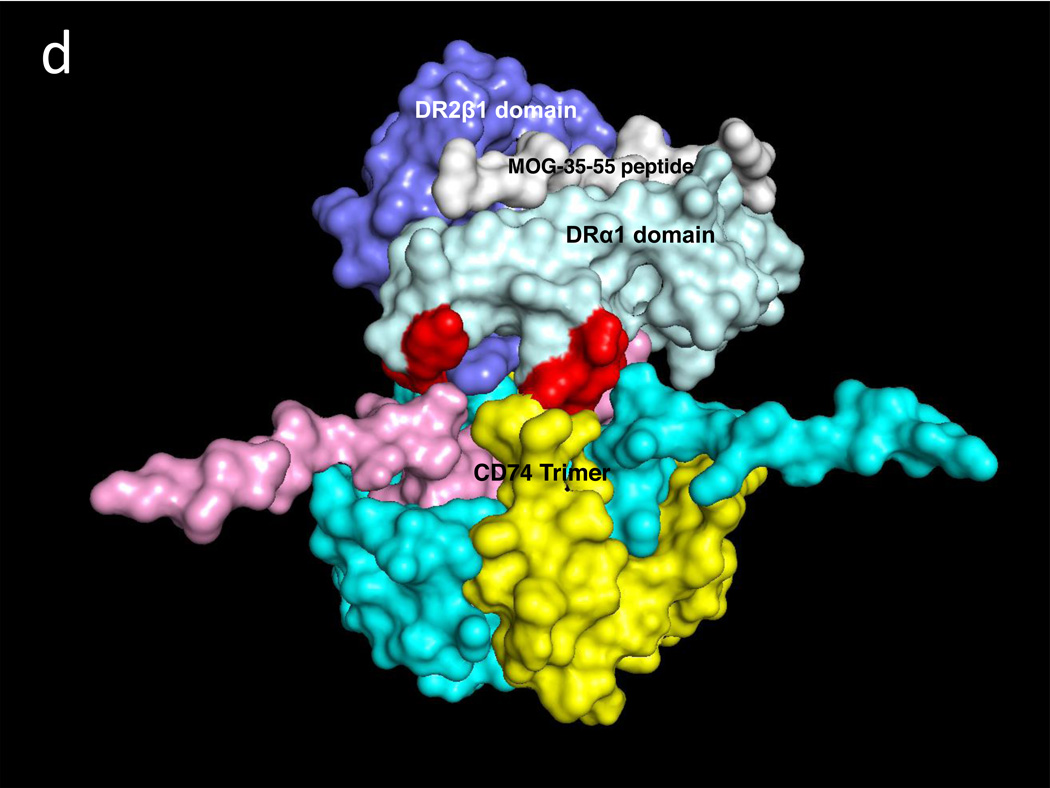
A similar docking approach carried out between CD74-TD and the DR2α1β1/hMOG-35-55 antagonist construct revealed two CD74-TD binding regions on the DRα1 domain that include residues F12, L14, C16, D17 & S19 and the 79YT80 residues, as well as a single CD74 binding site on the DR2β1 domain, residue Gln34 (Fig. 2a and Table 1). Fig. 2b shows the CD74-TD binding partners that include the same 118YGNMT122 and 179RHSLE183 residues present on each CD74 monomer that interface with the DR2α1β1/hMOG-35-55 construct shown in the same orientation as in Fig. 2a. Fig. 2c (top view) and Fig. 2d (side view) show the optimized docking between the CD74 trimer and the RTL1000 construct. Note that the covalently attached hMOG-35-55 peptide (in white) is in the binding groove on the opposite face of the DR2α1β1 construct (Fig. 2d) and is not involved in CD74 binding.
Fig. 2. Predicted binding interactions between the DR2α1β1/MOG-35-55 peptide construct (RTL1000) and the CD74-TD.
a. RTL1000 binding sites for the CD74-TD (Red) include the Phe12–Ser19 and Tyr79–Thr80 regions on the DRα1 domain (Light Blue), but only one residue (Gln34) on the DR2β1 domain (Dark Blue) that make contact with the CD74-TD. b. RTL1000 overlaid with N-terminal 118YGNMT122 and C-terminal 179RHSLE183 CD74 binding residues from each of the three CD74-TD monomers (A, Yellow, B, Pink and C, Teal). c. RTL1000 overlaid with one CD74-TD trimer (side view). d. RTL1000 bound to one CD74-TD (top view). Note that the MOG-35-55 peptide (White) in the RTL1000 binding groove does not interface with the CD74-TD
Collectively, these docking studies demonstrate common CD74 binding residues for three discrete sites on the hMIF trimer, each of which contains CD74 activation residues, and the MIF antagonist, RTL1000, that does not contain these CD74 activation residues. This proposed working model is supported by independent experimental data, including: 1) the requirement of trimerized MIF for CD74 activation and the positioning of CD74 activation residues as shown in Fig. 1a (El-Turk et al. 2012; Pantouris et al. 2015); 2) MIF binding and signaling activity in a CD74 1–148 construct (Leng et al. 2003) that includes the 118YGNMT122 MIF binding residues; 3) C terminal MIF residues 108WNN110 and 111STFA114 (shown in our model to bind CD74 residues 118YGNMT122 and 179RHSLE183, respectively) that stabilize MIF and are required for MIF enzymatic activity(El-Turk et al. 2008); 4) a competitive MIF binding inhibitor peptide from the CD74-TD α1-helix that contains the T122 residue (RMR, in preparation); and 5) a major CD74 binding region, residues 9–28 in the DRα1 domain shown in the model as 12F-S19; a minor DRα1 binding region shown in the model as 79YT80 and a minor DR2β1 binding region shown in the model as Q34 (RMR, in preparation) that support the major antagonist binding and inhibitory activity of the DRα1 with little binding to the DR2β1 moiety of RTL1000. However, our model showing docking of one CD74 trimer to a single class II DRα1β1/hMOG-35-55 construct differs from but is compatible with the model proposed by Jasanoff showing one CD74 trimer bound to three full length class II molecules (Jasanoff et al. 1998), due likely to exposure in our model of hydrophobic DRα1 and DR2β1 residues normally shielded by the α2 and β2 domains of full length class II.
The value of these docking models is to unify conceptually previous structural information addressing binding interactions and supportive functional data between MIF and CD74 and to provide a structural basis for the RTL1000 DR2α1β1/peptide as a pharmacologic antagonist that interferes with the MIF/CD74 signaling cascade. Taken together, our structural modeling predicts a new MIF(CD74)3 dodecamer that may provide the basis for increased MIF potency as a function of number of bound CD74 trimers and the requirement for ~3-fold excess RTL1000 to achieve full antagonism. These theoretical results will stimulate further experimentation to test the model and may promote the discovery or design of new compounds that could lead to new therapeutic strategies to treat MIF-mediated inflammatory disorders.
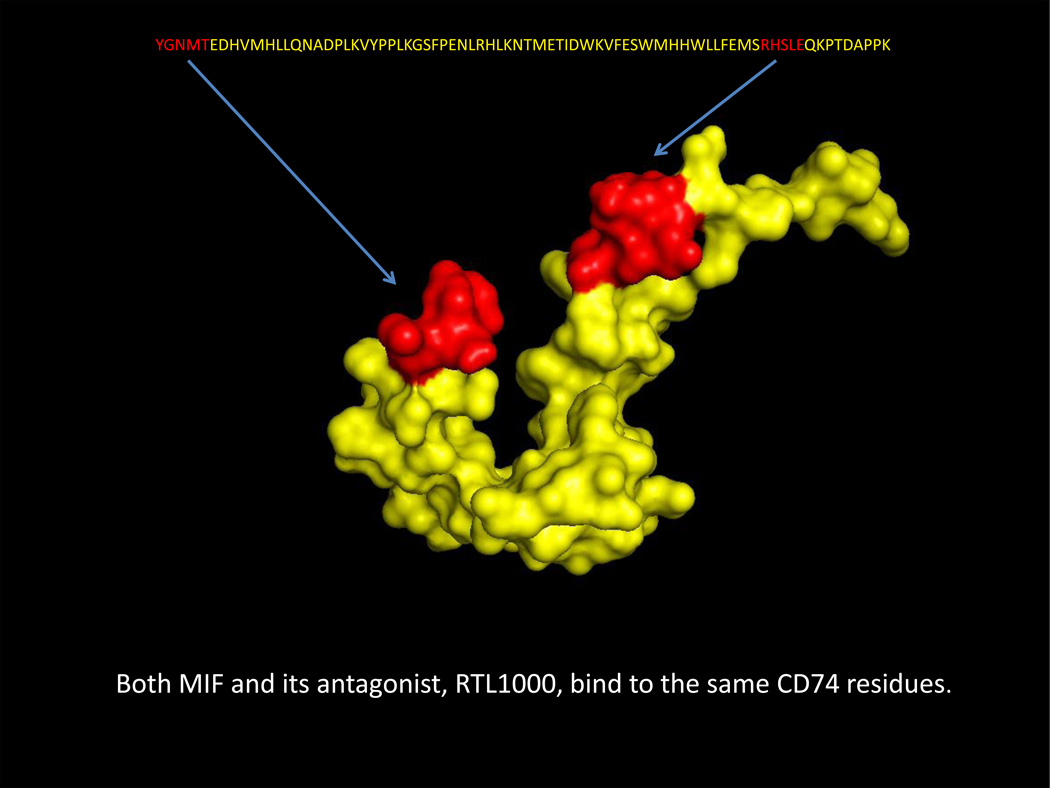
Fig. 3. Predicted common MIF and RTL1000 binding residues on CD74-TD monomers.
One CD74-TD monomer (shown in Yellow as in Fig. 1b, 1c, 1d, 2b, 2c and 2d with 118YGNMT122 N-terminal and 179RHSLE183 C-terminal residues highlighted in Red) is shown in the conformation predicted after MIF binding. These same CD74-TD residues are predicted to bind to the MIF antagonist, RTL1000
Acknowledgments
This work was supported by the National Institutes of Health grant NS047661 and National Multiple Sclerosis Society grant RG-5068-A-6 (AAV); National Institutes of Health grants AR049610 and AI042310 (LL, RB); National Multiple Sclerosis Society grant RG5272A1/T (GB); and the Department of Veterans Affairs, Veterans Health Administration, Office of Research and Development, Biomedical Laboratory Research and Development (AAV). The contents do not represent the views of the Department of Veterans Affairs or the United States Government.
Abbreviations
- MIF
macrophage migration inhibitory factor
- RTL1000
recombinant T cell receptor ligand
Footnotes
Compliance with Ethical Standards
Conflict of Interest: Drs. Vandenbark, Benedek, Meza-Romero and OHSU have a significant financial interest in Artielle ImmunoTherapeutics, Inc., a company that may have a commercial interest in the results of this research and technology. This potential conflict of interest has been reviewed and managed by the OHSU and VAPHCS Conflict of Interest in Research Committees. Drs. Leng and Bucala have no conflict of interest.
References
- Benedek G, et al. Partial MHC class II constructs inhibit MIF/CD74 binding and downstream effects. Eur J Immunol. 2013;43:1309–1321. doi: 10.1002/eji.201243162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox GM, et al. Macrophage migration inhibitory factor potentiates autoimmune-mediated neuroinflammation. J Immunol. 2013;191:1043–1054. doi: 10.4049/jimmunol.1200485. [DOI] [PubMed] [Google Scholar]
- El-Turk F, et al. The conformational flexibility of the carboxy terminal residues 105–114 is a key modulator of the catalytic activity and stability of macrophage migration inhibitory factor. Biochemistry. 2008;47:10740–10756. doi: 10.1021/bi800603x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Turk F, et al. Characterization of molecular determinants of the conformational stability of macrophage migration inhibitory factor: leucine 46 hydrophobic pocket. PLoS One. 2012;7:e45024. doi: 10.1371/journal.pone.0045024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieb G, Kim BS, Simons D, Bernhagen J, Pallua N. MIF and CD74 - suitability as clinical biomarkers. Mini Rev Med Chem. 2014;14:1125–1131. doi: 10.2174/1389557515666150203143317. [DOI] [PubMed] [Google Scholar]
- Jasanoff A, Wagner G, Wiley DC. Structure of a trimeric domain of the MHC class II-associated chaperonin and targeting protein Ii. EMBO J. 1998;17:6812–6818. doi: 10.1093/emboj/17.23.6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landsverk OJ, Bakke O, Gregers TF. MHC II and the endocytic pathway: regulation by invariant chain. Scand J Immunol. 2009;70:184–193. doi: 10.1111/j.1365-3083.2009.02301.x. [DOI] [PubMed] [Google Scholar]
- Leng L, et al. MIF signal transduction initiated by binding to CD74. J Exp Med. 2003;197:1467–1476. doi: 10.1084/jem.20030286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meza-Romero R, et al. HLA-DRalpha1 constructs block CD74 expression and MIF effects in experimental autoimmune encephalomyelitis. J Immunol. 2014;192:4164–4173. doi: 10.4049/jimmunol.1303118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morandi E, Tarlinton RE, Gran B. Multiple Sclerosis between Genetics and Infections: Human Endogenous Retroviruses in Monocytes and Macrophages Front. Immunol. 2015;6:647. doi: 10.3389/fimmu.2015.00647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantouris G, et al. An Analysis of MIF Structural Features that Control Functional Activation of CD74. Chem Biol. 2015;22:1197–1205. doi: 10.1016/j.chembiol.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce BG, Hourai Y, Weng Z. Accelerating protein docking in ZDOCK using an advanced 3D convolution library. PLoS One. 2011;6:e24657. doi: 10.1371/journal.pone.0024657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce BG, Wiehe K, Hwang H, Kim BH, Vreven T, Weng Z. ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics. 2014;30:1771–1773. doi: 10.1093/bioinformatics/btu097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell ND, Papenfuss TL, McClain MA, Gienapp IE, Shawler TM, Satoskar AR, Whitacre CC. Cutting edge: macrophage migration inhibitory factor is necessary for progression of experimental autoimmune encephalomyelitis. J Immunol. 2005;175:5611–5614. doi: 10.4049/jimmunol.175.9.5611. [DOI] [PubMed] [Google Scholar]
- Shi X, et al. CD44 is the signaling component of the macrophage migration inhibitory factor-CD74 receptor complex. Immunity. 2006;25:595–606. doi: 10.1016/j.immuni.2006.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starlets D, et al. Cell-surface CD74 initiates a signaling cascade leading to cell proliferation and survival. Blood. 2006;107:4807–4816. doi: 10.1182/blood-2005-11-4334. [DOI] [PubMed] [Google Scholar]
- Steinman L. Immunology of relapse and remission in multiple sclerosis. Annu Rev Immunol. 2014;32:257–281. doi: 10.1146/annurev-immunol-032713-120227. [DOI] [PubMed] [Google Scholar]
- Sun HW, Bernhagen J, Bucala R, Lolis E. Crystal structure at 2.6-A resolution of human macrophage migration inhibitory factor. Proc Natl Acad Sci U S A. 1996;93:5191–5196. doi: 10.1073/pnas.93.11.5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Sugimoto H, Nakagawa A, Tanaka I, Nishihira J, Sakai M. Crystal structure of the macrophage migration inhibitory factor from rat liver. Nat Struct Biol. 1996;3:259–266. doi: 10.1038/nsb0396-259. [DOI] [PubMed] [Google Scholar]
- Vandenbark AA, et al. A novel regulatory pathway for autoimmune disease: binding of partial MHC class II constructs to monocytes reduces CD74 expression and induces both specific and bystander T-cell tolerance. J Autoimmun. 2013;40:96–110. doi: 10.1016/j.jaut.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]