Abstract
SIZ1 is a small ubiquitin‐related modifier (SUMO) E3 ligase that mediates post‐translational SUMO modification of target proteins and thereby regulates developmental processes and hormonal and environmental stress responses in Arabidopsis. However, the role of SUMO E3 ligases in crop plants is largely unknown. Here, we identified and characterized two Glycine max (soybean) SUMO E3 ligases, GmSIZ1a and GmSIZ1b. Expression of GmSIZ1a and GmSIZ1b was induced in response to salicylic acid (SA), heat, and dehydration treatment, but not in response to cold, abscisic acid (ABA), and NaCl treatment. Although GmSIZ1a was expressed at higher levels than GmSIZ1b, both genes encoded proteins with SUMO E3 ligase activity in vivo. Heterologous expression of GmSIZ1a or GmSIZ1b rescued the mutant phenotype of Arabidopsis siz1‐2, including dwarfism, constitutively activated expression of pathogen‐related genes, and ABA‐sensitive seed germination. Simultaneous downregulation of GmSIZ1a and GmSIZ1b (GmSIZ1a/b) using RNA interference (RNAi)‐mediated gene silencing decreased heat shock‐induced SUMO conjugation in soybean. Moreover, GmSIZ1RNAi plants exhibited reduced plant height and leaf size. However, unlike Arabidopsis siz1‐2 mutant plants, flowering time and SA levels were not significantly altered in GmSIZ1RNAi plants. Taken together, our results indicate that GmSIZ1a and GmSIZ1b mediate SUMO modification and positively regulate vegetative growth in soybean.
Keywords: SIZ1, soybean, SUMO, SUMO E3 ligase, vegetative growth
Edited by: Zhongchi Liu, University of Maryland, USA
INTRODUCTION
Since the discovery of the SUMO peptide, researchers have established that SUMO conjugation to proteins (SUMOylation) profoundly influences biological processes including innate immunity, stress responses, DNA repair and transcriptional regulation (Enserink 2015). SUMOylation is a rapid and reversible post‐translational modification that affects protein–protein interactions, protein targeting, enzymatic activity, and protein stability (Cubeñas‐Potts and Matunis 2013). Although SUMOylation is widely known as a regulator of nuclear processes, growing evidence indicates that it also regulates non‐nuclear processes, such as channel activity, receptor and cytoskeletal functions, autophagy, and exocytosis (Gill 2004; Wasik and Filipek 2014).
SUMOs have a similar three‐dimensional structure to ubiquitin, but the amino acid sequences of these proteins share only approximately 20% similarity and the surface topology of SUMO is substantially different from that of ubiquitin (Müller et al. 2001). SUMO is translated as a pre‐protein (known as pre‐SUMO) and a SUMO‐specific cysteine protease (SUMO protease) deletes a short C‐terminal fragment (immediately downstream of a C‐terminal GG motif) to produce active mature SUMO (also known as free SUMO). SUMO conjugates to its target substrate in a stepwise manner via activation (E1), conjugation (E2), and ligation (E3) (Müller et al. 2001). SUMO is often conjugated to a ΨKXE/D motif (where Ψ is a large hydrophobic residue; K is lysine; X is any residue; and E/D is glutamic acid or aspartic acid) in substrates, resulting in an isopeptide bond between the C‐terminal G residue in SUMO and the K residue in the substrate (Johnson 2004). SUMO proteases cleave the isopeptide bond between SUMO and its substrate (Kim and Baek 2009).
SUMOylation has been implicated in the regulation of developmental, hormonal, and environmental responses in Arabidopsis, such as gametophyte development (Ling et al. 2012; Liu et al. 2014), embryogenesis (Saracco et al. 2007), photomorphogenesis (Sadanandom et al. 2015; Lin et al. 2016), flowering time (Murtas et al. 2003; Jin et al. 2008), cell proliferation (Huang et al. 2009; Ishida et al. 2009), abscisic acid (ABA) signaling (Miura et al. 2009; Zheng et al. 2012), gibberellic acid (GA) signaling (Kim et al. 2015), the salt stress response (Conti et al. 2008), thermal adaptation (Yoo et al. 2006; Miura et al. 2007), the drought stress response (Catala et al. 2007; Zhang et al. 2013), immune responses (Lee et al. 2007; Saleh et al. 2015), and nutrient (phosphate and nitrogen) starvation signaling (Miura et al. 2005; Park et al. 2011). The SUMO regulatory mechanism is conserved in Oryza sativa (rice), Zea mays (maize), Dendrobium (orchids), and Malus domestica (apple; Park et al. 2010; Liu et al. 2015; Augustine et al. 2016; Zhang et al. 2016). The SUMO E3 ligase OsSIZ1 regulates phosphate‐ and nitrogen‐dependent responses, spikelet fertility, and plant development in rice (Thangasamy et al. 2011; Wang et al. 2011, 2015). Moreover, the OsOTS1 SUMO protease positively regulates salt stress responses in rice (Srivastava et al. 2016). Mutations in an Arabidopsis SUMO E3 ligase, AtSIZ1, increase salicylic acid (SA) levels, resulting in reduced plant stature, constitutively activated immune responses, and early flowering (Lee et al. 2007; Jin et al. 2008; Miura et al. 2010). However, whether this regulatory mechanism is conserved in other plant species remains to be determined.
The function of SUMOylation in soybean, an important crop plant, is unknown. In this study, we identified and characterized two soybean SUMO E3 ligases, GmSIZ1a and GmSIZ1b. We demonstrated that both are bona fide SUMO E3 ligases that positively regulate vegetative growth in soybean. GmSIZ1a and GmSIZ1b are required for heat shock‐induced GmSUMO1 conjugation, implying that GmSIZ1a/b‐mediated SUMO modifications regulate heat stress responses in soybean as they do in Arabidopsis (Yoo et al. 2006). However, in contrast to Arabidopsis plants harboring a mutation in AtSIZ1, downregulation of GmSIZ1a/b did not affect flowering time and SA production. Thus, SUMO E3 ligases may have distinct regulatory roles in soybean.
RESULTS
Identification of genes encoding putative SUMO E3 ligases in soybean
To investigate the role of soybean SUMO E3 ligase, we searched the soybean genome database (http://www.phytozome.net) for SIZ1 homologs. We identified GmSIZ1a (Glyma12g07590, 880 aa) and GmSIZ1b (Glyma11g15880, 879 aa) (Figure S1A). The primary amino acid sequences of these proteins showed 96.2% identity to each other, but approximately 64% identity to AtSIZ1 (Figure S1B). Phylogenetic analysis of putative SIZ1/PIAS‐type SUMO E3 ligases from various plant species showed that GmSIZ1a and GmSIZ1b were closely related to AtSIZ1 in the dicot sub‐class of SUMO E3 ligases (Figure S1C). The gene structure of GmSIZ1a and GmSIZ1b was similar to that of AtSIZ1, although GmSIZ1a and GmSIZ1b had one fewer exon (17 vs. 18 in AtSIZ1) and different intron sizes (Figure S1D). GmSIZ1a and GmSIZ1b contained all five conserved domains found in AtSIZ1, including the N‐terminal scaffold attachment factor A/B/acinus/PIAS (SAP) domain; the plant homeodomain (PHD); the putative PINIT core domain; the SIZ/PIAS‐RING (SP‐RING) domain; and the SUMO‐interacting motif (SIM) (Figure 1A) (Miura et al. 2005), suggesting that GmSIZ1a and GmSIZ1b may have SUMO E3 ligase activity.
Figure 1.
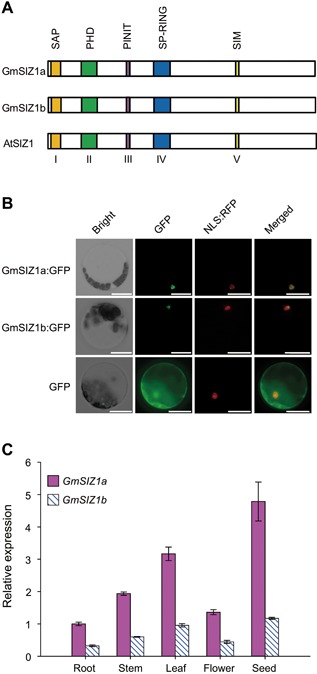
Expression pattern of GmSIZ1a and GmSIZ1b and protein subcellular localization (A) Schematic representation of GmSIZ1a, GmSIZ1b, and AtSIZ1, showing conserved domains in color. (B) Subcellular localization. GmSIZ1a:GFP or GmSIZ1b:GFP was transiently co‐expressed with NLS:RFP (nuclear localization signal:RFP; a nuclear marker) in Arabidopsis protoplasts. Green fluorescent protein (GFP) was used as a control. Bars = 20 μm. (C) Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) analysis of GmSIZ1a and GmSIZ1b expression in different tissues. Relative expression was normalized to that of GmUBI3, and data represent the mean ± SD, n = 3.
GmSIZ1a and GmSIZ1b are nuclear proteins
To determine the subcellular localization of GmSIZ1a and GmSIZ1b, we transiently expressed C‐terminal green fluorescent protein (GFP) fusions of these proteins in Arabidopsis protoplasts (Figure 1B). While GFP alone was localized to both the nucleus and cytosol (Figure 1B, bottom panel), GmSIZ1a:GFP and GmSIZ1b:GFP were exclusively localized to the nucleus (Figure 1B, upper and middle panel, respectively), suggesting that GmSIZ1a and GmSIZ1b are nuclear proteins.
Expression patterns of GmSIZ1a and GmSIZ1b
To characterize the expression profiles of GmSIZ1a and GmSIZ1b in different tissues of soybean, we analyzed GmSIZ1a and GmSIZ1b expression levels by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) (Figure 1C). Expression of both genes was higher in seeds and leaves than in other tissues examined. Next, we analyzed the promoter regions (1,500 bp upstream of the ATG translation start codon) of GmSIZ1a and GmSIZ1b using the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/search_CARE.html/), and identified several putative abiotic stress, biotic stress, and hormone response cis‐acting regulatory elements, including those related to heat, low‐temperature, defense, ABA, SA, jasmonic acid, and light (Table S1). This observation prompted us to determine the expression levels of GmSIZ1a and GmSIZ1b in response to SA, ABA, heat, dehydration, cold, and salt stress treatments (Figure 2). Consistent with previous reports, the controls GmPR1, GmHSF12, and GmDREB2 were induced by SA, heat, and dehydration treatment, respectively (Figure 2A–C; Chen et al. 2007; Sandhu et al. 2009; Chung et al. 2013). Under these conditions, transcript accumulation of GmSIZ1a and GmSIZ1b also increased significantly (Figure 2A–C). By contrast, cold, ABA, and NaCl treatment activated the expression of the control gene GmDREB2, but did not significantly affect transcript accumulation of GmSIZ1a and GmSIZ1b (Figure 2D–F). These results suggest that GmSIZ1a and GmSIZ1b function in certain biotic/abiotic stress responses.
Figure 2.
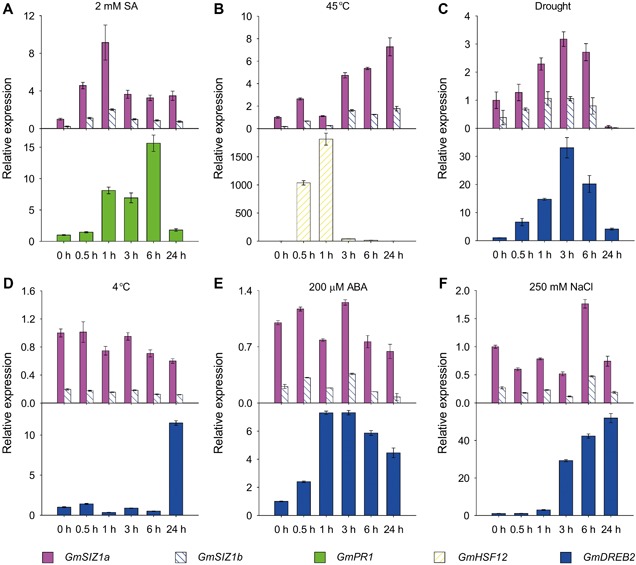
Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) analysis of GmSIZ1a and GmSIZ1b expression level in response to salicylic acid (SA), heat, drought, cold, abscisic acid (ABA) and NaCl treatments Fourteen‐day‐old soybean seedlings were treated with 2 mM SA (A), 45 °C heat stress (B), drought (C), 4 °C cold stress (D), 200 μM ABA (E), and 250 mM NaCl (F) for the indicated periods. GmPR1, GmDREB2, and GmHSF12 were used as stress‐ or hormone‐responsive positive controls. Relative expression was normalized to that of GmUBI3, and data represent the mean ± SD, n = 3.
Both GmSIZ1a and GmSIZ1b are functional SUMO E3 ligases
To explore the biochemical activity of GmSIZ1a and GmSIZ1b, we transformed Arabidopsis siz1‐2 plants with constructs expressing C‐terminal GFP fusions of GmSIZ1a or GmSIZ1b under the AtSIZ1 promoter (ProAtSIZ1:GmSIZ1a:GFP or ProAtSIZ1:GmSIZ1b:GFP, respectively), and three independent transgenic lines were used for further analysis (Figure S2A, B). Both GmSIZ1a and GmSIZ1b partially suppressed the dwarf phenotype of the Arabidopsis siz1‐2 plants (Figures 3A, B, S2C). Due to increased levels of SA in the siz1‐2 plants, the expression of PR genes and ISOCHORISMATE SYNTHASE1 (ICS1) is increased (Lee et al. 2007). The elevated PR and ICS1 gene expression level was almost completely rescued by the heterologous expression of GmSIZ1a or GmSIZ1b in siz1‐2 (Figure 3C). Moreover, the ABA‐sensitive seed germination phenotype of siz1‐2 (Miura et al. 2009) was suppressed by the expression of GmSIZ1a or GmSIZ1b (Figure 3D, E). These results indicate that both GmSIZ1a and GmSIZ1b are biologically functional proteins that may have SUMO E3 ligase activity.
Figure 3.
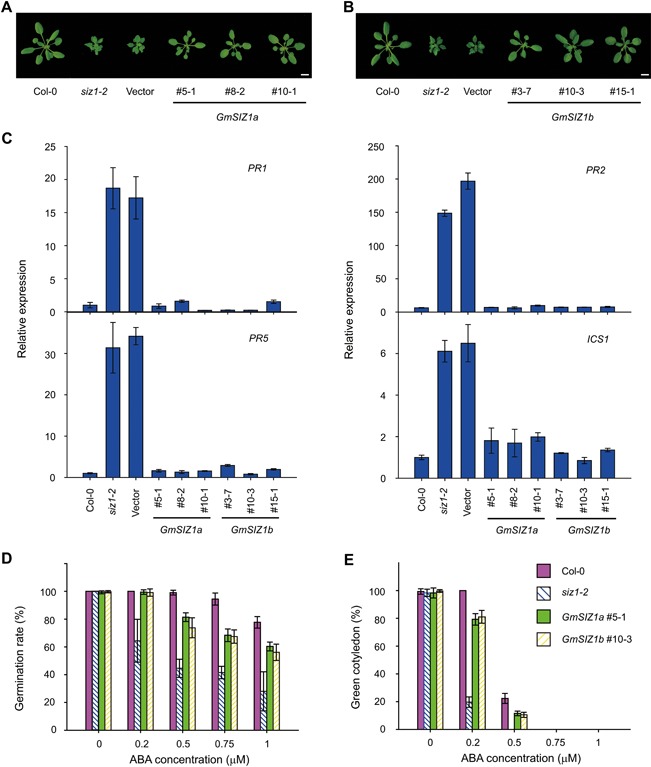
GmSIZ1a and GmSIZ1b rescue the Arabidopsis siz1‐2 mutant phenotype (A) and (B) Morphological phenotype of wild‐type, siz1‐2, and complementation plants. GmSIZ1a (#5‐1, #8‐2 and #10‐1) and GmSIZ1b (#3‐7, #10‐3 and #15‐1) indicate lines expressing GmSIZ1a:GFP or GmSIZ1b:GFP driven by the Arabidopsis SIZ1 promoter in the siz1‐2 background, respectively. Vector indicates empty vector transformed into siz1‐2 plants. Bars = 1 cm. (C) Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) analysis of Arabidopsis PR1, PR2, PR5, and ICS1 expression in Col‐0, siz1‐2, and complementation lines under normal conditions. Relative expression was normalized to that of AtUBC and data represent the mean ± SD, n = 3. (D) Seed germination rate of Col‐0, siz1‐2, GmSIZ1a #5‐1, and GmSIZ1b #10‐3 in the presence of the indicated concentrations of abscisic acid (ABA) at 4 days after planting. Data are the mean ± SD, n = 3. (E) Percentage of green cotyledons in Col‐0, siz1‐2, GmSIZ1a #5‐1, and GmSIZ1b #10‐3 seedlings at 7 days after planting in the presence of ABA. Data are the mean ± SD. Three biological replicates were performed.
The reduced SUMO E3 ligase activity in siz1 attenuates heat shock‐induced SUMO conjugation (Miura et al. 2005). This activity was restored by expression of GmSIZ1a or GmSIZ1b in siz1 (Figure 4A). We further analyzed the SUMO E3 ligase activity of GmSIZ1a and GmSIZ1b in vivo by transiently co‐expressing GmSIZ1a:GFP, GmSIZ1b:GFP, or AtSIZ1:GFP with FLAG:AtSUMO1 in Nicotiana benthamiana leaves as described previously (Park et al. 2010). Under non‐stress conditions, transient expression of AtSUMO1 alone did not promote SUMO conjugation, but co‐expression of AtSIZ1 with AtSUMO1 greatly induced SUMO conjugation. Similar to AtSIZ1, co‐expression of GmSIZ1a or GmSIZ1b with AtSUMO1 strongly promoted SUMO conjugation (Figure 4B). These results indicate that both GmSIZ1a and GmSIZ1b possess SUMO E3 ligase activity.
Figure 4.
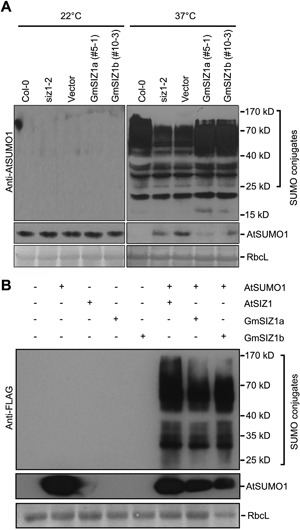
GmSIZ1a and GmSIZ1b exhibit SUMO E3 ligase activity (A) Expression of GmSIZ1a or GmSIZ1b suppresses impaired heat shock‐induced AtSUMO1 conjugation in siz1‐2. Fourteen‐day‐old long‐day‐grown seedlings were subjected to heat shock treatment (37°C) for 30 min, and the heat shock‐induced SUMO conjugates and free SUMO (AtSUMO1/2) were detected with anti‐AtSUMO1 antibody. 22 °C indicates the non‐stressed condition. Coomassie blue‐stained Rubisco large subunit (RbcL) was used as the loading control. (B) GmSIZ1a and GmSIZ1b induce the accumulation of AtSUMO1 conjugates in Nicotiana benthamiana. AtSIZ1:GFP, GmSIZ1a:GFP, or GmSIZ1b:GFP was transiently expressed in N. benthamiana with or without FLAG:AtSUMO1. Proteins were extracted from the leaves three days after infiltration, and the SUMO1 conjugates and free SUMO (AtSUMO1) were detected with anti‐FLAG antibody. Coomassie blue‐stained RbcL was used as a loading control.
GmSIZ1a and GmSIZ1b function as SUMO E3 ligases in soybean
To characterize the function of GmSIZ1a and GmSIZ1b in soybean, we simultaneously downregulated both GmSIZ1a and GmSIZ1b by RNA interference‐mediated gene silencing. We generated the GmSIZ1RNAi vector using shared sequences expected to silence both GmSIZ1a and GmSIZ1b simultaneously (Figures S1D, S3A). Three independent T3 GmSIZ1RNAi transgenic lines were obtained in the soybean cultivar Zhongdou 32 (35A3, 38A2 and 45B3) (Figure S3B–D). The expression of both GmSIZ1a and GmSIZ1b was significantly reduced in the GmSIZ1RNAi lines (Figure S3E).
To investigate whether the SUMO E3 ligase activity was reduced in the GmSIZ1RNAi plants, we analyzed heat shock‐induced SUMO conjugation. We attempted to use commercial anti‐SUMO1 antibody (Abcam, ab5316; a rabbit polyclonal antibody that reacts with Arabidopsis SUMO1/2) to detect the heat shock‐induced SUMO conjugation in Zhongdou 32, but we did not observe substantial levels of SUMO conjugates. The Soybean genome contains three identical SUMO isoforms, GmSUMO1, 2 and 3, which are closely related to AtSUMO2 (Figure S4A, B). A predicted 3D structure comparison suggested that AtSUMO1 and GmSUMO1 have similar structures, but may have different epitopes (Figure S4C). Therefore, we generated anti‐GmSUMO1 polyclonal antibody, and monitored SUMO conjugates under heat shock conditions (Figure 5A). Heat shock‐induced GmSUMO1 conjugates were substantially increased at 0.5 h after heat shock treatment, but gradually decreased at later time points (Figure 5A). However, anti‐GmSUMO1 antibody did not detect SUMO conjugates in Arabidopsis (Figure S4D).
Figure 5.
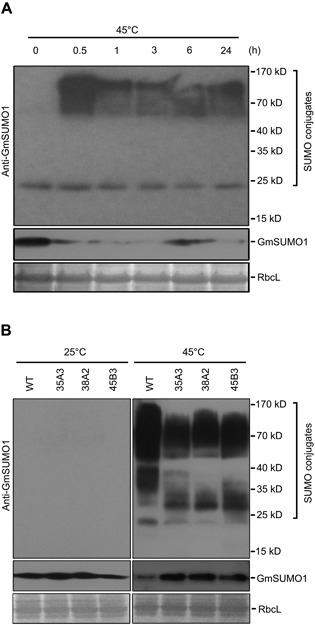
Downregulation of GmSIZ1a and GmSIZ1b causes reduced heat shock‐induced GmSUMO1 conjugation in soybean (A) Heat shock‐induced GmSUMO1 conjugates in soybean. Fourteen‐day‐old Zhongdou 32 (wild type) soybean seedlings were treated with heat shock (45 °C) for the indicated periods, and GmSUMO1 conjugates and free SUMO (GmSUMO1) were detected using anti‐GmSUMO1 antibody. Coomassie blue‐stained RbcL was used as a loading control. (B) GmSUMO1 conjugates and free SUMO (GmSUMO1) were detected with anti‐GmSUMO1 antibody under normal (25 °C) and heat shock (45 °C for 30 min) conditions. Proteins were extracted from 14‐day‐old wild‐type (WT) soybean seedlings and three independent GmSIZ1RNAi lines (35A3, 38A2 and 45B3). RbcL was used as the loading control.
In the absence of heat shock treatment, SUMO conjugates were hardly detected in GmSIZ1RNAi plants and the level of free SUMO was similar in wild‐type and GmSIZ1RNAi plants (Figure 5B, left panel). Under heat‐shock conditions, the SUMO conjugates were greatly increased in the wild type, but were increased to a lesser extent in GmSIZ1RNAi plants. In agreement with increased levels of SUMO conjugates, the level of free SUMO was lower in wild‐type plants than in GmSIZ1RNAi plants under heat‐shock conditions (Figure 5B). These results indicate that GmSIZ1a and GmSIZ1b facilitate SUMO conjugation in soybean.
GmSIZ1a and GmSIZ1b regulate vegetative growth in soybean
Mutations in Arabidopsis SIZ1 cause elevated SA levels, which results in dwarfism and early flowering (Lee et al. 2007; Jin et al. 2008). Similar to the Arabidopsis siz1‐2 plants, the GmSIZ1RNAi plants showed slightly reduced leaf size and plant height compared to those of the wild type (Figure 6A, B). However, downregulation of GmSIZ1a/b did not alter the flowering time (Figure 6C). To test if the GmSIZ1RNAi plants accumulated higher levels of SA, as did Arabidopsis siz1, we examined the SA levels in wild‐type and GmSIZ1RNAi leaves under normal conditions (Figure 6D). Surprisingly, wild‐type and GmSIZ1RNAi plants accumulated similar basal levels of SA. Moreover, in contrast to the constitutive activation of PR gene expression observed in Arabidopsis siz1‐2 pants (Lee et al. 2007), GmPR1 and GmPR10 expression was not induced in GmSIZ1RNAi plants, suggesting that downregulation of GmSIZ1a and GmSIZ1b expression does not cause a constitutive immune response (Figure 6E, F). Overall, our results indicate that GmSIZ1a and GmSIZ1b positively regulate vegetative growth.
Figure 6.
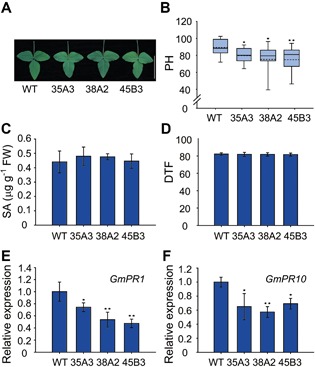
Downregulation of GmSIZ1a and GmSIZ1b causes reduced plant height and leaf size, but does not affect salicylic acid (SA) levels or flowering time in soybean (A) First trifoliate leaf of the wild type and three independent GmSIZ1RNAi lines (35A3, 38A2, and 45B3). Bar = 10 cm. (B) Plant height (PH) of 5‐week‐old soybean plants. (C) SA levels in 14‐day‐old wild‐type and GmSIZ1RNAi plants. Data represent the mean ± SD. (D) Flowering time (days to flower, DTF) of soybean plants. (E) and (F) quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) analysis of GmPR1 and GmPR10 in 14‐day‐old WT and GmSIZ1RNAi leaves. Relative expression was normalized to that of GmUBI3, and data represent the mean ± SD. Twenty soybean plants (Zhongdou 32 cultivars; WT) and each GmSIZ1RNAi transgenic line in the Zhongdou 32 background (35A3, 38A2 and 45B3) were grown in a greenhouse, and plant height and flowering time were recorded. (One‐way ANOVA test, **P < 0.01, *P < 0.05).
DISCUSSION
The functions of SUMO E3 ligases in the regulation of environmental stress responses and developmental processes have been extensively characterized in Arabidopsis (Park and Yun 2013). However, it was hitherto unknown whether SUMO E3 ligases have conserved functions in soybean. In this study, we identified two identical SIZ1 homologs in soybean, GmSIZ1a and GmSIZ1b, which localized to the nucleus (Figure S1A, B). Both proteins exhibited SUMO E3 ligase activity (Figures 3, 4) and promoted GmSUMO1 conjugation in soybean (Figure 5). Interestingly, downregulation of GmSIZ1a/b in soybean resulted in plants with reduced height and leaf size (Figure 6A, B). Thus, our study established that the SUMO E3 ligases GmSIZ1a and GmSIZ1b mediate SUMO modification of nuclear proteins and regulate vegetative growth in soybean.
In Arabidopsis, mutation of AtSIZ1 causes reduced plant stature (Catala et al. 2007). The dwarfism of siz1 plants is due to defects in cell division and expansion, which are caused by hyper‐accumulation of SA in the mutant (Miura et al. 2010). The rice Ossiz1 loss‐of‐function mutant also exhibits reduced plant height and shorter leaves, which are also likely attributable to defects in cell proliferation and expansion (Wang et al. 2011). Thus, it is possible that the reduced plant stature of GmSIZ1RNAi plants is also due to defects in cell division and expansion. In contrast to the Arabidopsis siz1 mutant, GmSIZ1RNAi plants did not accumulate a higher level of SA than the wild type (Figure 6D; Lee et al. 2007). Moreover, exogenous SA treatment stimulates shoot and root growth in soybean (Rivas‐San Vicente and Plasenciaa 2011). These results suggest that GmSIZ1a and GmSIZ1b regulate vegetative growth through an SA‐independent mechanism in soybean. It has been shown that AtSIZ1 is required for nitrogen assimilation and that application of exogenous ammonium rescues the dwarf phenotype of siz1‐2 plants (Park et al. 2011). By contrast, Ossiz1 rice mutant plants accumulate higher levels of nitrogen, suggesting that OsSIZ1 negatively regulates nitrogen assimilation in rice (Wang et al. 2015). Whether GmSIZ1a/b affects vegetative growth by regulating nitrogen assimilation remains to be determined.
SUMO modification also regulates flowering time in Arabidopsis. Mutations in SUMO E3 ligases (i.e., SIZ1 and HPY2/MMS21) or SUMO proteases (i.e., ESD4 and OTS1/2) cause early flowering in Arabidopsis (Murtas et al. 2003; Conti et al. 2008; Jin et al. 2008; Kwak et al. 2016). The early flowering phenotype of siz1 and esd4 plants under short‐day conditions is mainly due to hyper‐accumulation of SA (Jin et al. 2008; Villajuana‐Bonequi et al. 2014). In addition to Arabidopsis, SA promotes flowering in various plant species, such as Nicotiana tabacum (tobacco), members of the Lemnaceae family, and Helianthus annuus (sunflower). However, in the short‐day species Pharbitis nil, exogenous SA application promotes flowering only under nutrient deprivation conditions (Rivas‐San Vicente and Plasenciaa 2011). We found that downregulation of GmSIZ1a/b did not affect flowering time or SA levels in soybean (Figure 6). Thus, we speculate that GmSIZ1a/b modifies the nuclear localization of SUMO substrates that are not involved in the regulation of flowering time and SA accumulation. However, we cannot exclude the possibility that complete loss‐of‐function mutations of GmSIZ1a/b would alter flowering time and SA accumulation in soybean. Furthermore, GmSIZ1a/b may function redundantly with other SUMO E3 ligases to regulate flowering time and SA accumulation in soybean.
As plants are sessile organisms and are often exposed to abiotic stresses and pathogen attacks, they have evolved multi‐layered defense systems. SUMOylation plays a pivotal role in the regulation of abiotic stress responses (i.e., ABA signaling and salt, cold, heat and drought stress responses) and immune responses (Yoo et al. 2006; Catala et al. 2007; Lee et al. 2007; Miura et al. 2007, 2009; Conti et al. 2008; Zheng et al. 2012; Zhang et al. 2013). GmSIZ1a and GmSIZ1b expression was induced by SA, heat, and dehydration treatments (Figure 2A–C). Moreover, GmSIZ1a and GmSIZ1b were required for heat shock‐induced SUMO conjugation in soybean (Figure 5B). Downregulation of GmSIZ1a and GmSIZ1b did not cause constitutively elevated SA levels and PR gene expression in the absence of pathogen infection (Figure 6D–F). However, we cannot exclude the possibility that mutations in GmSIZ1a and GmSIZ1b cause higher levels of SA and PR gene expression under pathogen infection conditions than those observed in the wild type. Similar to SIZ1 in Arabidopsis, GmSIZ1a/b may also regulate heat, drought, and biotic stress responses in soybean. Although cold, ABA, and NaCl treatments did not affect the expression level of GmSIZ1a and GmSIZ1b, we cannot exclude the possibility that GmSIZ1a/b also regulate cold, ABA, and salt stress responses (Figure 2D–F), since SUMO modification regulates these stress responses at the post‐translational level (Miura et al. 2007, 2009; Conti et al. 2014). Given that SUMO modification is a key regulatory mechanism in biotic/abiotic stress responses in Arabidopsis, it would be of great interest to determine if GmSIZ1a/b regulate environmental stress responses in soybean.
MATERIALS AND METHODS
Plant materials and growth conditions
Soybean cultivar Glycine max L. Merr. cv. Zhongdou 32 was used in this study. Soybean seeds were germinated on filter paper moistened with deionized water for 1 d and then transferred to soil for further growth in the greenhouse (in the spring, under natural light plus incandescent light, at 25 °C to 30 °C) or in a plant growth room (16 h light (100 µmol m−2 s−1)/8 h darkness, at 25 °C). The Arabidopsis wild type and siz1‐2 (Salk_065397) plants used in this work were in the Columbia (Col‐0) background. The Arabidopsis plants were grown in a growth room under long‐day conditions (16 h light (100 µmol m−2 s−1 fluorescent lights)/8 h darkness) at 22 °C.
Bioinformatics analysis
Amino acid sequences of plant SIZ1 homologs were collected from the National Center for Biotechnology Information (NCBI) and Phytozome databases. ClustalW2 software was used to align these sequences, and the percent identity was determined and the divergence matrices were constructed using DNASTAR software (DNAStar Inc., Madison, WI, USA). The phylogenetic tree was constructed using the neighbor‐joining algorithm as instructed in MEGA 5.1 (Kumar et al. 2008). The homology model of the three‐dimensional (3D) structure was constructed using the Phyre server (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index) and the figures were generated using PyMOL (http://pymol.org/).
RNA isolation and real‐time reverse transcription‐PCR
RNA was isolated and qRT‐PCR was performed as described previously (Zhou et al. 2013). Briefly, total RNAs were isolated from Arabidopsis or soybean seedlings using TRIZOL reagents (RNAiso Plus, Code D9108B; TaKaRa). qRT‐PCR analyses were performed on an MX3000P QPCR system. Arabidopsis UBIQUITIN‐CONJUGATING ENZYME (UBC) or soybean UBIQUITIN‐LIKE PROTEIN3 (GmUBI3) was used as the internal control. The primers used for qRT‐PCR are listed in Table S2.
Plasmid construction
To generate p326‐GmSIZ1a/b:GFP, full‐length GmSIZ1a or GmSIZ1b cDNA without the termination codon was amplified using the gene‐specific primers GmSIZ1a/b‐F‐BamHI and GmSIZ1a/b‐R‐BamHI. The resulting products were inserted in frame at the BamHI site of the p326‐GFP vector (Zhou et al. 2013).
To generate pCambia1302‐ProAtSIZ1:GmSIZ1a/b:GFP, full‐length GmSIZ1a or GmSIZ1b cDNA without the termination codon was amplified using the gene‐specific primers GmSIZ1a/b‐F‐HindIII and GmSIZ1a/b‐R‐NcoI, and ligated into the pCambia1302 vector. pCambia1302‐GmSIZ1a/b:GFP plasmids were digested with XmaI, and the AtSIZ1 promoter (Jin et al. 2008) was inserted into the XmaI site of pCambia1302‐GmSIZ1a/b:GFP.
To generate pCambia3300‐GmSIZ1a/b:GFP, 326‐GmSIZ1a/b:GFP plasmids were digested with BamHI and the insert was ligated into the BamHI site of the pCambia3300‐GFP vector.
To generate pCambia3300‐GmSIZ1RNAi, pFGC1008 was digested with SacI/PmeI, and the insert was ligated into the SacI/PmeI sites of pCambia3300. A GmSIZ1a fragment was amplified with the GmSIZ1RNAi‐F and GmSIZ1RNAi‐R primers, and ligated into the AscI/SwaI and BamHI/SpeI sites of the pCambia3300‐RNAi vector.
To generate pET30a‐His:GmSUMO1, GmSUMO1 cDNA was amplified with the gene‐specific primers SUMO‐F‐BamHI and SUMO‐R‐XhoI, and inserted into the BamHI and XhoI sites of the pET30a vector. All primer sequences are listed in Table S2.
Subcellular localization
Plasmids were transformed into Arabidopsis protoplasts using polyethylene glycol (PEG)‐mediated DNA transfection (Jin et al. 2001). The fluorescence images were captured using a fluorescence microscope (Olympus BX53).
Antibody preparation
pET30a‐His:GmSUMO1 plasmid was transformed into the Escherichia coli BL21 (DE3) strain, and the expressed His‐GmSUMO1 fusion protein was purified with Ni Sepharose 6 Fast Flow resin (GE Healthcare). The purified full‐length soybean SUMO1 protein was used to generate anti‐GmSUMO1 rabbit polyclonal antibody.
Analysis of SUMO conjugation
Total proteins were extracted from Arabidopsis, N. benthamiana or soybean leaves using protein extraction buffer containing 150 mM Tris–HCl (pH 7.5), 150 mM NaCl, 5% (w/v) sodium dodecyl sulfate (SDS), 0.5% (v/v) NP40, 6 mM ethylenediaminetetraacetic acid (EDTA), 3 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF) and 30% (v/v) glycerol. The proteins were separated by sodium dodecyl sulfate‐polyacrylamide gel electrophoresis (SDS–PAGE) and detected with anti‐AtSUMO1 (Abcam, ab5316), anti‐FLAG (Sigma, F3165), or anti‐GmSUMO1 antibody.
Generation of transgenic plants
The pCambia3300‐GmSIZ1RNAi plasmid was transformed into Agrobacterium tumefaciens strain EHA105 using the freeze‐thaw method. Soybean transformation was carried out using the Agrobacterium‐mediated cotyledonary‐node method as described previously (Paz et al. 2004). Expression and integration of the bar marker gene in the GmSIZ1RNAi lines were confirmed based on leaf Glufosinate resistance, using bar gene detection strips (QuickStix for LibertyLink/bar, Envirologix, USA) and genomic DNA PCR. pCambia1302‐ProAtSIZ1:GmSUMO1a/b:GFP were introduced into A. tumefaciens strain GV3101, and transformed into Arabidopsis siz1‐2 using the floral‐dip method as previously described (Clough and Bent 1998).
SA measurement
Salicylic acid was extracted from 14‐day‐old long‐day‐grown soybean seedlings and quantified using a LECO Pegasus IV gas chromatography time‐of‐flight mass spectrometry (GC‐TOF/MS) system as previously described (Duan et al. 2012).
AUTHOR CONTRIBUTIONS
B.C. performed most of the research. X.K. generated the transgenic Arabidopsis plants and C.Z., S.S., Z.Z., Y.W. and X.L. performed some of the phenotypic analyses and designed the experiments. X.F.Z. and Y.H.J cloned the genes and constructed the plasmids. J.B.J. designed the research, supervised the study, and wrote the manuscript.
Supporting information
Additional Supporting Information may be found online in the supporting information tab for this article: http://onlinelibrary.wiley.com/doi/10.1111/jipb.12504/suppinfo
Figure S1. Amino acid sequence analysis of soybean GmSIZ1a and GmSIZ1b, and their gene structures
(A) Amino acid sequence alignment of GmSIZ1a, GmSIZ1b,and AtSIZ1. Identical and similar amino acid residues are highlighted with black and gray boxes, respectively. (B) Percentage of identity and divergence among GmSIZ1a, GmSIZ1b, and AtSIZ1 amino acid sequences. (C) Phylogenetic tree of SIZ1 homologs in various plant species. A neighbor‐joining tree was constructed based on an alignment of the complete protein sequences. The numbers on the side of each branch indicate bootstrap values from 1,000 replicates.The scale bar indicates the substitution rate per site. (D) Schematic representation of the GmSIZ1a, GmSIZ1b, and AtSIZ1 gene structures. The blue boxes represent exons and the green lines represent introns. The gray boxes represent UTR regions. PF1/PR1 and PF2/PR2 primer pairs were used for qRT‐PCRanalysis of GmSIZ1a and GmSIZ1b expression, respectively. Black line indicates the region used to generate GmSIZ1RNAi constructs.
Figure S2. Expression of GmSIZ1a or GmSIZ1b rescues the dwarf phenotype of Arabidopsis siz1‐2
(A) Presence of the siz1‐2 mutation was analyzed in complementation lines. GmSIZ1a (#5‐1, #8‐2, and #10‐1) and GmSIZ1b (#3‐7, #10‐3, and #15‐1) indicate the expression of GmSIZ1a:GFP or GmSIZ1b:GFP driven by the Arabidopsis SIZ1 promoter in siz1‐2, respectively. Vector indicates empty vector transformed into siz1‐2. Primer pair LB_a1 (LB)/RP was used to determine the presence of a T‐DNA insertion in the siz1 genetic background, and primer pair LP/RP was used to confirm homozygosity. (B) qRT‐PCR analysis of transgene expression in Col‐0, siz1‐2, the vector control, and GmSIZ1a/b transgenic plants.Relative expression was normalized to that of AtUBC, and data represent the mean ±SD, n=3. (C) Leaf length and rosette diameter of five‐week‐old long‐day‐grown plants. The lengths of leaves from 5 plants were measured.
Figure S3. Generation of GmSIZ1RNAi soybean transgenic plants
(A) Schematic representation of the GmSIZ1RNAi construct and nucleotide sequence alignment of the GmSIZ1a/b cDNA fragment, which was used to generate the GmSIZ1RNAi construct. (B) Glufosinate resistance of GmSIZ1RNAi (35A3, 38A2 and 45B3) leaves. The non‐transgenic soybean cultivar Zhoungdou32 (WT) was used as a negative control. Dashed white box indicates Glufosinate (200 gL−1 glufosinate ammonium, BAYER, Germany) painted leaf area. (C) A bar gene strip (QuickStix for LibertyLink/bar, Envirologix USA) was used to detect the presence of the bar gene in the transgenic plants. Upper and lower arrowheads indicate the control and positive line, respectively. (D) PCR analysis confirmed the presence of the bar gene in the transgenic plants.The Bar‐F and Bar‐R primers were used for the PCR analysis. (E) qRT‐PCR analysis of GmSIZ1a and GmSIZ1b levels in WT and GmSIZ1RNAi soybean transgenic lines. Relative expression was normalized to that of GmUBI3, and data represent the mean ± SD, n = 3. The numbers above the columns indicate the relative transcription level of GmSIZ1a or GmSIZ1b in the transgenic lines compared to in the wild type.
Figure S4. Amino acid sequence analysis and 3D structures of SUMO homologs
(A) Amino acid sequence alignment of SUMO homologs in Arabidopsis, soybean, and rice. Identical and similar amino acid residues are indicated with black and gray boxes, respectively. (B) Phylogenetic analysis of SUMO homologs in Arabidopsis, soybean, and rice. The neighbor‐joining tree was constructed based on an alignment of the complete protein sequences. The numbers on the side of each branch indicate bootstrap values from 1,000 replicates, and the scale bar indicates the substitution rate per site. (C) Three‐dimensional structure modeling of AtSUMO1 and GmSUMO1. (D) Detection of heat shock‐induced SUMO conjugates and free SUMO in soybean and Arabidopsis using anti‐GmSUMO1 antibody. Coomassie blue‐stained Rubisco large subunit (RbcL) was used as the loading control.
Table S1. Putative cis‐acting elements in the GmSIZ1a and GmSIZ1b promoters
Table S2. Primers used in this study
ACKNOWLEDGEMENTS
We thank Wei Wang, Zeliang Lian and Yuan Li for their excellent technical support. This work was supported by grants from the National Natural Science Foundation of China (31471363 for J.B.J.), the Ministry of Science and Technology of the People's Republic of China (2012CB114302 for J.B.J.), the National Transgenic Major Program (2009ZX08009‐087B for J.B.J. and 2009ZX08009‐132B for X.L.) and the Chinese Academy of Sciences (XDA08010105 for J.B.J.).
Cai B, Kong X, Zhong C, Sun S, Zhou XF, Jin YH, Wang Y, Li X, Zhu Z, Jin JB (2017) SUMO E3 Ligases GmSIZ1a and GmSIZ1b regulate vegetative growth in soybean. J Integr Plant Biol 59: 2–14
OnlineOpen, paid by authors
Online on Oct. 20, 2016
REFERENCES
- Augustine RC, York SL, Rytz TC, Vierstra RD (2016) Defining the SUMO system in maize: SUMOylation is up‐regulated during endosperm development and rapidly induced by stress. Plant Physiol 171: 2191–2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catala R, Ouyang J, Abreu IA, Hu Y, Seo H, Zhang X, Chua NH (2007) The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. Plant Cell 19: 2952–2966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Wang QY, Cheng XG, Xu ZS, Li LC, Ye XG, Xia LQ, Ma YZ (2007) GmDREB2, a soybean DRE‐binding transcription factor, conferred drought and high‐salt tolerance in transgenic plants. Biochem Biophys Res Commun 353: 299–305 [DOI] [PubMed] [Google Scholar]
- Chung E, Kim KM, Lee JH (2013) Genome‐wide analysis and molecular characterization of heat shock transcription factor family in Glycine max . J Genet Genomics 40: 127–135 [DOI] [PubMed] [Google Scholar]
- Clough SJ and Bent AF (1998) Floral dip: A simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Conti L, Nelis S, Zhang C, Woodcock A, Swarup R, Galbiati M, Tonelli C, Napier R, Hedden P, Bennett M, Sadanandom A (2014) Small ubiquitin‐like modifier protein SUMO enables plants to control growth independently of the phytohormone gibberellin. Dev Cell 28: 102–110 [DOI] [PubMed] [Google Scholar]
- Conti L, Price G, O'Donnell E, Schwessinger B, Dominy P, Sadanandom A (2008) Small ubiquitin‐like modifier proteases OVERLY TOLERANT TO SALT1 and −2 regulate salt stress responses in Arabidopsis . Plant Cell 20: 2894–2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubeñas‐Potts C, Matunis MJ (2013) SUMO: A multifaceted modifier of chromatin structure and function. Dev Cell 24: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan LX, Chen TL, Li M, Chen M, Zhou YQ, Cui GH, Zhao AH, Jia W, Huang LQ and Qi X (2012) Use of metabolomics approach to characterize Chinese medicinal material Huangqi. Mol Plant 5: 376–386 [DOI] [PubMed] [Google Scholar]
- Enserink JM (2015) Sumo and the cellular stress response. Cell Div 10: 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill G (2004) SUMO and ubiquitin in the nucleus: Different functions, similar mechanisms? Genes Dev 18: 2046–2059 [DOI] [PubMed] [Google Scholar]
- Huang L, Yang S, Zhang S, Liu M, Lai J, Qi Y, Shi S, Wang J, Wang Y, Xie Q, Yang C (2009) The Arabidopsis SUMO E3 ligase AtMMS21, a homologue of NSE2/MMS21, regulates cell proliferation in the root. Plant J 60: 666–678 [DOI] [PubMed] [Google Scholar]
- Ishida T, Fujiwara S, Miura K, Stacey N, Yoshimura M, Schneider K, Adachi S, Minamisawa K, Umeda M, Sugimoto K (2009) SUMO E3 ligase HIGH PLOIDY2 regulates endocycle onset and meristem maintenance in Arabidopsis . Plant Cell 21: 2284–2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin JB, Jin YH, Lee J, Miura K, Yoo CY, Kim WY, Van Oosten M, Hyun Y, Somers DE, Lee I, Yun DJ, Bressan RA, Hasegawa PM (2008) The SUMO E3 ligase, AtSIZ1, regulates flowering by controlling a salicylic acid‐mediated floral promotion pathway and through affects on FLC chromatin structure. Plant J 53: 530–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin JB, Kim YA, Kim SJ, Lee SH, Kim DH, Cheong GW, Hwang I (2001) A new dynamin‐like protein, ADL6, is involved in trafficking from the trans‐Golgi network to the central vacuole in Arabidopsis . Plant Cell 13: 1511–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ES (2004) Protein modification by SUMO. Annu Rev Biochem 73: 355–382 [DOI] [PubMed] [Google Scholar]
- Kim JH, Baek SH (2009) Emerging roles of desumoylating enzymes. Biochim Biophys Acta 1792: 155–162 [DOI] [PubMed] [Google Scholar]
- Kim SI, Park BS, Kim do Y, Yeu SY, Song SI, Song JT, Seo HS (2015) E3 SUMO ligase AtSIZ1 positively regulates SLY1‐mediated GA signalling and plant development. Biochem J 469: 299–314 [DOI] [PubMed] [Google Scholar]
- Kwak JS, Son GH, Kim SI, Song JT, Seo HS (2016) Arabidopsis HIGH PLOIDY2 sumoylates and stabilizes Flowering Locus C through its E3 ligase activity. Front Plant Sci 7: 530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Nam J, Park HC, Na G, Miura K, Jin JB, Yoo CY, Baek D, Kim DH, Jeong JC, Kim D, Lee SY, Salt DE, Mengiste T, Gong Q, Ma S, Bohnert HJ, Kwak SS, Bressan RA, Hasegawa PM, Yun DJ (2007) Salicylic acid‐mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J 49: 79–90 [DOI] [PubMed] [Google Scholar]
- Lin XL, Niu D, Hu ZL, Kim DH, Jin YH, Cai B, Liu P, Miura K, Yun DJ, Kim WY, Lin R, Jin JB (2016) An Arabidopsis SUMO E3 Ligase, SIZ1, negatively regulates photomorphogenesis by promoting COP1 activity. PLoS Genet 12: e1006016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Y, Zhang C, Chen T, Hao H, Liu P, Bressan RA, Hasegawa PM, Jin JB, Lin J (2012) Mutation in SUMO E3 ligase, SIZ1, disrupts the mature female gametophyte in Arabidopsis . PLoS ONE 7: e29470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Wang X, Su M, Yu M, Zhang S, Lai J, Yang C, Wang Y (2015) Functional characterization of DnSIZ1, a SIZ/PIAS‐type SUMO E3 ligase from Dendrobium . BMC Plant Biol 15: 225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Shi S, Zhang S, Xu P, Lai J, Liu Y, Yuan D, Wang Y, Du J, Yang C (2014) SUMO E3 ligase AtMMS21 is required for normal meiosis and gametophyte development in Arabidopsis . BMC Plant Biol 14: 153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, Ashworth EN, Bressan RA, Yun DJ, Hasegawa PM (2007) SIZ1‐mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis . Plant Cell 19: 1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Lee J, Jin JB, Yoo CY, Miura T, Hasegawa PM (2009) Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc Natl Acad Sci USA 106: 5418–5423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Lee J, Miura T, Hasegawa PM (2010) SIZ1 controls cell growth and plant development in Arabidopsis through salicylic acid. Plant Cell Physiol 51: 103–113 [DOI] [PubMed] [Google Scholar]
- Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, Raghothama KG, Baek D, Koo YD, Jin JB, Bressan RA, Yun DJ, Hasegawa PM (2005) The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci USA 102: 7760–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S, Hoege C, Pyrowolakis G, Jentsch S (2001) SUMO, ubiquitin's mysterious cousin. Nature Rev Mol Cell Biol 2: 202–210 [DOI] [PubMed] [Google Scholar]
- Murtas G, Reeves PH, Fu YF, Bancroft I, Dean C, Coupland G (2003) A nuclear protease required for flowering‐time regulation in Arabidopsis reduces the abundance of SMALL UBIQUITIN‐RELATED MODIFIER conjugates. Plant Cell 15: 2308–2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BS, Song JT, Seo HS (2011) Arabidopsis nitrate reductase activity is stimulated by the E3 SUMO ligase AtSIZ1. Nat Commun 2: 400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HC, Kim H, Koo SC, Park HJ, Cheong MS, Hong H, Baek D, Chung WS, Kim DH, Bressan RA, Lee SY, Bohnert HJ, Yun DJ (2010) Functional characterization of the SIZ/PIAS‐type SUMO E3 ligases, OsSIZ1 and OsSIZ2 in rice. Plant Cell Environ 33: 1923–1934 [DOI] [PubMed] [Google Scholar]
- Park HJ, Yun DJ (2013) New insights into the role of the small ubiquitin‐like modifier (SUMO) in plants. Int Rev Cell Mol Biol 300: 161–209 [DOI] [PubMed] [Google Scholar]
- Paz MM, Shou H, Guo Z, Zhang Z, Banerjee A, Wang K (2004) Assessment of conditions affecting Agrobacterium‐mediated soybean transformation using the cotyledonary node explant. Euphytica 136: 167–179 [Google Scholar]
- Rivas‐San Vicente M, Plasencia J (2011) Salicylic acid beyond defence: Its role in plant growth and development. J Exp Bot 62: 3321–3338 [DOI] [PubMed] [Google Scholar]
- Sadanandom A, Ádám É, Orosa B, Viczián A, Klose C, Zhang C, Josse EM, Kozma‐Bognár L, Nagy F (2015) SUMOylation of phytochrome‐B negatively regulates light‐induced signaling in Arabidopsis thaliana . Proc Natl Acad Sci USA 112: 11108–11113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A, Withers J, Mohan R, Marqués J, Gu Y, Yan S, Zavaliev R, Nomoto M, Tada Y, Dong X (2015) Posttranslational modifications of the master transcriptional regulator NPR1 enable dynamic but tight control of plant immune responses. Cell Host Microbe 18: 169–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu D, Tasma IM, Frasch R, Bhattacharyya MK (2009) Systemic acquired resistance in soybean is regulated by two proteins, orthologous to Arabidopsis NPR1. BMC Plant Biol 9: 105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saracco SA, Miller MJ, Kurepa J, Vierstra RD (2007) Genetic analysis of SUMOylation in Arabidopsis: Conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol 145: 119–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava AK, Zhang C, Yates G, Bailey M, Brown A, Sadanandom A (2016) SUMO is a critical regulator of salt stress responses in rice. Plant Physiol 170: 2378–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangasamy S, Guo CL, Chuang MH, Lai MH, Chen J, Jauh GY (2011) Rice SIZ1, a SUMO E3 ligase, controls spikelet fertility through regulation of anther dehiscence. New Phytol 189: 869–882 [DOI] [PubMed] [Google Scholar]
- Villajuana‐Bonequi M, Elrouby N, Nordström K, Griebel T, Bachmair A, Coupland G (2014) Elevated salicylic acid levels conferred by increased expression of ISOCHORISMATE SYNTHASE 1 contribute to hyperaccumulation of SUMO1 conjugates in the Arabidopsis mutant early in short days 4 . Plant J 79: 206–219 [DOI] [PubMed] [Google Scholar]
- Wang H, Makee K, Yan Y, Cao Y, Sun S, Xu G (2011) OsSIZ1 regulates the vegetative growth and reproductive development in rice. Plant Mol Biol Rep 29: 411–417 [Google Scholar]
- Wang H, Sun R, Cao Y, Pei W, Sun Y, Zhou H, Wu X, Zhang F, Luo L, Shen Q, Xu G, Sun S (2015) OsSIZ1, a SUMO E3 ligase gene, is involved in the regulation of the responses to phosphate and nitrogen in rice. Plant Cell Physiol 56: 2381–2395 [DOI] [PubMed] [Google Scholar]
- Wasik U, Filipek A (2014) Non‐nuclear function of sumoylated proteins. Biochim Biophys Acta 1843: 2878–2885 [DOI] [PubMed] [Google Scholar]
- Yoo CY, Miura K, Jin JB, Lee J, Park HC, Salt DE, Yun DJ, Bressan RA, Hasegawa PM (2006) SIZ1 small ubiquitin‐like modifier E3 ligase facilitates basal thermotolerance in Arabidopsis independent of salicylic acid. Plant Physiol 142: 1548–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang RF, Guo Y, Li YY, Zhou LJ, Hao YJ, You CX (2016) Functional identification of MdSIZ1 as a SUMO E3 ligase in apple. J Plant Physiol 198: 69–80 [DOI] [PubMed] [Google Scholar]
- Zhang S, Qi Y, Liu M, Yang C (2013) SUMO E3 ligase AtMMS21 regulates drought tolerance in Arabidopsis thaliana . J Integr Plant Biol 55: 83–95 [DOI] [PubMed] [Google Scholar]
- Zheng Y, Schumaker KS, Guo Y (2012) Sumoylation of transcription factor MYB30 by the small ubiquitin‐like modifier E3 ligase SIZ1 mediates abscisic acid response in Arabidopsis thaliana . Proc Natl Acad Sci USA 109: 12822–12827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XF, Jin YH, Yoo CY, Lin XL, Kim WY, Yun DJ, Bressan RA, Hasegawa PM, Jin JB (2013) CYCLIN H;1 regulates drought stress responses and blue light‐induced stomatalopening by inhibiting reactive oxygen species accumulation in Arabidopsis . Plant Physiol 162: 1030–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found online in the supporting information tab for this article: http://onlinelibrary.wiley.com/doi/10.1111/jipb.12504/suppinfo
Figure S1. Amino acid sequence analysis of soybean GmSIZ1a and GmSIZ1b, and their gene structures
(A) Amino acid sequence alignment of GmSIZ1a, GmSIZ1b,and AtSIZ1. Identical and similar amino acid residues are highlighted with black and gray boxes, respectively. (B) Percentage of identity and divergence among GmSIZ1a, GmSIZ1b, and AtSIZ1 amino acid sequences. (C) Phylogenetic tree of SIZ1 homologs in various plant species. A neighbor‐joining tree was constructed based on an alignment of the complete protein sequences. The numbers on the side of each branch indicate bootstrap values from 1,000 replicates.The scale bar indicates the substitution rate per site. (D) Schematic representation of the GmSIZ1a, GmSIZ1b, and AtSIZ1 gene structures. The blue boxes represent exons and the green lines represent introns. The gray boxes represent UTR regions. PF1/PR1 and PF2/PR2 primer pairs were used for qRT‐PCRanalysis of GmSIZ1a and GmSIZ1b expression, respectively. Black line indicates the region used to generate GmSIZ1RNAi constructs.
Figure S2. Expression of GmSIZ1a or GmSIZ1b rescues the dwarf phenotype of Arabidopsis siz1‐2
(A) Presence of the siz1‐2 mutation was analyzed in complementation lines. GmSIZ1a (#5‐1, #8‐2, and #10‐1) and GmSIZ1b (#3‐7, #10‐3, and #15‐1) indicate the expression of GmSIZ1a:GFP or GmSIZ1b:GFP driven by the Arabidopsis SIZ1 promoter in siz1‐2, respectively. Vector indicates empty vector transformed into siz1‐2. Primer pair LB_a1 (LB)/RP was used to determine the presence of a T‐DNA insertion in the siz1 genetic background, and primer pair LP/RP was used to confirm homozygosity. (B) qRT‐PCR analysis of transgene expression in Col‐0, siz1‐2, the vector control, and GmSIZ1a/b transgenic plants.Relative expression was normalized to that of AtUBC, and data represent the mean ±SD, n=3. (C) Leaf length and rosette diameter of five‐week‐old long‐day‐grown plants. The lengths of leaves from 5 plants were measured.
Figure S3. Generation of GmSIZ1RNAi soybean transgenic plants
(A) Schematic representation of the GmSIZ1RNAi construct and nucleotide sequence alignment of the GmSIZ1a/b cDNA fragment, which was used to generate the GmSIZ1RNAi construct. (B) Glufosinate resistance of GmSIZ1RNAi (35A3, 38A2 and 45B3) leaves. The non‐transgenic soybean cultivar Zhoungdou32 (WT) was used as a negative control. Dashed white box indicates Glufosinate (200 gL−1 glufosinate ammonium, BAYER, Germany) painted leaf area. (C) A bar gene strip (QuickStix for LibertyLink/bar, Envirologix USA) was used to detect the presence of the bar gene in the transgenic plants. Upper and lower arrowheads indicate the control and positive line, respectively. (D) PCR analysis confirmed the presence of the bar gene in the transgenic plants.The Bar‐F and Bar‐R primers were used for the PCR analysis. (E) qRT‐PCR analysis of GmSIZ1a and GmSIZ1b levels in WT and GmSIZ1RNAi soybean transgenic lines. Relative expression was normalized to that of GmUBI3, and data represent the mean ± SD, n = 3. The numbers above the columns indicate the relative transcription level of GmSIZ1a or GmSIZ1b in the transgenic lines compared to in the wild type.
Figure S4. Amino acid sequence analysis and 3D structures of SUMO homologs
(A) Amino acid sequence alignment of SUMO homologs in Arabidopsis, soybean, and rice. Identical and similar amino acid residues are indicated with black and gray boxes, respectively. (B) Phylogenetic analysis of SUMO homologs in Arabidopsis, soybean, and rice. The neighbor‐joining tree was constructed based on an alignment of the complete protein sequences. The numbers on the side of each branch indicate bootstrap values from 1,000 replicates, and the scale bar indicates the substitution rate per site. (C) Three‐dimensional structure modeling of AtSUMO1 and GmSUMO1. (D) Detection of heat shock‐induced SUMO conjugates and free SUMO in soybean and Arabidopsis using anti‐GmSUMO1 antibody. Coomassie blue‐stained Rubisco large subunit (RbcL) was used as the loading control.
Table S1. Putative cis‐acting elements in the GmSIZ1a and GmSIZ1b promoters
Table S2. Primers used in this study


