Abstract
D satellite RNA (satRNA) is a strain of cucumber mosaic virus (CMV) satRNA that induces an epidemic lethal disease in tomato. No natural resistance or tolerance has ever been found. Previously, we demonstrated the involvement of programmed cell death in disease development. Here, transgenic tomato plants expressing animal antiapoptotic genes bcl-xL and ced-9 were generated through agrobacterium-mediated transformation. High expression of bcl-xL or ced-9 affected plant growth and seed development. Inoculation of seedlings with CMV/D satRNA at T1 and T2 generations resulted in delayed cell-death symptoms or absence of symptoms. The degree of symptom suppression was correlated with increasing expression levels of the transgenes. Survival rates were compared among inoculated transgenic lines expressing bcl-xL, ced-9, and bcl-xL (G138A), a loss-of-function mutant of bcl-xL. More than 80% of the bcl-xL and ced-9 T1 transgenic lines showed higher survival rates than the average for bcl-xL (G138A) transgenic lines. Total RNA extracted from surviving plants contained D satRNA, indicating systemic accumulation of D satRNA. Thus, expression of bcl-xL and ced-9 improved tolerance to, rather than resistance to, CMV/D satRNA infection. In addition, expression of bcl-xL and ced-9 specifically abrogated the formation of necrotic lesions, but not other symptoms, in tomato leaves during chilling at 4°C. At 7°C, temperature-induced leaf senescence was dramatically delayed in bcl-xL and ced-9 transgenic plants, and high levels of anthocyanins accumulated, possibly limiting oxidative stress. Hence, expression of these animal antiapoptotic genes improved plant survival under abiotic or biotic stress.
Cucumber mosaic virus (CMV) is an isometric plant virus with a tripartite plus-sense RNA genome (1). It has the broadest host range of any known virus, infecting >1,200 species of plants (2). The symptoms induced by CMV infection in different hosts are sometimes modified by specific molecular parasites, the nonencoding satellite RNAs (satRNAs). D satRNA is one strain of CMV satRNA that benefits most of its host plants by attenuating CMV symptoms. However, it induces systemic necrosis in tomato and some accessions of Solanum lycopersicoides (P.X., unpublished data) and causes an epidemic lethal disease in tomato in the field. This disease has caused catastrophic tomato loss (3–6). No natural resistance or tolerance in tomato has been reported. Physiological and cell biological studies showed that multiple defense responses are induced during the disease development, whereas classic markers of programmed cell death (PCD) are found during the development of systemic necrosis (7, 8).
PCD is an essential process for plant growth and development. However, it also may result from abiotic or biotic stress and cause necrotic diseases in plants (reviewed in ref. 9). Although progress has been made in understanding plant PCD, little is known about its regulation and execution, especially compared with animal apoptosis (9). Apoptosis is a type of PCD with specific biochemical characteristics. It involves cascades of caspases and Bcl-2 family members for execution and regulation (10). The Bcl-2 family has been implicated in pivotal decision points regarding cell survival in animals (11). Bcl-xL is an antiapoptotic member of the Bcl-2 family. Overexpression of bcl-xL extends cell survival against apoptotic signals induced by a variety of treatments including viral infection, UV and γ-radiation, heat shock, and agents that promote formation of free radicals. Ced-9 is a Bcl-2 analog from Caenorhabditis elegans that promotes cell survival by binding to Ced-4 and preventing the activation of Ced-3 caspase (12). Core mammalian PCD regulators are not present in plants at the primary sequence level. So far, no analogs of Bcl-2 family members or caspases have been reported in plants except for the existence of genes encoding some metacaspases (13). Metacaspases are proteins distantly related to caspases, containing a caspase-like domain. They are found in plants, fungi, and protozoa. However, their biochemical roles in plant PCD are still unknown (13). Conversely, several research groups have expressed some animal cell death-regulation genes in plants and found the function of their encoded proteins conserved in plants. For example, when bcl-2 proapoptotic member bax was constitutively or transiently expressed in plants, PCD was induced (14, 15). The expression of antiapoptotic genes such as iap, bcl-2, bcl-xL, and ced-9 in tobacco plants suppressed the extensive cell death caused by necrotrophic fungal pathogens and also enhanced resistance to some abiotic stresses such as wounding, salt, cold, UV-B, and paraquat treatment (16, 17). In addition, the expression of a baculovirus antiapoptotic gene p35 in tomato plants suppressed the cell death caused by a fungal toxin and the infection of certain bacterial or fungal pathogens (18, 19). These studies suggest the conservation of PCD regulation across the kingdoms.
The ability of antiapoptotic genes to suppress complex virus-induced cell death in plants varies. Various researchers found that the progression of tobacco mosaic virus-induced hypersensitive-response cell death was affected but not completely suppressed by the expression of these genes, and virus resistance was compromised (16, 20–22). The tobacco cell death caused by tomato spotted wilt virus infection was abrogated and was associated with enhanced virus resistance mediated by these antiapoptotic proteins (16). Here we report the generation of transgenic tomato plants expressing bcl-xL and ced-9 and the improved tolerance to CMV/D satRNA infection in these plants. Additionally, tomato is a tropical plant and very sensitive to low temperature. Given that PCD occurs in low-temperature-treated tobacco suspension cells (23), it seems possible that chilling injury in tomato plants may involve PCD. Thus, we tested cold tolerance of these transgenic tomato plants and show here that both Bcl-xL and Ced-9 prevent cold-induced tomato cell death.
Materials and Methods
Constructs and Plant Transformation. The cDNA clones of bcl-xL, bcl-xL (G138A), and ced-9 in the pPTN binary vector were obtained from M. B. Dickman (University of Nebraska, Lincoln) (16). Clone bcl-xL (G138A) contains one loss-of-function substitution at codon 138 in its encoded protein. Agrobacterium tumefaciens strain LBA4044 was transformed with these constructs and selected on LB medium supplemented with streptomycin (50 mg/liter), spectinomycin (100 mg/liter), and rifampicin (50 mg/liter). The cotyledon explants of tomato (Lycopersicon esculentum, cv. MicroTom) seedlings were transformed, and plants were regenerated with a published protocol for tomato agrobacteria transformation (24).
Expression Analysis of Transgenes. Regenerated plants were moved from magenta boxes to soil pots, and leaf tissues were harvested. Total RNA was extracted with the Tri-reagent RNA extraction kit (Molecular Research Center, Cincinnati). The plasmids containing the cDNAs of transgenes were digested with EcoRI, and released DNA fragments were gel-purified with a QIAquick gel extraction kit (Qiagen) and used as templates for probing. The probes for transgene mRNAs were labeled with [α-32P]-dCTP by random priming with Ready-To-Go DNA-labeling beads (Amersham Pharmacia). RNA gel blot, hybridization, and exposure were performed as described (8). Genomic DNA was extracted from the plants expressing transgenes with Plant DNAzol reagent (Molecular Research Center), and Southern blot was performed by following the standard protocols of Sambrook et al. (25). Seeds were harvested from the T0 tomato plants having one or two copies of the transgene.
Inoculation Test. The seeds from different T0 transgenic tomato plants were planted in magenta boxes. A small piece of cotyledon explant was cut from each seedling and cultured on callus induction medium supplemented with the selective antibiotic kanamycin (100 mg/liter) (24). Callus formation ratio was used to determine segregation in the T1 generation. In the meantime, 40–80 T1 plants from each line and from nontransgenic tomato plants were inoculated at the two-leaf stage with CMV/D satRNA as described (7). Symptoms were checked daily, and survival rate (number of surviving/number of infected plants) for each line was recorded. Seeds were harvested from the surviving T1 plants, the inoculation test was repeated for the T2 generations, and one of the bcl-xL lines was further inoculated to the T4 generation.
Detection of Viral RNAs. Total RNA from the systemic leaves of surviving plants was extracted with the Tri-reagent RNA extraction kit. The accumulation of viral RNAs was detected by electrophoresis in agarose gel stained with ethidium bromide and by reverse transcription (RT)-PCR. Approximately 10 μg of total RNA was taken for RT. The following primers were used for RT to produce cDNAs of CMV RNA3 and D satRNA: 5′-GGCTGCAGTGGTCTCCTT-3′ for RNA 3 and 5′-GGGGTCTAGACCCGGGTCCTGCAG-3′ for D satRNA. SuperScript reverse transcriptase (Invitrogen) was used in the reaction by following the manufacturer's protocol with the adjustment of the reaction temperature to 50°C. The following 5′-end primers, respectively, combined with the 3′-end primers mentioned above were used for a thermocycling reaction to generate the DNA fragments: 5′-GATAAGAAGCTTGTTTCGC-3′ for RNA3 and 5′-GGGAATTCATTTAGGTGACACTATAGTTTTGTTTG-3′ for D satRNA. The reaction system contained 1 μl of RT product, 1 μM primers, 0.2 mM dNTP, 4 mM MgCl2 buffer (Idaho Technology, Salt Lake City), and 0.2 unit of TaqDNA polymerase, and the program consisted of 35 cycles of 94°C for 15 s, 55°C for 15 s, and 72°C for 15 s in a thermocycling machine (RapidCycler, Idaho Technology). The amplified DNA products were detected by electrophoresis in agarose gels followed by ethidium bromide staining and visualization with UV light.
Cold Treatment. T1 plants (30- to 40-day-old) from seven transgenic lines as well as nontransgenic tomato plants were placed in growth chambers. Cold treatments were performed by adjusting the chamber temperatures to either 4°C or 7°C for 2–10 wk. For one bcl-xL line, the cold treatment was repeated to the T4 generation. A duplicate set of plants was maintained at normal growth conditions (28°C daytime, 20°C nighttime).
Anthocyanin Measurement. Approximately 200 mg of leaf tissues were harvested, fully covered, and extracted with acidic methanol (5 ml) in the dark at 4°C. The acidic methanol was changed four times, once every 12 h, and the extracts were pooled to estimate total anthocyanins by spectrometry analysis at 528 nm (26).
Results
Generation of Transgenic Tomato Plants. The expression of bcl-xL, ced-9, and bcl-xL (G138A) in putative transgenic plants was analyzed by RNA blot analysis. Expression among the regenerated plants was detected for ≈70% of the bcl-xL lines, 58% of the ced-9 lines, and 60% of the bcl-xL (G138A) lines. Phenotype and the presence of the transgene were not significantly correlated for the T0 plants. The copy number of each transgene in these plants was further estimated by Southern blot (data not shown). The transgenic lines with one or two copies of the transgene were chosen for seed increase and further inoculation tests.
Results from a segregation analysis for the transgenes in tomato indicated that most T0 plants were regenerated from chimeric origins (data not shown), which likely caused the confusion in correlating phenotype with transgenes at T0. Therefore, identification of transgenic lines and inoculation tests were carried out on the T1 and T2 generations.
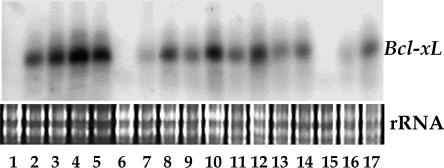
Expression of Antiapoptotic Gene bcl-xL Affects Plant Development. During the screening for pure transgenic lines, the transgene bcl-xL cosegregated with a phenotype of reduced plant size. We show an RNA blot for progeny of line bcl-xL.1.1#37 (Fig. 1; RNA sample order was random). The RNA was derived from tissues that were harvested from plants at the three-leaf stage, when no difference in phenotype was seen. Approximately 6–7 wk later, plants with high expression of bcl-xL were stunted, whereas the plants with relatively low expression were intermediate in size compared with the larger plants, which showed no transgene expression (Fig. 2). None or very few viable seeds were formed in the plants with high levels of bcl-xL. Similar phenotype segregation also was seen for the ced-9 transgenic lines. In the plants with extremely high expression of bcl-xL, flower structure was abnormal and fruits were seedless. Little difference was observed between nontransgenic and transgenic plants with high expression of bcl-xL (G138A) (data not shown).
Fig. 1.
Northern blot assay for the expression of bcl-xL in the progeny plants of line bcl-xL.1.1#37. Plants 1, 6, and 15 showed no expression, whereas 7, 9, 11, 13, 14, 16, and 17 showed relatively low expression. The other plants showed high expression.
Fig. 2.
Phenotype comparison among tomato plants expressing bcl-xL and nontransgenic plants. (A) Plants with high expression of bcl-xL. (B) Plants with relatively low expression. (C) Plants with no transgene and nontransgenic plants.
Expression of Antiapoptotic Genes bcl-xL and ced-9 Resulted in Improved Tolerance to CMV/D satRNA Infection. In inoculation tests, all nontransgenic tomato plants died from the infection with CMV/D satRNA at 1–2 wk after inoculation (Fig. 3A). Most of the bcl-xL (G138A) transgenic plants developed systemic necrosis (Fig. 3B). Three conditions were seen in the infected progeny plants of bcl-xL transgenic lines: systemic necrosis, delayed partial necrosis, or no symptoms (Fig. 3C). Similar results were obtained for ced-9-transformed plants (Fig. 3D).
Fig. 3.
Symptoms in transgenic tomato plants infected with CMV/D satRNA. (A) Nontransgenic plants with systemic necrosis. (B) Plants expressing bcl-xL (G138A) with systemic necrosis. (C) Plants expressing bcl-xL, with no symptoms or with delayed symptoms (arrow). (D) Plants expressing ced-9, with no symptoms or with delayed cell death in the stem (arrow).
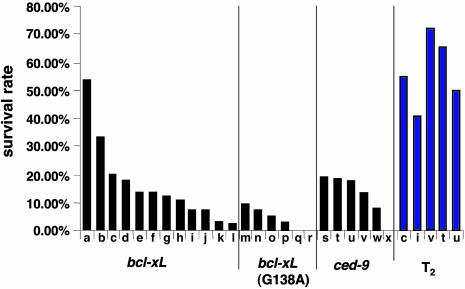
Survival rates at the T1 generation varied among different bcl-xL and ced-9 transgenic lines and were correlated with the expression level of the transgenes (data not shown). Overall, >80% of these lines showed a higher survival rate than the average for bcl-xL (G138A) lines (Fig. 4). A few lines of bcl-xL (G138A) transgenic tomato showed <10% survival rate (Fig. 4), but none of these lines had stable or increased survival rates at the T2 generation (data not shown). Because most analyzed T0 plants were regenerated from chimeric origins, the inoculation tests were continued to the T2 generation. Survival rates were dramatically increased in several bcl-xL and ced-9 transgenic lines (Fig. 4), among which only one bcl-xL heterozygous line produced enough seeds for continuous inoculation tests at the T3 and T4 generations. Southern blot analysis indicated a single copy in the genomic DNA for this line (data not shown). The survival rate at a given generation was stable when measured repeatedly with different seed lots. Hence, these data suggest that the expression of animal antiapoptotic genes bcl-xL and ced-9 suppresses CMV/D satRNA-induced PCD.
Fig. 4.
Survival rates of transgenic tomato plants infected with CMV/D satRNA. Each black column indicates the percentage survival after inoculation of 40–80 plants from each line at the T1 generation. The blue columns indicate the percentage survival of the corresponding lines at the T2 generation. The bcl-xL transgenic lines (a–l); bcl-xL (G138A) lines (m–r); and ced-9 lines (s–x).
These transgenic plants were also inoculated with CMV alone. Both transgenic and nontransgenic tomato plants developed mosaic symptoms, shoestring leaves, and numerous side shoots at 7 days postinoculation. In CMV/D satRNA-infected non-transgenic tomato plants, these typical CMV symptoms are normally attenuated or not detectable, although PCD is induced later in infection. In contrast, the surviving T1 bcl-xL transgenic plants showed no symptoms. To determine whether virus resistance or tolerance was involved, the accumulation of D satRNA was detected by electrophoresis in agarose gels. For confirmation, D satRNA sequences were amplified by RT-PCR, and the expression of transgenes was confirmed by RNA blot analysis. Fig. 5 shows results from one inoculation test. All of the nine surviving plants expressed transgene bcl-xL (Fig. 5A), and D satRNA accumulated in the systemic leaves of eight plants (Fig. 5B), indicating that the expression of bcl-xL had no effect on systemic infection by D satRNA or its function in attenuating CMV symptoms.
Fig. 5.
D satRNA accumulation in surviving plants expressing bcl-xL. (A) Northern blot showing the expression of bcl-xL in nine surviving plants. Negative control from a nontransgenic plant (lane 1); total RNA samples from nine surviving plants with no symptoms (lanes 2–10). (B) Total RNA extracts from the systemic leaves of transgenic tomato plants inoculated with CMV/D satRNA. A 1-kb ladder (lane 1); bcl-xL (G138A) transgenic plant with epinastic leaf symptoms (lane 2); bcl-xL (G138A) transgenic plant with systemic necrosis symptoms (lane 3); and the bcl-xL transgenic plants corresponding to the nine surviving plants in A (lanes 4–12). Arrow indicates the D satRNA band.
Expression of bcl-xL or ced-9 Enhanced Tomato Tolerance to Low-Temperature Stress. In addition to virus-induced PCD, we also looked at the effect of these antiapoptotic genes on an abiotic stress, low-temperature-induced cell death. Tomato is a tropical crop, very susceptible to chilling injury. When 1-mo-old non-transgenic tomato plants were transferred to 4°C with normal lighting conditions, necrotic lesions appeared in the leaves in ≈3 or 4 days and became more severe with time, as shown in Fig. 6Aa. The plants completely collapsed at ≈9 wk after the temperature switch. However, transgenic tomato plants expressing bcl-xL or ced-9 showed strong tolerance to chilling stress at 4°C. Although leaves of transgenic plants appeared to be extensively curled, they remained green and exhibited no lesions (Fig. 6A c–e). The development of these plants slowed at 4°C, but they continued to grow and produce flowers and fruits. All five tested lines, some with high and others with relatively low expression of bcl-xL (Fig. 6A c and d, respectively) and ced-9 (Fig. 6Ae), showed tolerance to cold. The high-expressing plants, although stunted, were most effectively protected (Figs. 6A d and e). Plants expressing a high level of the mutant bcl-xL (G138A) showed a slight delay in lesion formation, but nevertheless developed leaf lesions from the cold injury (Fig. 6Ab) and eventually died.
Fig. 6.
Effects of cold treatment at 4°C for 2 wk. (A) Comparison of symptoms among nontransgenic and transgenic tomato plants. Nontransgenic plant (a); bcl-xL (G138A) transgenic plant (b); transgenic plant with relatively low expression of bcl-xL (c); transgenic plant with high expression of bcl-xL (d); and transgenic plant with high expression of ced-9 (e). (B) Northern blot for the expression of bcl-xL in the plants for cold treatment. Lanes 1, 2, 5, and 6 were from plants A a–d; lanes 3 and 4 were from other cold-tolerant plants expressing bcl-xL. (C) Northern blot for the expression of ced-9 in the plants for cold treatment. Plant in Aa (lane 1); plant in Ae (lane 3); and other plants expressing ced-9 (lanes 2 and 4–6).
Tolerance to 7°C was tested by using 6-wk-old nontransgenic and transgenic tomato plants. After the temperature shift (4 wk), all of the plants produced purple pigment in the leaves while yellowish patches were also developing in the older leaves of the nontransgenic plants. Within another 3–4 wk at 7°C, the leaves of the nontransgenic plants turned yellow and yellowish red, which is typical of stress-induced senescence (Fig. 7Aa). However, transgenic tomato plants expressing bcl-xL remained purple for ≈5 mo and were still able to develop flowers and small fruits (Fig. 7A b and c). A similar phenomenon was seen when ced-9 transgenic plants were subjected to the same treatment (data not shown). As was observed in the 4°C tests, high expression of mutant bcl-xL (G138A) delayed senescence slightly but provided no long-term protection.
Fig. 7.
Effects of cold treatment at 7°C for 2 mo. (A) Comparison of symptoms among nontransgenic and transgenic tomato plants. Nontransgenic plant undergoing senescence (a); plant expressing a low level of bcl-xL, with purple pigments (b); plant expressing a high level of bcl-xL, with purple pigments (c). (B) Anthocyanin analysis. NG and TG, nontransgenic and transgenic plants expressing bcl-xL from growth chamber under normal temperature; NC and TC, nontransgenic and transgenic plants expressing bcl-xL at 7°C.
The purple color observed in the transgenic plants expressing bcl-xL and ced-9 at 7°C suggested the accumulation of anthocyanins. Total amounts of anthocyanins were measured in nontransgenic and transgenic tomato leaves before and after temperature switch. There was no difference between the samples from nontransgenic and bcl-xL transgenic tomato plants under normal growth conditions, but cold treatment at 7°C increased the accumulation of anthocyanins (Fig. 7B). Increases of anthocyanin content (5- to 6-fold) were found in bcl-xL transgenic plants after 2 mo of cold treatment at 7°C, whereas only 1- to 2-fold increases occurred in nontransgenic plants (Fig. 7B).
Discussion
D satRNA attenuates CMV symptoms in most plant hosts but induces PCD and causes a lethal disease in tomato plants (6). From an evolutionary perspective, it is unusual for a parasite to destroy its plant host and helper virus and eventually itself. It is almost certain that the CMV/D satRNA relationship developed in a host other than tomato, where the satRNA attenuates viral symptoms and viral titer, and behaves more like a mutualist for the plant host. Here, we demonstrate that the expression of antiapoptotic genes imported from other kingdoms suppressed tomato PCD and altered the necrogenic satRNA phenotype in nontransgenic tomato into a more attenuating satRNA in transgenic plants. This finding suggests an accidental initiation of an intrinsic cell death program by D satRNA in tomato and closely related solanaceous species.
Virus-induced cell death is not always an accident. In animals, apoptosis is usually an important factor in host defense that limits the replication and spread of certain viruses, although in some situations apoptosis can contribute to pathogenesis. For example, an animal coxsackievirus adopts apoptosis for efficient virus egress, which can be suppressed by overexpression of bcl-xL without affecting virus replication (27). In plants, a necrotic disease caused by tomato spotted wilt virus was abrogated in tobacco expressing antiapoptotic genes, and virus resistance resulted (16). An opposite result was obtained from the interaction between tobacco mosaic virus and N gene-containing tobacco plants expressing caspase inhibitor P35 or antiapoptotic genes bcl-xL or ced-9. Cell death progression was affected but not completely suppressed, and the resistance to tobacco mosaic virus was compromised, indicating the involvement of cell death in tobacco mosaic virus resistance (21, 22). Hence, the consequences of inhibiting virus-induced PCD in plant necrotic diseases vary with the role of cell death in host defense or viral pathogenesis.
Growth and development of tomato are very sensitive to low temperatures. The symptoms of chilling are premature senescence, discoloration, and necrotic lesions. Swollen nuclei with fragmented chromatin and nuclear DNA fragmentation were also associated with chilling injury, indicating an induction of PCD (23, 28). Here, expression of bcl-xL and ced-9 successfully abrogated low-temperature-associated necrotic lesion formation, further confirming that an active cell death process is triggered by chilling. Additionally, we found that, under cold treatment at 7°C, premature senescence was dramatically delayed and anthocyanins accumulated to high levels in transgenic tomato plants. Leaf senescence is a highly regulated, ordered series of events. The most obvious sign of senescence is leaf yellowing, the stage when PCD occurs (29). Exposure to adverse temperatures can initiate the onset of senescence (30), which we observed for nontransgenic tomato plants. Rather than turning yellow, transgenic plants turned purple with high levels of anthocyanins. Anthocyanins are end products of the flavonoid pathway (31). Low temperature has been shown to induce anthocyanin synthesis in some plant species, although high levels of anthocyanin generally are achieved by accumulation over time rather than by induction to a high level of synthesis (31). Therefore, it is possible that low temperature initiates leaf senescence and anthocyanin synthesis in tomato plants, and as senescence continues, anthocyanins are degraded in nontransgenic plants. The expression of bcl-xL or ced-9 protects against PCD, which results in anthocyanin accumulation in the long-lived cells. The accumulation of anthocyanin in transgenic tomato plants at 4°C was not as obvious as that at 7°C. This finding may be the result of a strict temperature range that triggers anthocyanin synthesis (32). Reactive oxygen species are known to be generated by chilled plants as well as in leaf senescence (33, 34). Reactive oxygen species are recognized as signaling molecules in activating PCD during normal development and during the responses to abiotic and biotic stress. Anthocyanins, as strong antioxidants, have been suggested as reactive oxygen species scavengers in plant tissues (33). Therefore, the accumulation of anthocyanins in the leaves of transgenic tomato plants at low temperature may further protect plants from reactive oxygen species damage and enhance cell survival.
The results reported in this study indicate a potential agricultural value for these transgenic tomato plants. These plants also provide a good system for further study on plant PCD. Not much is known about the pathways in plant cell death regulation and execution. This report and related work of others (16–19, 21) suggest that some molecular components involved in the regulation of animal or nematode PCD are functionally conserved in plants, and the plant PCD caused by different stress factors such as CMV/D satRNA and low temperature may have a common regulation mechanism or have cross-talk. The loss-of-function mutation in Bcl-xL at amino acid 138 acted similarly in tomato PCD and in animal PCD, although the slight effect of this mutant in plant PCD was found and also reported previously (16). This finding indicates that the essential domain in Bcl-xL for inhibiting PCD is conserved across kingdoms. Because the biochemical functions of these gene products in plants are unknown, we cannot exclude the possibility that their multiple biological activities interrupt and inhibit different PCD regulation pathways. It is not fully understood how Bcl-xL and Ced-9 inhibit cell death and promote cell survival in animal cells. An abundance of Bcl-xL can sequester caspase activators or proapoptotic members of the Bcl-2 family and protect the cells from apoptosis, which is critical for the protective role by Bcl-xL against Sindbis virus-induced apoptosis (35). Although no caspases have been identified in plants, caspase-like protease activities have been detected in several plant PCD systems, and two types of metacaspases were found in Arabidopsis (13, 36). We detected a big increase of caspase-like protease activity just before the initiation of systemic necrosis in CMV/D satRNA-infected tomato plants, but the expression of P35, the inhibitor of some caspases, had no effect on CMV/D satRNA or chilling-induced PCD (unpublished data). Thus, the direct target(s) of Bcl-xL or Ced-9 in plants may shed light on the mechanisms of CMV/D satRNA or low-temperature-induced tomato PCD.
PCD is an essential process during plant growth and development. It was not surprising to see that expression of antiapoptotic genes caused changes in tomato growth. Similar defects also were observed in transgenic tobacco plants with high expression of bcl-xL and bcl-2 (16). It remains to be analyzed how these antiapoptotic genes interrupt plant growth. So far, no bcl-2 homologous genes have been found in plants. The subcellular localization of Bcl-xL in the transgenic tobacco cells is similar to animal cells, but a high level of Bcl-xL also is found in plastids and the soluble cytoplasm in tobacco suspension cells (17). Thus, Bcl-xL may share similarities in its inhibitory mechanisms of apoptosis, but may also have some different features in affecting plant cell homeostasis and lead to some developmental defects.
In conclusion, heterozygous expression of bcl-xL and ced-9 in tomato plants enhanced plant survival by inhibiting PCD induced by both biotic and abiotic stress, including virus infection and chilling, and also affects plant development. These transgenic plants not only provide a good system for plant PCD research but also have a potential value for agricultural applications.
Acknowledgments
We thank Dr. Marty B. Dickman for providing the bcl-xL, ced-9, and bcl-xL (G138A) constructs and Drs. Elison B. Blancaflor, Luiz M. Marquez, and Deyu Xie for reviewing the manuscript. This study was supported by the Samuel Roberts Noble Foundation.
Author contributions: P.X. and M.J.R. designed research; P.X. and S.J.R. performed research; P.X. and S.J.R. analyzed data; P.X. wrote the paper; and M.J.R. supervised all research and edited the manuscript and figures.
Abbreviations: satRNA, satellite RNA; CMV, cucumber mosaic virus; RT, reverse transcription; PCD, programmed cell death.
References
- 1.Palukaitis, P., Roossinck, M. J., Dietzgen, R. G. & Francki, R. I. B. (1992) Adv. Virus Res., 41, 281-348. [DOI] [PubMed] [Google Scholar]
- 2.Edwardson, J. R. & Christie, R. G. (1991) in CRC Handbook of Viruses Infecting Legumes (CRC, Boca Raton, FL), pp. 293-319.
- 3.Grieco, F., Lanave, C. & Gallitelli, D. (1997) Virology 229, 166-174. [DOI] [PubMed] [Google Scholar]
- 4.Jordá, C., Alfaro, A., Aranda, M. A., Moriones, E. & García-Arenal, F. (1992) Plant Dis. 76, 363-366. [Google Scholar]
- 5.Kaper, J. M. & Tousignant, M. E. (1977) Virology 80, 186-195. [DOI] [PubMed] [Google Scholar]
- 6.García-Arenal, F. & Palukaitis, P. (1999) in Satellites and Defective Viral RNAs, eds. Vogt, P. K. & Jackson, A. O. (Springer, Berlin), pp. 37-63.
- 7.Xu, P. & Roossinck, M. J. (2000) Plant Cell 12, 1079-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu, P., Blancaflor, E. B. & Roossinck, M. J. (2003) Mol. Plant–Microbe Interact. 16, 467-476. [DOI] [PubMed] [Google Scholar]
- 9.Hoeberichts, F. A. & Woltering, E. J. (2002) BioEssays 25, 47-57. [DOI] [PubMed] [Google Scholar]
- 10.Steller, H. (1995) Science 267, 1445-1449. [DOI] [PubMed] [Google Scholar]
- 11.Schendel, S. L., Montal, M. & Reed, J. C. (1998) Cell Death Differ. 5, 372-380. [DOI] [PubMed] [Google Scholar]
- 12.Lutz, R. J. (2000) Cell Survival Apoptosis 28, 51-56. [Google Scholar]
- 13.Uren, A. G., O'Rourke, K., Aravind, L., Pisabarro, M. T., Seshagiri, S., Koonin, E. V. & Dixit, V. M. (2000) Mol. Cell 6, 961-967. [DOI] [PubMed] [Google Scholar]
- 14.Lacomme, C. & Cruz, S. S. (1999) Proc. Natl. Acad. Sci. USA 96, 7956-7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawai-Yamada, M., Jin, L., Yoshinaga, K., Hirata, A. & Uchimiya, H. (2001) Proc. Natl. Acad. Sci. USA 98, 12295-12300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dickman, M. B., Park, Y. K., Oltersdorf, T., Li, W., Clemente, T. & French, R. (2001) Proc. Natl. Acad. Sci. USA 98, 6957-6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiao, J., Mitsuhara, I., Yazaki, Y., Sakano, K., Gotoh, Y., Miura, M. & Ohashi, Y. (2002) Plant Cell Physiol. 43, 992-1005. [DOI] [PubMed] [Google Scholar]
- 18.Hansen, G. (2000) Mol. Plant–Microbe Interact. 13, 649-657. [DOI] [PubMed] [Google Scholar]
- 19.Lincoln, J. E., Richael, C., Overduin, B., Smith, K., Bostock, R. & Gilchrist, D. G. (2002) Proc. Natl. Acad. Sci. USA 99, 15217-15221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.del Pozo, O. & Lam, E. (1998) Curr. Biol. 8, 1129-1132. [DOI] [PubMed] [Google Scholar]
- 21.del Pozo, O. & Lam, E. (2003) Mol. Plant–Microbe Interact. 16, 485-494. [DOI] [PubMed] [Google Scholar]
- 22.Mitsuhara, I., Malik, K. A., Miura, M. & Ohashi, Y. (1999) Curr. Biol. 1999, 775-778. [DOI] [PubMed] [Google Scholar]
- 23.Koukalová, B., Kovarík, A., Fajkus, J. & Siroky, J. (1997) FEBS Lett. 414, 289-292. [DOI] [PubMed] [Google Scholar]
- 24.Fillatti, J. J., Kiser, J., Rose, R. & Comai, L. (1987) Bio/Technology 5, 729-730. [Google Scholar]
- 25.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 26.Xie, D.-Y., Sharma, S. B., Paiva, N. L., Ferreira, D. & Dixon, R. A. (2003) Science 299, 396-399. [DOI] [PubMed] [Google Scholar]
- 27.Carthy, C. M., Yanagawa, B., Luo, H., Granville, D. J., Yang, D., Cheung, P., Cheung, C., Esfandiarei, M., Rudin, C. M., Thompson, C. B., et al. (2003) Virology 313, 147-157. [DOI] [PubMed] [Google Scholar]
- 28.Kratsch, H. A. & Wise, R. R. (2000) Plant Cell Environ. 23, 337-350. [Google Scholar]
- 29.Yen, C.-H. & Yang, C.-H. (1998) Plant Cell Physiol. 39, 922-927. [DOI] [PubMed] [Google Scholar]
- 30.Thomas, H. & Stoddart, J. L. (1980) Ann. Rev. Plant Physiol. 31, 83-111. [Google Scholar]
- 31.Chalker-Scott, L. (1999) Photochem. Photobiol. 70, 1-9. [Google Scholar]
- 32.Christie, P. J., Alfenito, M. R. & Walbot, V. (1994) Planta 194, 541-549. [Google Scholar]
- 33.Nagata, T., Todoriki, S., Masumizu, T., Suda, I., Furuta, S., Du, Z. & Kikuchi, S. (2003) J. Agric. Food Chem. 51, 2992-2999. [DOI] [PubMed] [Google Scholar]
- 34.Feild, T. S., Lee, D. W. & Holbrook, N. M. (2001) Plant Physiol. 127, 566-574. [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng, E. H. Y., Levine, B., Boise, L. H., Thompson, C. B. & Hardwick, J. M. (1996) Nature 379, 554-557. [DOI] [PubMed] [Google Scholar]
- 36.Epple, P., Mack, A. A., Morris, V. R. F. & Dangl, J. L. (2003) Proc. Natl. Acad. Sci. USA 100, 6831-6836. [DOI] [PMC free article] [PubMed] [Google Scholar]