Abstract
Actin-related protein (Arp) 2/3 complex stimulates formation of actin filaments at the leading edge of motile cells. Nucleation of filaments depends on hydrolysis of ATP bound to Arp2. Here we report crystal structures of Arp2/3 complex with bound ATP or ADP. The nucleotides are immobilized on the face of subdomains 3 and 4 of Arp2, whereas subdomains 1 and 2 are flexible and absent from the electron density maps. This flexibility may explain why Arp2 does not hydrolyze ATP until the complex is activated. ATP stabilizes a relatively closed conformation of Arp3 with the γ-phosphate bridging loops from opposite sides of the cleft. ADP binds Arp3 in a unique conformation that favors an open cleft, revealing a conformational change that may occur in actin and Arps when ATP is hydrolyzed and phosphate dissociates. These structures provide the an opportunity to compare all nucleotide-binding states in an actin-related protein and give insights into the function of both the Arp2/3 complex and actin.
Hydrolysis of ATP bound to actin and dissociation of the γ-phosphate play an important role in actin filament elongation and disassembly (1). The bound nucleotide affects the rates of monomer association and dissociation at both ends of the filament, as well as the affinity of regulatory proteins for actin monomers and filaments (2). For example, the actin monomer binding proteins profilin and thymosin-β4 have higher affinities for ATP-actin than ADP-actin (3, 4, 5). Nucleotide-dependent changes in subunit structure allow actin-binding proteins to distinguish between old and newly assembled lengths of filament, providing a conformational timer that regulates disassembly of filamentous actin (1).
High-resolution structures of actin are known from crystals of actin monomers complexed with other proteins or small molecules that prevent polymerization (6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16). Each of these structures contains an adenine nucleotide bound in the cleft between the two domains of actin. The nucleotide is ATP or adenosine 5′-β,γ-imidotriphosphate in most of the structures, whereas two contain bound ADP (10, 13). In all but one of the actin monomer structures, the cleft is tightly closed on the nucleotide, regardless of the number of phosphates on the adenine nucleotide or the identity of the proteins complexed to actin. The β-actin:profilin: ATP complex is the exception; the structure of this complex has been solved in both the closed state and an open state in which the two domains swing apart by ≈10° (6, 15).
Arp2/3 complex consists of seven protein subunits, including two actin-related proteins, Arp2 and Arp3 (17). Arp2/3 complex is concentrated at the leading edge of motile cells and in vitro initiates new actin filaments by forming 70° branches on the sides of preexisting mother filaments (18, 19, 20, 21, 45). The structure of nucleotide-free bovine Arp2/3 complex showed minimal contacts between Arp2 and Arp3 compared with the close association of adjacent subunits in the helical actin filament (22). This separation of the Arps was postulated to account for the inactivity of the crystallized complex.
Activation of the Arp2/3 complex is a multistep process in which the complex binds an existing (mother) filament and a nucleation-promoting factor, such as WASp or Scar, that brings along an actin monomer (23). These interactions are thermodynamically coupled, suggesting that both mother filaments and WASp favor the same conformation (24). The active conformation is thought to result from a conformational change that brings the Arps together like two successive subunits in an actin filament (22). This active Arp dimer is postulated to bind the actin monomer associated with the nucleation-promoting factor and to initiate the growth of a new filament in the barbed-end direction. Some evidence suggests that filament initiation requires hydrolysis of ATP by Arp2, whereas other evidence suggests that hydrolysis is associated with debranching of the dendritic network (25, 26). In the presence of a nucleation-promoting factor, actin monomer, and mother filament, Arp2 hydrolyzes ATP with a rate constant >0.05 s-1 (27). No hydrolysis by Arp2 has been detected in the inactive complex (27, 28). Arp3 binds ATP, but appreciable hydrolysis of the nucleotide by this subunit was not detected in either the active or inactive Arp2/3 complex (27, 28).
To investigate the role of nucleotide in the function of the complex, we soaked ATP or ADP into nucleotide-free crystals and determined the structures to a 2.6-Å resolution. These structures provide insight into Arp2/3 complex function and the actin family in general.
Methods
Nucleotide-free Arp2/3 complex purified from bovine thymus (29, 30) was crystallized by vapor diffusion vs. 0.1 M Hepes (pH 7.5), 0.2 M KSCN, and 16% PEG 8000, a modification of Robinson et al. (22), and seeded into Arp2/3 saturated in 0.1 M Hepes (pH 7.5), 0.2 M KSCN, and 8% PEG 8000. Nucleotide-bound complex was made by adding to the mother liquor CaCl2 to 2 mM and ATP or ADP to 2 mM and incubating at 4°C for 2 days. The crystals were transferred to 0.1 M Hepes (pH 7.5), 0.2 M KSCN, 16% PEG 8000, and 10% glycerol for 12 h and frozen in liquid nitrogen. Diffraction data were collected at beamline X9B of Brookhaven National Laboratory (Upton, NY). The space group (P212121) of Arp2/3 complex soaked in ATP or ADP was identical to nucleotide-free complex (Table 1). The volume of the unit cell in the ADP-soaked crystals was very similar to the apo crystals, whereas the c-axis was ≈2.6% smaller for the ATP-soaked crystals. Data were integrated and scaled by using denzo and hkl2000 (31). The diffraction limit was 2.55 Å for each of the structures. Using the Arp2/3 apo-complex as a starting model, we carried out rigid-body refinement (cns) (32), allowing each of the individual subunits and the two halves of the Arp3 subunit to move independently. The Rfree after this initial round of refinement was 29.0% for the ADP complex and 31.5% for the ATP complex. Fo - Fc maps generated at this stage showed strong density for the nucleotides in both Arp2 and Arp3. Coordinates and parameter files for ATP and ADP were obtained by using prodrg (33). Subsequent rounds of refinement included the nucleotides and were carried out by using cns. The program o was used to rebuild the models manually after each round of refinement (34). Water molecules were added near the end of the refinement by using weighted Fo - Fc density maps and a σ cutoff of 2.5. The final model of the ADP complex has 1,692 residues, 2 molecules of ADP, 1 calcium ion, 314 water molecules, and an Rfree of 25.9%. The final ATP model has 1,671 residues, 2 molecules of ATP, 2 calcium ions, 181 water molecules, and an Rfree of 27.4%. The coordinates have been deposited in the Protein Data Bank under ID codes 1TYQ and 1U2V.
Table 1. Data collection and refinement statistics.
| ATP | ADP | |
|---|---|---|
| Space group | P212121 | P212121 |
| Unit cell dimensions, Å | ||
| a | 111.4 | 111.3 |
| b | 129.3 | 129.3 |
| c | 199.7 | 204.7 |
| Resolution maximum, Å | 2.55 | 2.55 |
| Total observations | 330,991 | 415,278 |
| Unique observations | 72,816 | 90,833 |
| Completeness, % | 79.2 (68.8) | 99.8 (99.4) |
| I/σ | 21.4 (5.0) | 24.1 (6.8) |
| Rsym, % | 9.3 (35.8) | 5.8 (23.2) |
| Refinement statistics | ||
| Rcryst,* % | 23.0 | 22.3 |
| Rfree,† % | 27.4 | 25.9 |
| Bond angle rms deviation, ° | 1.28 | 1.28 |
| Bond length rms deviation, Å | 0.0070 | 0.0064 |
| Nonhydrogen atoms | 13,925 | 13,589 |
| Average B-factor, Å2 | ||
| Main chain | 49.0 | 45.8 |
| Side chain | 49.4 | 46.2 |
| Nucleotide (Arp3) | 51.7 | 64.1 |
| Nucleotide (Arp2) | 61.1 | 79.2 |
Values for the outermost shell (2.64-2.55 Å) are shown in parentheses.
Rcryst = Σ(Fo - Fc)/Σ(Fo)
Rfree = Σ(Fo - Fc)/Σ(Fo) for 5% of data not used in refinement. Reflections for Rfree were selected randomly and were not identical to the reflections used for the refinement of the apo-complex (PDB entry 1K8K)
Results and Discussion
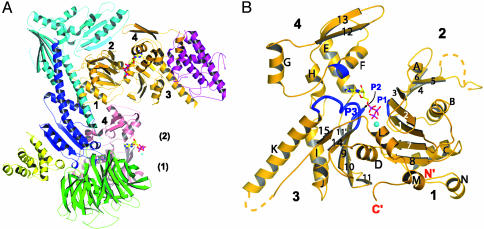
Effects of Nucleotide Binding on Arp2/3 Complex. The structures of Arp2/3 complex with bound ATP (Fig. 1A) or ADP are similar to complex without nucleotide (apo-complex). Each structure has the same arrangement of seven subunits. Most of subdomains 1 and 2 of Arp2 are disordered in all three structures. The nucleotides bind to both Arps and exhibit strong electron density (Fig. 2). In each Arp, the adenine and ribose rings of the nucleotide fill a notch between subdomains 3 and 4, as observed in actin (Fig. 1B). This arrangement places the phosphates between two loops (P1 and P2) from opposite sides of the cleft in Arp3 while leaving them exposed to solvent on the face of subdomains 3 and 4 in Arp2 (Fig. 1 A).
Fig. 1.
Ribbon diagram of Arp2/3 complex and Arp3 with bound Ca2+-ATP. (A) Arp2/3 complex with bound Ca2+-ATP. Orange, Arp3; pink, Arp2; green, p40; cyan, p34; magenta, p21; blue, p20; yellow, p16. The subdomains of the Arps are numbered. Numbers in parenthesis indicate approximate location of Arp2 subdomains 1 and 2. (B) Ribbon diagram of Arp3 with secondary structure labeled. Regions that contact the nucleotide are highlighted in blue. The polypeptide not modeled because of disorder is indicated with a dashed line. Note that Arp3 is reoriented relative to its representation in A.
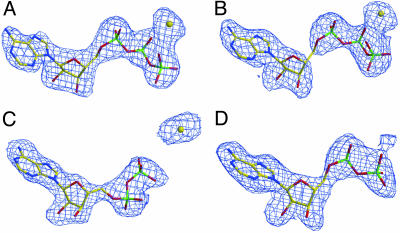
Fig. 2.
Fo - Fc simulated annealing omit map of nucleotides and calcium ion. (A) ATP-Arp3 contoured at 3σ. (B) ATP-Arp2 contoured at 2σ. (C) ADP-Arp3 contoured at 3σ. (D) ADP-Arp2 contoured at 2σ.
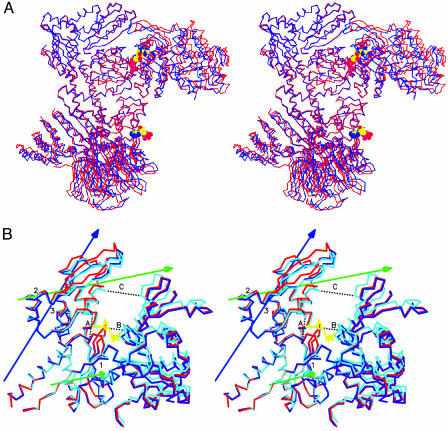
The nucleotide-binding cleft of Arp3 closes upon ATP binding, causing conformational changes that propagate throughout the complex (Fig. 3A). The p21 subunit binds subdomains 3 and 4 of Arp3 and tracks with them as the cleft closes. ATP binding does not change the positions of Arp2 and p34 relative to Arp3 subdomains 1 and 2. A unit consisting of p16, p40, and a portion of p20 rotates counterclockwise, bringing p40 closer to Arp2 and bending the long C-terminal helix of p20. Because there is no obvious structural link between this change and the closure of Arp3 within the same complex, we attributed this motion to a compensation for closing of a neighboring symmetry-related Arp2/3 complex. The conformational changes associated with ADP binding to Arp3 are modest compared with those caused by ATP. Neither nucleotide causes significant structural changes upon binding to Arp2.
Fig. 3.
Stereo figure showing conformational changes cased by ATP binding. (A) Overlay of subdomains 1 and 2 of Arp3 from the apo-(red) and ATP (blue) complexes. (B) Overlay of Arp3 based on the superposition of the entire Arp3 subunits from Apo-containing (red), ADP-containing (blue), and ATP-containing (cyan) complexes. ATP is shown in yellow. dyndom-defined rotation axes required to bring the apo-Arp3 in alignment with each of the nucleotide-containing structures are blue for ADP-Arp3 and green for ATP-Arp3. The dotted black lines indicate distances measured to compare cleft closure. Lengths in Å of A-C are as follows: apo-Arp3, 9.84, 8.47, and 16.08; ADP-Arp3, 8.28, 8.84, and 15.02; ATP-Arp3, 8.35, 6.75, and 13.10; actin (PDB entry 1ATN), 8.35, 5.49, and 10.36. The coordinate error estimate by Luzzati plot is 0.36 Å for the ADP complex and 0.38 Å for the ATP complex (44).
The program dyndom (35) identified rotation axes that describe the conformational changes caused by nucleotide binding to Arp3. In the ATP complex, the first axis (1 in Fig. 3B) runs through subdomain 3 and is parallel to the z axis in the standard orientation of actin. Subdomains 1 and 2 rotate 4.1° about this axis toward subdomains 3 and 4, accounting for much of the closure of the cleft in the ATP complex. A similar scissors-like motion occurs in normal mode analysis of the actin monomer and in the transition between open and tight conformations of β-actin in β-actin:profilin:ATP structures (6, 15, 36).
A second rotation axis in ATP-Arp3 (2 in Fig. 3B) runs through subdomain 4, offset ≈45° from the plane of the page. Rotation about this axis is tied to the insertion of the adenine ring and ribose in the notch between subdomains 3 and 4. ATP binding results in an 8.5° rotation about this axis, causing helix αF, β12, and β13 from subdomain 4 to tilt downward toward subdomain 3. This decreases the width of the notch between subdomains 3 and 4 by 1.5 Å (distance A in Fig. 3B). ADP binding to Arp3 causes a 6.5° rotation around a similar subdomain 4 axis (3 in Fig. 3B), narrowing the notch by 1.6 Å.
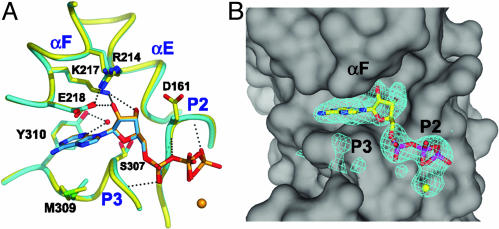
Nucleotide Interactions with Arp2. ADP and ATP adopt identical conformations when bound to Arp2, making van der Walls and hydrogen-bonding interactions that are conserved in actin (Fig. 4A). The adenine and ribose rings bind in a deep notch at the interface of subdomains 3 and 4. The bottom face of the adenine ring interacts with the β14/αK loop (P3) of subdomain 3. Gly-306 and Ser-307 lie directly under the ring. The side chain of conserved Met-309 forms the remainder of this platform. In actins and Arp3, the side chain of this methionine bends into an L-shape to avoid clashing with the β15/αL loop. In Arp2, much of the β15/αL loop is disordered, allowing Met-309 to adopt a more extended conformation that expands the hydrophobic platform for the adenine. The back edge of the adenine ring (N1-C2-N3) interacts with a tyrosine (Y310 in Arp2) in Arp2 and actins. A water bridges the tyrosine hydroxyl group and N3 of the adenine ring in the ADP complex. This water-mediated hydrogen bond is also present in many actin-nucleotide structures.
Fig. 4.
ATP binding to subdomains 3 and 4 of Arp2. (A) Overlay of Arp2 from ATP (yellow) and ADP (cyan) complexes based on the superposition of the entire large domain. (B) Surface representation highlighting the hydrophobic notch between subdomains 3 and 4. The electron density from a 2σ Fo - Fc omit map is shown for ATP and Ca2+.
The top of the pocket for the adenine ring is formed by the C-terminal end of helix αF and the side chain of Glu-218, a residue within helix αF. The aliphatic arm of Glu-218 stretches over the top face of the ring, and the carboxyl group hydrogen bonds to the O2′ of the ribose. The glutamate carboxyl is also hydrogen-bonded to a conserved arginine (Arg-214) from the previous turn of the helix. A conserved arginine makes the same interaction in α- and β-actin.
The bottom edge of the ribose ring (O1′) makes van der Waals contacts with the P3 loop, and the face interacts with a surface formed by Lys-217 and Glu-218 from αF, Gly-197 from αE, Ser-307 from the P3 loop, and Gly-160 and Asp-161 from β9 (Fig. 4A). The ribose hydroxyls are anchored by hydrogen bonds to Lys-217 and Glu-218. Because the small domain of Arp2 is disordered, the phosphates of bound ADP and ATP are stabilized solely by interactions with the P2 and P3 loops. Accordingly, the average B-factor of the phosphates in ATP-Arp2 increases by 20 Å2 compared with ATP-Arp3, whereas the average B-factor of the entire nucleotide is only 10 Å2 higher.
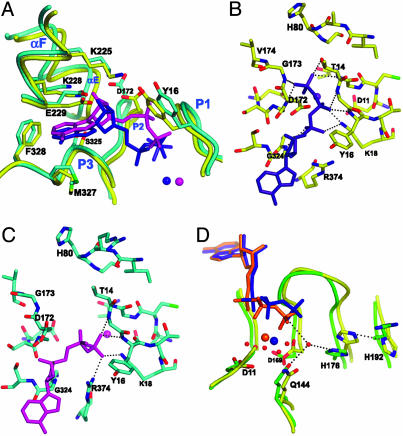
Nucleotide Interactions with Arp3. The nucleotide-binding clefts of actin and Arp2 are nearly identical, whereas several residues differ in Arp3 (37). The aromatic residue at the back of the subdomain 3/4 notch is tyrosine in Arp2 and actins from most species but is phenylalanine in Arp3. In Arp2 and actins, this tyrosine coordinates a water that contacts the adenine. The absence of a hydroxyl group at this position in Arp3 may result in a less optimal nucleotide binding. The residue corresponding to Arg-214 of Arp2 is a lysine in Arp3. Arp2 Arg-214 bends back toward subdomain 4 to interact with Glu-229, whereas in ATP-Arp3 Lys-225 stretches across the cleft to hydrogen bond to Tyr-16 in the P2 loop, connecting subdomains 1 and 4 (Fig. 5A). This interaction is not seen in any of the actin structures.
Fig. 5.
Nucleotide binding to Arp3. (A) Overlay of Arp3 from ATP (yellow) and ADP-Arp2/3 (cyan) complexes based on the superposition of the entire Arp3 subunit (as in Fig. 3B). ADP is shown in magenta, and ATP is shown in blue. (B) Interactions of ATP phosphates with Arp3. (C) Interactions of ADP phosphates with Arp3. (D) Overlay of actin (PDB entry 1NMD, green) and ATP-Arp3 (yellow) based on superposition of the entire molecules. ATP and divalent cation from the actin structure are orange, ATP and Ca2+ from Arp3 are blue. Select water molecules from 1NMD are modeled as red spheres.
Whereas ATP adopts the same conformation when bound to Arp2, Arp3, and most actin structures, ADP bound to Arp3 has a unique conformation in which the O5′-C5′-C4′-C3′ torsion angle flips from 177° to 292° (Fig. 5A). The α- and β-phosphorus atoms of ADP move 3.96 Å and 1.64 Å, respectively, from their positions in ATP-Arp3.
In ATP-Arp3, nine hydrogen bonds stabilize the phosphates (Fig. 5B). The α-phosphate hydrogen bonds to the backbone amide of Gly-324 in the P3 loop of subdomain 3 and Lys-18 in the P1 loop of subdomain 1. The β-phosphate is hydrogen-bonded to Lys-18 and two backbone amides of the P1 loop. The γ-phosphate hydrogen bonds to both the P1 and P2 loops, forming a link between the two domains that is not possible when ADP is bound. In the ADP structure, the β-phosphate twists into a new position, maintaining all of its hydrogen bonds with the P1 loop and making a new hydrogen bond to Arg-374 (Fig. 5C). The α-phosphate of ADP swings up in the cleft (toward Asp-172) relative to its position in the ATP complex. The position of the ADP phosphates is incompatible with the closed conformation seen in the ATP structure.
Because Arp3 has been shown to hydrolyze ATP only inefficiently, it is uncertain whether the ADP-bound conformation is directly relevant to understanding the function of Arp2/3 complex. However, the conformational responses of Arp3 to different nucleotides may be relevant to all of the proteins within the actin family. This is discussed in detail below.
Calcium Binding. ATP-Arp3, ADP-Arp3, and ATP-Arp2 each showed strong electron density beneath the nucleotide phosphates within their nucleotide-binding clefts. Calcium was modeled into this density based on its location relative to the nucleotide and by analogy to the position of the divalent in actin crystal structures. Omit maps of the calcium ions showed strong positive peaks (7.1, 11.45, and 12.6 σ), but we could not rule out the possibility that they were due to water, because they are similar in magnitude to peaks from the most well ordered water molecules from each structure. In ATP-Arp3, the modeled calcium is 2.83 Å from the γ-phosphate and 2.88 Å from the γ-phosphate (Fig. 5D). These distances are longer than in actin-ATP or other typical O-Ca interactions (≈2.3 Å) (38). In actin, the calcium contacts the protein through a network of waters at the bottom of the cleft. Although some weak electron density is visible near the modeled calcium ion in Arp3, no peaks met the criteria we used to identify individual water molecules. However, actin residues that make water-mediated contacts with calcium are conserved in the Arps: Gln-144 from αD (sub-2/3 interface), Asp-169 from β9, and Asp-11 from β1.
The presumed calcium peak is at the same location in ATP-Arp2 as in ATP-Arp3, but the B-factor of the modeled divalent is much higher (86.8 Å2 in ATP-Arp2 vs. 70.4 Å2 in ATP-Arp3). In crystals soaked in ADP and CaCl2, the presumed calcium peak is absent from Arp2 but present in the Arp3 site (B-factor = 77.0 Å2). The calcium modeled in this site does not contact the ADP. It shifts away from the phosphates and toward Asp-11 in subdomain 1 (Fig. 5C). The torsional flip in ADP places the β-phosphate higher in the cleft and even further from the Ca2+. These observations suggest that charge neutralization by the divalent cation may not be necessary for nucleotide binding once the γ-phosphate is released.
Adenine and Ribose Track with the Large Domain. ATP and ADP are closely associated with the notch between subdomains 3 and 4 of Arps, irrespective of the behavior of subdomains 1 and 2. The most extreme example is Arp2, where nucleotides bind even though subdomains 1 and 2 are disordered. In Arp3, the nucleotide sticks in the notch even as the cleft opens. The distance from the ribose to the back wall of the notch (ribose O3 to Gly-197 of αE) is ≈4.6 Å for both ADP and ATP, whereas the distance from the ribose C3 to subdomain 1 (Tyr-16 CA) is 1.7 Å greater in ADP-Arp3. ATP is also closely associated with subdomains 3 and 4 in the open conformation of β-actin bound to profilin (PDB entry 1HLU). These observations suggest that much of the binding energy for the nucleotide in actin and Arps derives from interactions with subdomains 3 and 4. Furthermore, the asymmetrical mode of binding suggests the following sequence of nucleotide exchange: (i) the cleft opens with the nucleotide bound to subdomains 3 and 4; (ii) the nucleotide dissociates; (iii) the new nucleotide binds subdomains 3 and 4; and (iv) the cleft closes.
Low ATPase Activity and Flexibility in Arp2. We were surprised that nucleotide binding did not stabilize subdomains 1 and 2 of Arp2, because nucleotide protects actin from denaturation (39). However, the absence of density for these domains in the new structures may explain why both ATP hydrolysis by Arp2 and actin filament nucleation by the complex depend on activating interactions with other proteins. Binding an actin monomer, mother filament, and activator likely stabilizes subdomains 1 and 2 of Arp2 around ATP in a closed cleft, allowing Arp2 to be catalytically active.
Steric constraints may account for the persistent disorder of Arp2 subdomains 1 and 2. In a model of subdomains 1 and 2 of Arp based on a closed actin structure, the top of subdomain 2 clashes with the bottom of subdomain 3 from Arp3. A disordered segment at the end of an extended helix in Arp3 (αK) projects toward the expected location of subdomain 2 of Arp2 and may keep Arp2 from closing on the nucleotide until the complex is activated.
The disorder in Arp2 may also be due to flexibility inherent to actin and Arps that has not been detected by crystallography. In our structures, Arp2 subdomains 3 and 4 are held tightly by the complex, whereas subdomains 1 and 2 are free to adopt multiple conformations within a large solvent channel (Fig. 6, which is published as supporting information on the PNAS web site). By modeling actin into the Arp2 site in the complex, we estimate that no atom in subdomains 1 or 2 of Arp2 is closer than 6 Å to a symmetry-related molecule. Freedom from intermolecular packing contacts (present in all reported actin crystals) may allow subdomains 1 and 2 of Arp2 to exhibit their inherent flexibility. Such flexibility could contribute to the low rate of ATP hydrolysis in monomeric actin (40). Polymerization may stabilize the interaction of subdomains 1 and 2 with ATP bound to subdomains 3 and 4, promoting ATP hydrolysis. However, stabilization of the two domains cannot be the sole requirement for hydrolysis, because in many of the actin-ATP structures the nucleotide is not hydrolyzed in the crystal.
Low ATPase Activity of Arp3. These structures reveal three factors that may contribute to inefficient ATP hydrolysis by Arp3 compared with (activated) Arp2 (25, 26) and polymerized actin (41). His-176 from β10 is conserved in actin and most Arps and is postulated to activate a water molecule for nucleophilic attack on the γ-phosphate (16) (Fig. 5D). In Arp3, a neighboring residue, His-192, may compromise activation by hydrogen-bonding to His-176 and holding its imidazole ring in a catalytically unproductive conformation. Both actin and Arp2 have a conserved arginine in this position that cannot hydrogen-bond to the proposed catalytic histidine. Mutation of this arginine (Arg-177) to histidine in zebrafish actin causes abnormal heart development (42). However, ATP hydrolysis is reported to be normal in this mutant, so Arg-177 may affect some other activity. Additional assays are required to evaluate the role of these residues in nucleotide binding and hydrolysis.
Incomplete closure of the Arp3 cleft around ATP is a second factor that may contribute to inefficient ATP hydrolysis. The P1 and P2 loops are closer together in closed ATP-actin structures than in ATP-Arp3 (Fig. 3B). The γ-phosphate of ATP hydrogen bonds to the amide backbone of Asp-172 and Gly-173 in the P2 loop of Arp3 (Fig. 5B) but to residues equivalent to Gly-173 and Val-174 on the far side of the loop in actin. This bond ratcheting moves the P1 and P2 loops closer together and pushes the phosphates deeper into the cleft (Fig. 5D). High-resolution structures of actin suggest that catalysis depends on a network of ordered water molecules in the bottom of the cleft coordinated by the divalent metal (16). We expect that hydrolysis requires at least transient adoption of a tightly closed state with the phosphates deep in the cleft. This conformation may be inaccessible in Arp3.
The position of Arp3 in a newly formed branch may be a third factor limiting ATP hydrolysis. Arp3 and Arp2 are thought to be the first two subunits of the daughter filament, with the first actin subunit making a short-pitch contact to Arp2 (22, 45). This actin may promote hydrolysis of ATP bound to Arp2, whereas Arp2 may lack the ability to stimulate hydrolysis of ATP bound to Arp3. Most residues at the proposed Arp2-Arp3 interface are conserved but distinct from those at the actin-Arp2 interface (37).
Nucleotide State and Conformation in Actin. The nucleotide state of actin determines its affinity both for the ends of filaments and for several actin-binding proteins. The structural basis for these differential affinities is still being debated. Although the cleft has been proposed to open and close depending on the nucleotide state, all ADP-actin crystal structures (10, 13) and all but one ATP/adenosine 5′-β,γ-imidotriphosphate-actin structure (6) have a tightly closed cleft. In the structure of TMR-labeled actin with bound ADP, dissociation of the γ-phosphate allows Ser-14 from P2 to swing into the cleft, initiating a series of discrete structural changes, but the cleft remains closed. On the other hand, reconstructions of actin filaments from electron micrographs are consistent with the nucleotide cleft being more open with ADP than ATP (43). It has been suggested that in ADP-actin structures, the nucleotide-binding state is uncoupled from the cleft conformation, perhaps because of crystal packing interactions and/or covalent modification by rhodamine (43).
We compared the structures of nucleotide-free ATP- and ADP-Arp2/3 complex and found that the cleft of Arp3 is capable of differential closure in response to binding ADP or ATP. Whereas Arp3 closes nearly completely with bound ATP, ADP causes little closure. Given these observations and the high structural and sequence similarity of the Arps and actin, serious consideration should be given to the possibility that ADP-containing actin also adopts an open conformation. The differences we observed in the specific interactions of the nucleotides suggest that after nucleotide hydrolysis and release, lost bridging interactions of the γ-phosphate and the repositioning of the α and β phosphate may allow ADP-bound actin to adopt an open conformation.
Implications for Nucleation Activity. ATP binding to Arp2/3 complex favors a conformation different from that of the apo-complex but does not bring Arp2 and Arp3 together like two successive subunits in an actin filament, the conformation expected to support nucleation of a daughter filament. In the model of active Arp2/3 complex with Arp2 and Arp3 oriented like two successive subunits in an actin filament, the DNase binding loop of Arp2 clashes with p21 (22). Closure of the cleft of Arp3 around ATP did not relieve this clash. These observations are consistent with experiments showing that ATP binding and hydrolysis are necessary but not sufficient for nucleation (27). In both structures reported here, the conformation of Arp2/3 complex is inactive toward nucleotide hydrolysis and nucleation. We note that soaking the nucleotides into the apo-crystals may prevent us from observing conformational changes that break crystal contacts, and that structures obtained by cocrystallizing the complex with nucleotide may show significant differences. However, we have recently cocrystallized Arp2/3 complex with both ADP and ATP in conditions nearly identical to those used here, suggesting that the cocrystallized structures will not be significantly different from the soaked crystals.
Although more work is needed to determine how the conformational changes reported here are involved in the function of Arp2/3 complex, these structures provide a glimpse of flexibility and provide valuable insight into how nucleotide binding drives conformational changes.
Supplementary Material
Acknowledgments
We thank Steve Almo and colleagues at Brookhaven National Laboratory for synchrotron beam time and help in collection strategies; Senyon Choe, Witek Kwiatkowski, and others at the Structural Biology Laboratory of the Salk Institute for Biological Studies; Kirsi Turbedsky, Robert Robinson, and Donald Kaiser for help with the Arp2/3 complex protein and crystals; and Rebecca Page for help with the Arp3 conformational changes. R.S.L. thanks Claire Waterman-Storer for her forbearance in providing time to complete this work. This work was supported by National Institutes of Health Research Grants GM26338 and GM066311 (to T.D.P.) and a Jane Coffin postdoctoral fellowship (to R.S.L.).
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 1TYQ (Arp2/3 with bound ATP) and 1U2V (Arp2/3 with bound ADP)].
References
- 1.Pollard, T. D., Blanchoin, L. & Mullins, R. D. (2000) Annu. Rev. Biophys. Biomol. Struct. 29, 545-576. [DOI] [PubMed] [Google Scholar]
- 2.Pollard, T. D. (1986) J. Cell Biol. 103, 2747-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carlier, M. F., Ressad, F. & Pantaloni, D. (1999) J. Biol. Chem. 274, 33827-33830. [DOI] [PubMed] [Google Scholar]
- 4.Safer, D. & Nachmias, V. T. (1994) BioEssays 16, 473-479. [DOI] [PubMed] [Google Scholar]
- 5.Blanchoin, L. & Pollard, T. D. (1998) J. Biol. Chem. 273, 25106-25111. [DOI] [PubMed] [Google Scholar]
- 6.Chik, J. K., Lindberg, U. & Schutt, C. E. (1996) J. Mol. Biol. 263, 607-623. [DOI] [PubMed] [Google Scholar]
- 7.Graceffa, P. & Dominguez, R. (2003) J. Biol. Chem. 278, 34172-34180. [DOI] [PubMed] [Google Scholar]
- 8.Head, J. F., Swamy, N. & Ray, R. (2002) Biochemistry 41, 9015-9020. [DOI] [PubMed] [Google Scholar]
- 9.Hertzog, M., van Heijenoort, C., Didry, D., Gaudier, M., Coutant, J., Gigant, B., Didelot, G., Preat, T., Knossow, M., Guittet, E. & Carlier, M. F. (2004) Cell 117, 611-623. [DOI] [PubMed] [Google Scholar]
- 10.Kabsch, W., Mannherz, H. G., Suck, D., Pai, E. F. & Holmes, K. C. (1990) Nature 347, 37-44. [DOI] [PubMed] [Google Scholar]
- 11.Klenchin, V. A., Allingham, J. S., King, R., Tanaka, J., Marriott, G. & Rayment, I. (2003) Nat. Struct. Biol. 10, 1058-1063. [DOI] [PubMed] [Google Scholar]
- 12.McLaughlin, P. J., Gooch, J. T., Mannherz, H. G. & Weeds, A. G. (1993) Nature 364, 685-692. [DOI] [PubMed] [Google Scholar]
- 13.Otterbein, L. R., Graceffa, P. & Dominguez, R. (2001) Science 293, 708-711. [DOI] [PubMed] [Google Scholar]
- 14.Otterbein, L. R., Cosio, C., Graceffa, P. & Dominguez, R. (2002) Proc. Natl. Acad. Sci. USA 99, 8003-8008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schutt, C. E., Myslik, J. C., Rozycki, M. D., Goonesekere, N. C. & Lindberg, U. (1993) Nature 365, 810-816. [DOI] [PubMed] [Google Scholar]
- 16.Vorobiev, S., Strokopytov, B., Drubin, D. G., Frieden, C., Ono, S., Condeelis, J., Rubenstein, P. A. & Almo, S. C. (2003) Proc. Natl. Acad. Sci. USA 100, 5760-5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Machesky, L. M., Atkinson, S. J., Ampe, C., Vandekerckhove, J. & Pollard, T. D. (1994) J. Cell Biol. 127, 107-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amann, K. J. & Pollard, T. D. (2001) Nat. Cell Biol. 3, 306-310. [DOI] [PubMed] [Google Scholar]
- 19.Blanchoin, L., Amann, K. J., Higgs, H. N., Marchand, J. B., Kaiser, D. A. & Pollard, T. D. (2000) Nature 404, 1007-1011. [DOI] [PubMed] [Google Scholar]
- 20.Mullins, R. D., Heuser, J. A. & Pollard, T. D. (1998) Proc. Natl. Acad. Sci. USA 95, 6181-6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Welch, M. D., DePace, A. H., Verma, S., Iwamatsu, A. & Mitchison, T. J. (1997) J. Cell Biol. 138, 375-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson, R. C., Turbedsky, K., Kaiser, D. A., Marchand, J. B., Higgs, H. N., Choe, S. & Pollard, T. D. (2001) Science 294, 1679-1684. [DOI] [PubMed] [Google Scholar]
- 23.Higgs, H. N. & Pollard, T. D. (2001) Annu. Rev. Biochem. 70, 649-676. [DOI] [PubMed] [Google Scholar]
- 24.Marchand, J. B., Kaiser, D. A., Pollard, T. D. & Higgs, H. N. (2001) Nat. Cell Biol. 3, 76-82. [DOI] [PubMed] [Google Scholar]
- 25.Dayel, M. J., Holleran, E. A. & Mullins, R. D. (2001) Proc. Natl. Acad. Sci. USA 98, 14871-14876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le Clainche, C., Pantaloni, D. & Carlier, M. F. (2003) Proc. Natl. Acad. Sci. USA 100, 6337-6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dayel, M. J. & Mullins, R. D. (2004) PLoS Biol. 2, E91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Clainche, C., Didry, D., Carlier, M. F. & Pantaloni, D. (2001) J. Biol. Chem. 276, 46689-46692. [DOI] [PubMed] [Google Scholar]
- 29.Higgs, H. N., Blanchoin, L. & Pollard, T. D. (1999) Biochemistry 38, 15212-15222. [DOI] [PubMed] [Google Scholar]
- 30.Higgs, H. N. & Pollard, T. D. (2000) J. Cell Biol. 150, 1311-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307-326. [DOI] [PubMed] [Google Scholar]
- 32.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 33.Schuettelkopf, A. W. & van Aalten, D. M. F. (2004) Acta Crystallogr. D 60, in press. [DOI] [PubMed]
- 34.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110-119. [DOI] [PubMed] [Google Scholar]
- 35.Hayward, S. & Berendsen, H. J. (1998) Proteins 30, 144-154. [PubMed] [Google Scholar]
- 36.Tirion, M. M. & ben-Avraham, D. (1993) J. Mol. Biol. 230, 186-195. [DOI] [PubMed] [Google Scholar]
- 37.Beltzner, C. C. & Pollard, T. D. (2004) J. Mol. Biol. 336, 551-565. [DOI] [PubMed] [Google Scholar]
- 38.Yang, W., Lee, H. W., Hellinga, H. & Yang, J. J. (2002) Proteins 47, 344-356. [DOI] [PubMed] [Google Scholar]
- 39.Kinosian, H. J., Selden, L. A., Estes, J. E. & Gershman, L. C. (1993) J. Biol. Chem. 268, 8683-8691. [PubMed] [Google Scholar]
- 40.Brenner, S. L. & Korn, E. D. (1980) J. Biol. Chem. 255, 841-844. [PubMed] [Google Scholar]
- 41.Blanchoin, L. & Pollard, T. D. (2002) Biochemistry 41, 597-602. [DOI] [PubMed] [Google Scholar]
- 42.Wen, K. K. & Rubenstein, P. A. (2003) J. Biol. Chem. 278, 48386-48394. [DOI] [PubMed] [Google Scholar]
- 43.Sablin, E. P., Dawson, J. F., VanLoock, M. S., Spudich, J. A., Egelman, E. H. & Fletterick, R. J. (2002) Proc. Natl. Acad. Sci. USA 99, 10945-10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luzzati, V. (1952) Acta Crystallogr. 5, 802-810. [Google Scholar]
- 45.Volkmann, N., Amann, K. J., Stoilova-McPhie, S., Egile, C., Winter, D. C., Hazelwood, L., Heuser, J. E., Li, R., Pollard, T. D. & Hanein, D. (2001) Science 293, 2456-2459. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.