Abstract
MeDALL (Mechanisms of the Development of ALLergy; EU FP7‐CP‐IP; Project No: 261357; 2010–2015) has proposed an innovative approach to develop early indicators for the prediction, diagnosis, prevention and targets for therapy. MeDALL has linked epidemiological, clinical and basic research using a stepwise, large‐scale and integrative approach: MeDALL data of precisely phenotyped children followed in 14 birth cohorts spread across Europe were combined with systems biology (omics, IgE measurement using microarrays) and environmental data. Multimorbidity in the same child is more common than expected by chance alone, suggesting that these diseases share causal mechanisms irrespective of IgE sensitization. IgE sensitization should be considered differently in monosensitized and polysensitized individuals. Allergic multimorbidities and IgE polysensitization are often associated with the persistence or severity of allergic diseases. Environmental exposures are relevant for the development of allergy‐related diseases. To complement the population‐based studies in children, MeDALL included mechanistic experimental animal studies and in vitro studies in humans. The integration of multimorbidities and polysensitization has resulted in a new classification framework of allergic diseases that could help to improve the understanding of genetic and epigenetic mechanisms of allergy as well as to better manage allergic diseases. Ethics and gender were considered. MeDALL has deployed translational activities within the EU agenda.
Keywords: asthma, IgE, multimorbidity, polysensitization, rhinitis
Abbreviations
- AD
atopic dermatitis
- AIRWAYS ICPs
Integrated care pathways for airway diseases (EIP on AHA)
- AMIC
Asthma Multicentre Infant Cohort Study
- BAMSE
Barn Allergi Milj. Stockholm Epidemiologi Projektet
- BIB
Born in Bradford
- CC16
club cell secretory protein
- CQ
Core Questionnaire
- DARC
Danish Allergy Research Centre
- ECA
Environment and Childhood Asthma
- ECRHS
European Community Respiratory Health Survey
- EDEN
Etude des Déterminants pré et post natals du développement et de la santé de l’ Enfant
- EFA
European Federation of Allergy and Airways Diseases Patients’ Associations
- EGEA
Epidemiological study on the Genetics and Environment of Asthma, bronchial hyperresponsiveness and atopy
- EIP on AHA
European Innovation Partnership on Active and Healthy Ageing
- ENRIECO
Environmental Health Risks in European Birth Cohorts
- ESCAPE
European Study of Cohorts for Air Pollution Effects
- EU
European Union
- GA2LEN
Global Allergy and Asthma European Network
- GARD
WHO Global Alliance against Chronic Respiratory Diseases
- GINIplus
German Infant study on the influence of Nutrition Intervention PLUS environmental and genetic influences on allergy development
- HDM
House dust mite
- IgE
Immunoglobulin E
- INMA
Infancia y medio ambiente
- LISAplus
Lifestyle factors on the development of the Immune System and Allergies in East and West Germany PLUS the influence of traffic emissions and genetics
- MAAS
Manchester Asthma and Allergy Study
- mAb
monoclonal antibody
- MAS
German Multicenter Allergy Study
- MeDALL
Mechanisms of the Development of ALLergy
- MHC
major histocompatibility complex
- NCD
Noncommunicable disease
- PARIS
Pollution and Asthma Risk: an Infant Study
- PIAMA
The Prevention and Incidence of Asthma and Mite Allergy
- RHEA
Mother–Child cohort in Crete
- Robbic
Rome and Bologna birth cohorts
- SPT
skin prick test
- TCR
T‐cell receptor
- Treg
T regulatory cell
- WAO
World Allergy Organization
Allergic diseases such as asthma, rhinitis and eczema are complex and are associated with allergen‐specific IgE and nonallergic mechanisms 1. They often coexist in the same subject (multimorbidity) 2 and are multifactorial, with genetic, lifestyle and environmental components. These interactions start early in life, develop during infancy and childhood 3 and may persist throughout life.
Allergic diseases are not separate diseases, but are linked by complex and insufficiently defined interrelationships across the life cycle. There is a growing interest to apply the systems approach proposed in systems biology to complex chronic diseases 4. The 7th Framework Programme of the EU promoted research to develop systems medicine in order to better understand chronic diseases. MeDALL (Mechanisms of the Development of ALLergy; EU FP7‐CP‐IP; Project No: 261357; 2010–2015) has attempted to better understand the complex links of allergic diseases at the clinical and mechanistic levels 1, 5.
MeDALL completed its project in May 2015, and the present paper reports the results and achievements published by the consortium to date.
Innovative approach
MeDALL was one of the first EU projects to adopt a systems medicine approach for the understanding of complex NCDs 1, 5. It has generated novel knowledge on the mechanisms of initiation of allergy from early childhood to young adulthood. MeDALL has linked epidemiological, clinical and basic research 6. It was based on a novel, stepwise, large‐scale and integrative approach led by a network of all necessary experts. The following steps were proposed and developed during the project:
Identification of ‘classical’ and ‘novel’ phenotypes in existing birth cohorts (Table 1).
Discovery of the relevant mechanisms in IgE‐associated allergic diseases in existing longitudinal birth cohorts.
Validation and redefinition of the classical and novel phenotypes of IgE‐associated allergic diseases.
Translational integration of systems biology outcomes into health care, including societal aspects.
Table 1.
MeDALL dual approach
| Hypothesis‐driven approach: The identification of classical phenotypes was based on experts’ criteria following a review of the literature aiming at the definition of IgE‐associated allergic diseases. |
| Data‐driven approach: To identify the novel phenotypes, the children from the birth cohorts were analysed using hypothesis‐free methods by cluster analysis. |
The strategy of MeDALL was based on information and samples (already existing in the repository and acquired during the project) obtained from a large network of 14 ongoing birth cohorts.
Major methodological achievements
Development of a knowledge management platform
Systems medicine involves the large‐scale integration of existing knowledge with newly acquired multidimensional data. The MeDALL knowledge management platform was developed to empower all partners with open sharing and access to all the data and information collected, as well as to all the experimental and computational procedures. The MeDALL knowledge base integrates historical and newly collected data from around 44 000 participants on 398 clinical and phenotypic attributes (harmonized from 7495 individual cohort variables) and 160 different follow‐ups at 25 different time points between pregnancy and age 20. It also contains information about available blood samples that are stored in the individual biobanks of the different MeDALL partners.
Information on 863 genes involved in allergy (283 from a systematic literature review and 580 from automatic text mining) is integrated with data on protein–protein interactions, transcriptional regulation, miRNA regulation and signalling pathways from public databases. It is directly connected to the omics data generated within MeDALL.
Omics data produced or made available within MeDALL include 23 000 historical GWAS, 9500 epigenetics, 2000 proteomics, 750 transcriptomics, IgE microarrays (4000 subjects) and individual estimates of ambient air pollution exposure (10 000 children) using the land use model (ESCAPE) 7, 8, 9, 10.
The newly performed epigenetics, proteomics, transcriptomics and IgE microarray data are integrated into the MeDALL knowledge base which currently includes 3292 IgE chips, 2173 DNA methylations, 1427 biomarkers and 723 transcription experiments. The availability of longitudinal samples of the same individual is a unique resource.
Development of the harmonized MeDALL‐Core Questionnaire (in eight languages) 11
Numerous birth cohorts have been initiated throughout the world using heterogeneous methods to assess the incidence, course and risk factors for asthma and allergies 12. One of the major achievements of MeDALL was the development of the harmonized MeDALL‐Core Questionnaire (MeDALL‐CQ), used historically in 14 cohorts and prospectively in 11 11. The harmonization of standardized core questions was accomplished in four steps: (i) collection of previous questions from 14 European birth cohorts, (ii) consensus on core questionnaire items, (iii) translation and back‐translation of the harmonized English MeDALL‐CQ into eight languages and (iv) implementation of the three core questionnaires MeDALL‐CQ (two for parents of children aged 4–9 and 14–18 and one for adolescents aged 14–18). The harmonized MeDALL follow‐up leads to more comparable data across different cohorts and offers the possibility to validate the results of former single cohort analyses.
Worldwide interest has been expressed in the MeDALL‐CQs: future follow‐up assessments of the Cincinnati Childhood Allergy and Air Pollution Study (CCAAPS) birth cohort 13 and of the large Japanese national birth cohort Japan Environment and Children's Study (JECS), which recently recruited 100 000 parent–child pairs (www.env.go.jp/en/chemi/hs/jecs/).
Development of a database of pooled cohorts using a harmonized questionnaire 12
MeDALL birth cohorts include AMICS‐Menorca** 14, BAMSE** 15, BIB* 16, 17, DARC** 18, ECA** 19, EDEN* 20, GINI plus** 21, 22, INMA* Gipuzkoa, Sabadell, Valencia 23, LISA plus** 24, MAS 25, 26, 27, 28, PARIS* 29, PIAMA 30, RHEA*31 and ROBIC* 32.
MeDALL included the birth cohorts with historical databases and harmonized follow‐up 11. They were integrated in a knowledge management system 33 (Table 2).
Table 2.
Pooled MeDALL database
| • AMICS‐M: data from 482 integrated participants with up to 12 follow‐ups. |
| • BAMSE: data from 4089 integrated participants with up to five follow‐ups. |
| • BiB: data from 13 565 (2594 in MeDALL) integrated participants with up to seven follow‐ups. |
| • DARC: data from 562 integrated participants with up to nine follow‐ups. |
| • ECA: data from 3754 integrated participants with up to six follow‐ups. |
| • EDEN: data from 1140 integrated participants with up to six follow‐ups. |
| • GINI: data from 5991 integrated participants with up to eight follow‐ups. |
| • LISA: data from 3095 integrated participants with up to nine follow‐ups. |
| • INMA‐Gipuzkoa: data from 406 integrated participants with up to four follow‐ups. |
| • INMA‐Sabadell: data from 772 integrated participants with up to seven follow‐ups. |
| • INMA‐Valencia: data from 855 integrated participants with up to six follow‐ups. |
| • MAS: data from 1314 integrated participants with up to 19 follow‐ups. |
| • PARIS: data from 1549 integrated participants with up to 10 follow‐ups. |
| • PIAMA: data from 3963 integrated participants with up to 12 follow‐ups. |
| • RHEA: data from 1336 integrated participants with up to six follow‐ups. |
| • ROBBIC‐Bologna: data from 434 integrated participants with up to five follow‐ups. |
| • ROBBIC‐Roma: data from 694 integrated participants with up to six follow‐ups. |
A joint NIH‐MeDALL workshop on birth cohorts in allergy and asthma was organized in September 2012 to address a wide research agenda, including potential new study designs and the harmonization of existing birth cohort data 6.
The MeDALL database with its new central database is the starting point for future common and sustainable research initiatives in asthma and allergy. It can be extended with other epidemiologic studies such as birth and patient cohorts.
Development of a new allergen microarray technology
The ‘MeDALL allergen‐chip’ is a collection of 170 allergen molecules for the reliable detection of allergen‐specific antibody signatures. When compared to the traditional ImmunoCAP, this system shows a higher sensitivity. The new tool has been integrated in clinical work within MeDALL and beyond, allowing a hitherto not available precision regarding the mapping of sensitization profiles down to the level of disease‐causing allergens and the monitoring of the early evolution of the allergic immune response 34, 35, 36, 37. The MeDALL allergen‐chip is a customized chip by ThermoFisher, which includes a large collection of allergens produced by the MeDALL consortium.
Applying bioinformatics to develop the systems medicine approach
The working hypothesis underlying systems medicine approaches is that biological function and dysfunction in disease develop from the interplay between spatial and temporal processes involving multiple components interacting in complex networks. The ultimate goal of systems medicine is therefore to understand, as fully as possible, this integrative process and to identify directions to investigate the development of more efficient clinical diagnostics and therapies. In MeDALL, we have integrated bioinformatics with more traditional methods (classical and novel phenotypes). We have used machine‐learning methods as an unsupervised strategy to identify novel phenotypes and have developed an in silico model of multimorbidity.
Applying machine‐learning methods to identify novel phenotypes
In MeDALL, we used an unsupervised approach to identify novel phenotypes. At variance with previous studies that applied this method to one single disease, we assessed asthma, rhinitis and eczema together in the same models 5. We included 17 209 children at 4 years and 14 585 at 8 years from seven birth cohorts. At each age period, we performed partitioning cluster analysis, according to the distribution of 23 variables covering: symptoms ‘ever’ and ‘in the last 12 months’; doctor diagnosis; age of onset and treatments for asthma, rhinitis and eczema; IgE sensitization; weight; and height. The analysis used repeated latent class analysis and self‐organizing maps.
Development of a bioinformatic model of multimorbidity of allergic diseases
An in silico study based on the analysis of the topology of the protein interaction network was performed to characterize the molecular mechanisms of multimorbidity of asthma, eczema and rhinitis. As a first step, proteins associated with either disease were identified using data mining approaches, and their overlap was calculated. Secondly, the functional interaction network was built, allowing the identification of cellular pathways involved in allergic multimorbidity. Finally, a network‐based algorithm generated a ranked list of newly predicted multimorbidity‐associated proteins (Aguilar, submitted).
Novel findings
Literature review on phenotypes and course of allergic diseases in children
A large heterogeneity of allergic phenotypes exists and no systematic review had been carried out on phenotype classification or multimorbidity. MEDLINE was searched up to December 2012 to identify relevant original studies in children. From a total of 13 767 citations, 197 met the criteria for inclusion, of which 54% were cohorts. The review showed that studies reporting the phenotypes of IgE‐associated diseases in children are heterogeneous and often lack objective measures. The knowledge on multimorbidity was mostly restricted to links between asthma and rhinitis 38.
Classical and novel phenotypes reveal the importance of multimorbidity
The term ‘multimorbidity’ is more appropriate than comorbidity because the primary allergic disease is poorly known and the allergy march accounts for few patients 39. Although the concept of multimorbidity of allergic diseases has been known for years 40, MeDALL is the first population study to have assessed allergic multimorbidity of allergic diseases using the dual approach: hypothesis driven 41 and data driven (unsupervised cluster analyses) 42. It is also the first to have quantified the net excess of multimorbidity 41.
Classical epidemiological methods enabled a precise analysis of the multimorbidity of asthma, rhinitis and eczema 41. The absolute excess of any multimorbidity was 1.6% for children aged 4 and 2.2% for children aged 8. 44% of the observed multimorbidity at the age of 4 and 50.0% at the age of 8 were not a result of chance. Children with multimorbidities at 4 years had an increased risk of having multimorbidity at 8 years. The coexistence of eczema, rhinitis and asthma in the same child is more common than expected by chance alone, suggesting that these diseases share causal mechanisms. For children without multimorbidity at 4 years, 38% of the multimorbidity at the age of 8 was attributable to the presence of IgE sensitization at the age of 4. On the other hand, the use of machine‐learning methods 42 showed that 30 to 40% of children at the age of 4 to 8 belong to a multimorbidity cluster. This cluster included 99% of children exhibiting multimorbidity with classical models. Although IgE sensitization is independently associated with excess multimorbidity, its presence at 4 years accounted for only 38% of the new multimorbidity at the age of 8, suggesting that IgE sensitization can no longer be considered the common causal mechanism of multimorbidity for these diseases (Fig. 1).
Figure 1.
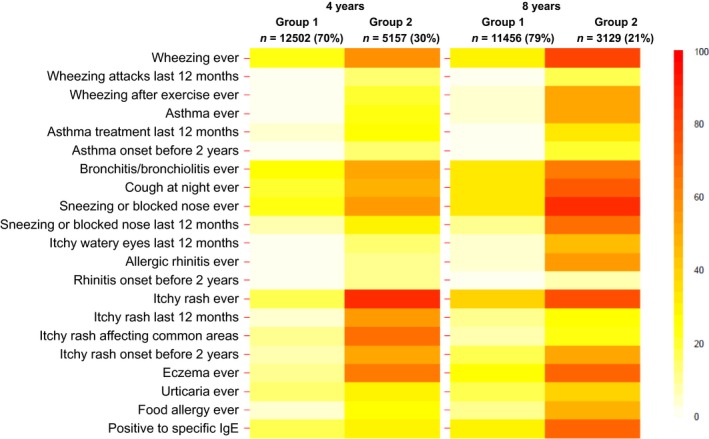
Prevalence* of symptoms of asthma, rhinitis and eczema according to the two groups identified in cluster analysis, at 4 and 8 years (Garcia‐Aymerich et al. 42).
Monosensitization and polysensitization of distinct allergy phenotypes
The concept of monosensitization and polysensitization has been previously proposed 43, 44, 45, but never formally evaluated due to the lack of samples in a population‐based study and inadequate methods, making it impossible to study a wide array of allergens. This became possible in MeDALL. In the BAMSE cohort (Sweden), the same 2607 children were tested at least twice at 4, 8 and 16 years 46. Results confirm that monosensitization and polysensitization represent two different phenotypes of IgE‐associated diseases. These results were also confirmed in the MeDALL cohorts 2 and in patient cohorts in children 47. In the EGEA study, the monosensitized and polysensitized MeDALL phenotypes 2 were confirmed in adults 48.
Effect of pollutants in the development of allergic diseases
Traffic‐related air pollution (nitrogen dioxide (NO2), particulate matter < 2.5 μm (PM2.5), < 10 μm (PM10) and PM2.5 absorbance (‘soot’)) was studied. In four cohorts, residential exposure to traffic‐related air pollution at the birth address and follow‐up addresses was examined 10, 49, 50. Exposure to nitrogen dioxide (NO2) and PM2.5 absorbance early in life is a risk factor for the development of asthma through childhood and adolescence, particularly after the age of 4 49. Analyses of other environmental risk factors for allergy‐related disease (e.g. second‐hand tobacco smoke, moulds, dampness) are ongoing.
Gender switch during puberty
Longitudinal gender‐specific analyses of 18 852 children participating in PIAMA, BAMSE, LISAplus, GINIplus, DARC and MAS showed considerable changes in the sex‐specific occurrence of asthma and allergic rhinitis at around puberty. The strong male predominance of asthma or rhinitis prevalence in prepubertal childhood declined as the teenagers grew older (manuscripts in preparation). Our hypothesis that by adulthood the gender imbalance in the prevalence is switching to a clear female predominance of asthma and rhinitis will be examined with the next follow‐up data of MeDALL cohorts in adulthood.
Omic data for allergy molecular fingerprints and phenotype handprints
In the discovery phase of targeted proteomics, levels of a large panel of proteins via multiplexing and ELISAs were measured in four cohorts. In the replication phase, top biomarkers of interest were measured in seven cohorts. The analyses identified a potential novel biomarker of asthma and the role of systemic inflammation in multimorbidities in early childhood. Results were consistent using the classical and novel phenotypes (manuscript in preparation). Following a hypothesis‐driven approach, MeDALL data for CC16 (club cell secretory protein) were included in a large international collaborative study. Low CC16 levels at the age of 4 predicted subsequent FEV1 deficits up to the age of 16 in the BAMSE cohort, and these data were in line with independent results from the CRS (USA) and MAAS (UK) cohorts 50.
An epigenome‐wide association study on asthma was completed and followed up by a large replication study across all MeDALL cohorts (Xu et al., 2016, in preparation). Based on an a priory hypothesis, strong and consistent effects of maternal smoking on the child's whole‐blood DNA methylome at the of age 0, 4/5 and 8 were observed and the results published as part of the PACE consortium (Joubert et al., submitted). Moreover, the effects of traffic‐related air pollution on DNA methylation have been investigated (Gruzieva et al., 2016, in preparation).
Expression profiling of the identified genes is currently ongoing in these projects, based on the transcriptomic profiling elaborated at single‐ and multicohort levels, for single disease phenotypes and multimorbidities. An exhaustive portfolio of significant and statistically validated genes, molecular interactions and pathways is available. A set of overlapping genes defines molecular fingerprints related to asthma, rhinitis and eczema, blood cell activity, immune and antigen response and receptor activity (Lemonnier et al., in preparation). Integrative analyses of large‐scale transcriptomics and epigenomics datasets with protein expression levels enabled the discovery of an initial allergy phenotype handprint (Ballereau et al., in preparation).
In comprehensive integrative analyses, protein levels were analysed in relation to both genetic variation and methylation profiles in their encoding genes. Multiple CpG sites from 25 genes were found for possible mediation of effects of genetic variation on protein levels. By identifying a set of putatively functional SNPs and CpG methylation sites, these results may provide specific loci to be investigated in association studies. A focused integrated analysis in childhood asthma was completed with genetic–epigenetic–protein data for the asthma biomarker chitinase‐3‐like protein 1 (CHI3L1/YKL‐40. It showed that CHI3L1 genetic variation affects circulating YKL‐40 by regulating its gene methylation profiles (Guerra S et al., in preparation).
Mechanistic experimental studies in animal studies and in vitro to complement MeDALL data
A major achievement has been the construction of transgenic mice in which the TCR reacts to the relevant Der p 1 allergen of house dust mite (HDM). A fine mapping of the T‐cell response to HDM in vivo was possible. This was achieved by performing the microarray analysis of CD4 T cells obtained from these TCR transgenic mice, essentially unravelling a novel IL‐21‐producing T‐cell subset involved in asthma development 51. We also validated the effect of farm exposure on the development of allergy and asthma. Mice exposed to farm dust fail to mount allergy and asthma to HDM due to a downregulation of epithelial cell activation and a subsequent lack of dendritic cell activation. The pathway involved the ubiquitin‐modifying enzyme A20. Genetic polymorphisms of A20 were associated with the risk of developing asthma in a cohort study of children in rural Germany 52. The latter study also involved innovative air–liquid interface cultures of epithelial cells obtained by bronchoscopy. In air–liquid interface cultures of asthmatics, A20 levels were reduced compared with healthy controls.
In allergic patients, the immune profile of the tonsils represents the atopic status of patients, with low expression of Th1. Cellular and molecular mechanisms of tolerance induction to food and aeroallergens occur in human tonsils, suggesting that they represent a suitable lymphatic organ for direct immune interventions 53, 54, 55. In addition to Treg cells, IL‐10‐producing regulatory B cells have been demonstrated with a potent anti‐inflammatory activity.
Cytokine‐producing memory B‐cell subsets exist in humans and may show different pro‐inflammatory, anti‐inflammatory as well as immune effector and immune regulatory functions. We identified novel molecules that play a role in B‐cell regulation and demonstrate that functional human B‐cell subsets show in vivo clonal expansion during allergen tolerance, such as allergen immunotherapy and high‐dose allergen exposure 56, 57. Moreover, Treg cells are reduced during asthma exacerbations as markers of virus‐induced asthma exacerbations, which are of great interest for the development of future biomarkers 58. To better analyse cytokine‐producing immune cell subsets, we developed an assay that allows the purification of any single and several cytokine‐secreting cell subsets as well as their characterization 59.
The bronchial epithelial layer serves as the first site of exposure to inhaled allergens, dust particles, pollutants or microorganisms. Consequently, bronchial epithelial cells are at the forefront of tissue defense and the innate immune response, preventing the invasion of tissues as a physical barrier. Tight junctions (TJs), located at the most apical region of the lateral cell membrane, seal the epithelium and form an essential part of the barrier between inner tissues and the external environment. They can be regulated by cytokines of the allergy pathways 60, 61. Many efforts have been made to improve the barrier integrity, and it was recently demonstrated in vitro that the administration of CpG‐DNA could be useful for restoring impaired epithelial barriers 62.
Novel classification of allergic diseases: the MeDALL hypothesis 2
IgE sensitization should be considered as a qualitative (IgE response) and quantitative trait (monosensitization and polysensitization), as important clinical and immunological differences exist between monosensitized and polysensitized subjects (Table 3). The integration of multimorbidities and polysensitization has resulted in a new classification framework of allergic diseases 2, 47, which could help to improve the understanding of genetic and epigenetic mechanisms of allergy and better manage allergic diseases (Tables 4 and 5).
Table 3.
Differences between monosensitized and polysensitized subjects
| Polysensitization, as compared to nonsensitization and monosensitization, is associated with 2: |
| • A higher frequency of family history of allergy (asthma and rhinitis) 92. |
| • A higher prevalence of asthma and rhinitis symptoms. |
| • A higher prevalence of multimorbidity. |
| • A higher level of specific IgE and a higher level of total IgE compared to monosensitized. |
| • A broader sensitization to different allergens. |
| • The persistence of allergic diseases with a lower probability of remission of IgE sensitization and clinical allergy. |
Table 4.
Novel classification of IgE‐mediated diseases 2
| 1. Nonsensitized asymptomatic individuals. |
| 2. IgE response restricted to one environmental allergen with no family history: low IgE responders (the number of components and level of IgE). |
| • Nonsymptomatic subjects who are unlikely to develop symptoms over time. |
| • Symptomatic subjects (symptoms similar to polysensitized subjects). |
| 3. Polyclonal IgE response to environmental allergens with family history: high IgE responders: Most subjects are symptomatic, with an early life onset, a high rate of multimorbidities and persistence of the disease over time. |
| 4. Nonallergic polyclonal IgE without family history: late‐onset disease and local polyclonal IgE. |
| 5. Intermediate phenotypes. |
| • Polyclonal IgE response without family history. The role of cofactors (pollutants, viruses) needs to be better understood. |
| • IgE response restricted to few allergens. |
Table 5.
Implications of the novel definition of IgE‐associated allergic diseases 2
| Subphenotyping of allergic diseases for precision medicine: Phenotyping subtypes can be used to characterize allergic diseases, severity and progression and may help identify unique targets for prevention and treatment. |
| Clinical practice: An updated definition provides a framework to inform decisions relating to treatment priorities and to indicate need for improvement in health care and delivery through better organization for prediction, diagnosis and treatment. The prediction of allergic disease trajectories in preschool children is essential. |
| Clinical trials: Clarity on definitions is essential for clinical trials, evaluating efficacy and safety. The stratification of patients by sensitization and multimorbidity is essential in allergen immunotherapy (both for treatment and for prevention). |
| Research on mechanisms and genetics: The new definition is likely to change the concepts of the mechanisms of allergic disease and to propose novel mechanisms related to shared and unique genetic factors 93, 94, 95. |
| Population studies: In longitudinal epidemiological population studies, standardized definitions are required to be able to compare cohorts across time and place and to develop dynamic models capturing risk factors that predict transitions through different stages of health. |
| Public health planning: For public health purposes, a comprehensive definition is needed (i) to identify the prevalence, burden and costs incurred by all phenotypes, (ii) to improve quality of care and optimize healthcare planning and policies and (iii) to model the economic and social benefits of population‐level preventive policies to improve respiratory health of the current generation of children and thus of future generations. |
| Social welfare planning: For social welfare purposes, a phenotypic definition is also needed to predict the burden and costs at an early age in order to model the individual and collective economic and social benefits of specific interventions. |
| Applicability to high‐ and low‐income countries: A uniform allergy definition should be applicable to the local and geographical conditions of all countries, phenotypes, risk factors, availability and affordability to treatment differing widely around the world. This would help to better understand which preventive measures are most effective in specific environments and interactions with parasitic diseases in particular. |
| Development of novel preventive approaches and therapies: Detailed cellular and molecular phenotyping is needed to identify novel primary and secondary prevention strategies, as well as new targets for the development of novel therapies. Ultimately, novel therapies studied in clinical trials should help define IgE‐mediated pathways and determine the importance of the intervention in large patient populations or in subpopulations of patients based on the concept of distinct phenotypes. The life course approach of allergic diseases is of great interest because it may lead to preventive strategies for health promotion and preventive policy measures. |
Clinical impact
Many MeDALL data have been translated into clinical practice, and a meeting at the European Parliament was organized by EFA (European Federation of Allergy and Airways Diseases Patients’ Association) to conclude the project (27 May 2015). In particular, MeDALL results have an impact on precision medicine as they improve the stratification of allergic preschool children for diagnosis, prognosis and allergen‐specific immunotherapy. Multimorbidity and IgE polysensitization are markers of persistence of disease.
Childhood asthma prediction models
Early identification of children at risk of developing asthma at school age is crucial, but the usefulness of childhood asthma prediction models in clinical practice is still unclear. We systematically reviewed all existing prediction models to identify preschool children with asthma‐like symptoms at risk of developing asthma at school age. Some models were able to better predict asthma development and others to better rule it out. However, the predictive performance in both aspects simultaneously stood out in neither of the models. This study suggests that the prediction of asthma development is difficult 63, possibly because of interactions with viral infections.
Prediction of the persistence of allergic diseases at 4 years
Polysensitization and/or multimorbidity at 4 years predicts the persistence of allergy later in life 36, 37, 41. This result is of importance for the parents of affected children. ‘Will my child grow out of his or her allergy?’ is the invariable question raised by the parents. Polysensitization to the major birch pollen allergen in combination with allergens of the same protein family predicted future birch pollen allergy much better than IgE to the birch pollen allergen extract itself. The same was true for polysensitization to different cat and dog allergens in relation to the development of cat and dog allergy, respectively. The MeDALL results are the first that may propose a simple answer to the practising physician if the results are confirmed elsewhere.
Stratification of patients for the initiation of allergen‐specific immunotherapy
The results of the MeDALL (monosensitization–polysensitization, multimorbidity) study, the systematic review on prediction models as well as recent studies in patients with allergic rhinitis 64 and in the EGEA cohort 48 may help to propose a stratification of patients for treatment and future randomized control trials 65. Moreover, the molecular sensitization profiles determined with the MeDALL chip may be the basis for the stratification for allergen‐specific immunotherapy 65.
Patient empowerment
The goal and rationale of patient involvement in medical decisions is patient empowerment. Empowered patients know their disease. Since its inception, MeDALL has worked for and with patients for their empowerment in the project. EFA, one of the MeDALL partners, was present at the beginning and end of the project.
Ethics
A specific WP was dedicated to ethics in MeDALL to manage ethical issues throughout the project. In addition, the ethics WP prepared practical information on regulatory issues for exchanging biological samples and attached data for the relevant partners of MeDALL. This was made available as a practical web‐based tool, as a complementary development of the site hSERN.eu developed by GA²LEN 66, 67. The conditions for exchanges of samples and data regarding a few countries (www.hsern.eu) were integrated online on the hSERN website. The communication of results and disclosure of incidental findings in longitudinal paediatric research have been innovative MeDALL initiatives 68.
MeDALL at the cross‐roads of the EU and WHO political agenda
European Innovation Partnership on Active and Healthy Ageing (EIP on AHA) and WHO
The NCD WHO research agenda indicated that birth cohorts were needed for the understanding of the early determinants of chronic respiratory diseases for innovative health promotion strategies 69. MeDALL is in line with this agenda.
The WHO Global Alliance against Chronic Respiratory Diseases (GARD) action plan 70, 71, 72 was the model of AIRWAYS ICPs (integrated care pathways for airway diseases) 73, 74, 75, a new initiative of the EIP on AHA. This was jointly organized by MeDALL, the Reference Site Network of the EIP on AHA (DG Santé and DG Connect) 76 and WHO GARD (Fig.2).
Figure 2.
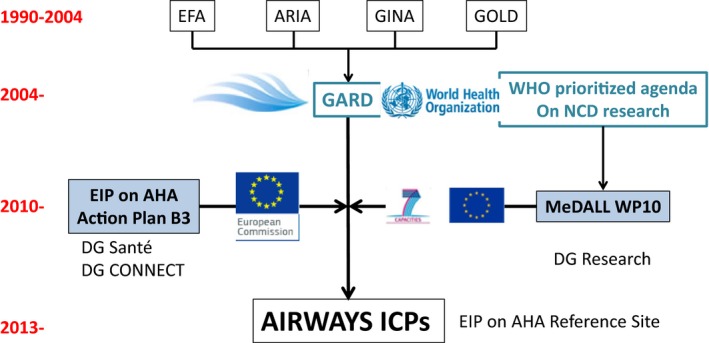
MeDALL interactions between EU and WHO (from Ref. 74). GARD, Global Alliance against Chronic Respiratory Diseases; NGO, nongovernmental organization; GO, governmental organization; MOH, Ministry of Health; NCD, noncommunicable disease.
MeDALL has interacted with the EIP on AHA 76, 77, 78 via several actions. Synergies have been achieved between MeDALL and AIRWAYS ICPs – the model of chronic diseases of Area 5 of the B3 Action Plan of the EIP on AHA 73 – due to their burden, mortality and comorbidities 71 as well as their early development 79, 80. AIRWAYS ICPs has strategic relevance to the European Union Health Strategy, adding value to existing public health knowledge.
Impact of MeDALL on the EU policies in the early diagnosis and management of allergic diseases in active and healthy ageing
The leading priority for the Polish Presidency of the Council of the European Union (2011) was to reduce health inequalities across European societies and, within its framework, to concentrate on the prevention and control of respiratory diseases in children in order to promote AHA 79, 80. The clinical implications of MeDALL reinforce the priority of the EU and suggest solutions for implementation.
Pre‐ and perinatal events play a fundamental role in health, the development of diseases and ageing 79, 80. The developmental determinants of NCDs in ageing were reinforced during the Cyprus Presidency of the EU Council (2012) 81. A meeting was convened by the Reference Site of Languedoc Roussillon 76 on the early determinants of active and healthy ageing (NIH, EIP on AHA, MeDALL) 82. These concepts have been considered in the senioral policy of Poland 83. Moreover, the project results were presented on the 28th of May 2015 at the European Parliament in Brussels to an audience composed of policy makers, healthcare professionals, researchers and patients’ representatives.
Translation into policies
The first results of ‘The Finnish Allergy Programme 2008–2018’ 84, supported by MeDALL data, indicate that allergy burden can be reduced with relatively simple means. This has been endorsed by the Norwegian Allergy Health Programme and, along with the Finnish programme, will serve as a platform for other countries (Oslo, November 2014) 85.
Gaps and the future
The European Commission considers MeDALL to be a success story. A summary can be found on the Horizon 2020 website. It will also be present in the future Health Success Stories Brochure, scheduled to be launched within the next few months.
As other similar projects, MeDALL has involved a huge multidisciplinary effort by a large international network of partners. MeDALL has been made possible thanks to previous consortiums (such as GA2LEN, ENRIECO and CHICOS) 12 that have paved the way of pooling and integrating national or local birth cohorts. Each of these consortiums has experienced the challenge of sustaining the network. MeDALL includes over 44 000 children recruited at birth for the study of the most common chronic disease (allergy). The power of the study is sufficient for the assessment of primary diseases (asthma, rhinitis and eczema), but it is at the limit for multimorbidity, in particular at the early stages of the disease (4 years) 41 and for the discovery of biomarkers. In studies of multimorbid NCDs (e.g. COPD, cardiovascular diseases, diabetes), the power of population cohorts will be sufficient to assess established diseases. However, cohorts are likely to fail in the identification of early multimorbid diseases, their causality and discovery of biomarkers. Thus, the data of MeDALL are generalizable to multimorbid NCDs across the life cycle. Funding from the EU Structure and Cohesion Funds have been obtained by the Région Languedoc Roussillon in the frame of MACVIA‐LR 76, 78 to maintain the database until new funding from the EU or other sources is available for other projects.
Despite the successful experience of MeDALL to integrate birth cohorts of asthma and allergy, we really do need to escalate the level of integration. A new methodology should be proposed to combine the strengths and weaknesses of the birth cohorts, possibly enriching them with patient cohorts 47, 86, 87, registered data in primary care 88, clinical trial databases 89 and/or internet based studies. In very young children, similar severe asthma phenotypes exist in patient cohorts of persistent recurrent wheezers 86 and in cohorts in the general population 90, suggesting the possibility of pooling both types of cohorts. However, ethical issues are of great importance in pooling such studies.
Developing a systems medicine approach to complex diseases is a phenomenal challenge. MeDALL was used as a model of systems medicine and has initiated a common language for the assessment of all noncommunicable diseases 91. The novel trend for the management of NCDs is evolving towards integrative, holistic approaches. NCDs are intertwined with ageing. To tackle NCDs in their totality in order to reduce their burden and societal impact, it has been proposed that NCDs should be considered as a single expression of disease with different risk factors and entities. An innovative integrated health system built around systems medicine and strategic partnerships is proposed to combat NCDs 4, 91.
Conflicts of interest
The authors declare no conflict of interest.
Acknowledgments
The authors would like to thank the members of the Scientific and Ethics Advisory Boards who made invaluable comments during the project duration: P Burney (Imperial College, London, UK), SGO Johansson (Karolinska Institute, Stockholm, Sweden), J Kiley (NHLBI, Bethesda, USA), P Sterk (Amsterdam University, NL), M Van Hague (Karolinska Institute, Stockholm, Sweden), M Yazdanbakhsh (Leiden University, NL), K Dierickx (Leuven, Belgium), T Garani‐Papadatos (Athens, Greece) and M Sutrop (Riga, Estonia).
Bousquet J, Anto JM, Akdis M, Auffray C, Keil T, Momas I, Postma DS, Valenta R, Wickman M, Cambon‐Thomsen A, Haahtela T, Lambrecht BN, Lodrup Carlsen KC, Koppelman GH, Sunyer J, Zuberbier T, Annesi‐Maesano I, Arno A, Bindslev‐Jensen C, De Carlo G, Forastiere F, Heinrich J, Kowalski ML, Maier D, Melén E, Palkonen S, Smit HA, Standl M, Wright J, Asarnoj A, Benet M, Ballardini N, Garcia‐Aymerich J, Gehring U, Guerra S, Hohman C, Kull I, Lupinek C, Pinart M, Skrindo I, Westman M, Smagghe D, Akdis C, Albang R, Anastasova V, Anderson N, Bachert C, Ballereau S, Ballester F, Basagana X, Bedbrook A, Bergstrom A, von Berg A, Brunekreef B, Burte E, Carlsen KH, Chatzi L, Coquet JM, Curin M, Demoly P, Eller E, Fantini MP, Gerhard B, Hammad H, von Hertzen L, Hovland V, Jacquemin B, Just J, Keller T, Kerkhof M, Kiss R, Kogevinas M, Koletzko S, Lau S, Lehmann I, Lemonnier N, McEachan R, Mäkelä M, Mestres J, Minina E, Mowinckel P, Nadif R, Nawijn M, Oddie S, Pellet J, Pin I, Porta D, Rancière F, Rial‐Sebbag A, Saeys Y, Schuijs MJ, Siroux V, Tischer CG, Torrent M, Varraso R, De Vocht J, Wenger K, Wieser S, Xu C. Paving the way of systems biology and precision medicine in allergic diseases: the MeDALL success story. Allergy 2016; 71: 1513–1525
Edited by: Thomas Bieber
References
- 1. Bousquet J, Anto J, Auffray C, Akdis M, Cambon‐Thomsen A, Keil T et al. MeDALL (Mechanisms of the Development of ALLergy): an integrated approach from phenotypes to systems medicine. Allergy 2011;66:596–604. [DOI] [PubMed] [Google Scholar]
- 2. Bousquet J, Anto JM, Wickman M, Keil T, Valenta R, Haahtela T et al. Are allergic multimorbidities and IgE polysensitization associated with the persistence or re‐occurrence of foetal type 2 signalling? The MeDALL hypothesis Allergy 2015;70:1062–1078. [DOI] [PubMed] [Google Scholar]
- 3. Martinez FD. Genes, environments, development and asthma: a reappraisal. Eur Respir J 2007;29:179–184. [DOI] [PubMed] [Google Scholar]
- 4. Bousquet J, Anto JM, Sterk PJ, Adcock IM, Chung KF, Roca J et al. Systems medicine and integrated care to combat chronic noncommunicable diseases. Genome Med 2011;3:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Anto JM, Pinart M, Akdis M, Auffray C, Bachert C, Basagana X et al. Understanding the complexity of IgE‐related phenotypes from childhood to young adulthood: a Mechanisms of the Development of Allergy (MeDALL) seminar. J Allergy Clin Immunol 2012;129:943–954. [DOI] [PubMed] [Google Scholar]
- 6. Bousquet J, Gern JE, Martinez FD, Anto JM, Johnson CC, Holt PG et al. Birth cohorts in asthma and allergic diseases: report of a NIAID/NHLBI/MeDALL joint workshop. J Allergy Clin Immunol 2014;133:1535–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Molter A, Simpson A, Berdel D, Brunekreef B, Custovic A, Cyrys J et al. A multicentre study of air pollution exposure and childhood asthma prevalence: the ESCAPE project. Eur Respir J 2015;45:610–624. [DOI] [PubMed] [Google Scholar]
- 8. Fuertes E, MacIntyre E, Agius R, Beelen R, Brunekreef B, Bucci S et al. Associations between particulate matter elements and early‐life pneumonia in seven birth cohorts: results from the ESCAPE and TRANSPHORM projects. Int J Hyg Environ Health 2014;217:819–829. [DOI] [PubMed] [Google Scholar]
- 9. Pedersen M, Giorgis‐Allemand L, Bernard C, Aguilera I, Andersen AM, Ballester F et al. Ambient air pollution and low birthweight: a European cohort study (ESCAPE). Lancet Respir Med 2013;1:695–704. [DOI] [PubMed] [Google Scholar]
- 10. Eeftens M, Beelen R, de Hoogh K, Bellander T, Cesaroni G, Cirach M et al. Development of land use regression models for PM(2.5), PM(2.5) absorbance, PM(10) and PM(coarse) in 20 European study areas; results of the ESCAPE project. Environ Sci Technol 2012;46:11195–11205. [DOI] [PubMed] [Google Scholar]
- 11. Hohmann C, Pinart M, Tischer C, Gehring U, Heinrich J, Kull I et al. The development of the MeDALL core questionnaires for a harmonized follow‐up assessment of eleven European birth cohorts on asthma and allergies. Int Arch Allergy Immunol 2014;163:215–224. [DOI] [PubMed] [Google Scholar]
- 12. Bousquet J, Anto J, Sunyer J, Nieuwenhuijsen M, Vrijheid M, Keil T et al. Pooling birth cohorts in allergy and asthma: European Union‐funded initiatives–a MeDALL, CHICOS, ENRIECO, and GA(2)LEN joint paper. Int Arch Allergy Immunol 2013;161:1–10. [DOI] [PubMed] [Google Scholar]
- 13. Sucharew H, Ryan PH, Bernstein D, Succop P, Khurana Hershey GK, Lockey J et al. Exposure to traffic exhaust and night cough during early childhood: the CCAAPS birth cohort. Pediatr Allergy Immunol 2010;21(Pt 1):253–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Garcia Algar O, Pichini S, Basagana X, Puig C, Vall O, Torrent M et al. Concentrations and determinants of NO2 in homes of Ashford, UK and Barcelona and Menorca, Spain. Indoor Air 2004;14:298–304. [DOI] [PubMed] [Google Scholar]
- 15. Wickman M, Kull I, Pershagen G, Nordvall SL. The BAMSE project: presentation of a prospective longitudinal birth cohort study. Pediatr Allergy Immunol 2002;13(Suppl 15):11–13. [DOI] [PubMed] [Google Scholar]
- 16. Wright J, Small N, Raynor P, Tuffnell D, Bhopal R, Cameron N et al. Cohort profile: the Born in Bradford multi‐ethnic family cohort study. Int J Epidemiol 2013;42:978–991. [DOI] [PubMed] [Google Scholar]
- 17. Raynor P. Born in Bradford, a cohort study of babies born in Bradford, and their parents: protocol for the recruitment phase. BMC Public Health 2008;8:327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Host A, Halken S, Jacobsen HP, Christensen AE, Herskind AM, Plesner K. Clinical course of cow's milk protein allergy/intolerance and atopic diseases in childhood. Pediatr Allergy Immunol 2002;13(Suppl 15):23–28. [DOI] [PubMed] [Google Scholar]
- 19. Lodrup Carlsen KC. The environment and childhood asthma (ECA) study in Oslo: ECA‐1 and ECA‐2. Pediatr Allergy Immunol 2002;13(Suppl 15):29–31. [DOI] [PubMed] [Google Scholar]
- 20. Slama R, Thiebaugeorges O, Goua V, Aussel L, Sacco P, Bohet A et al. Maternal personal exposure to airborne benzene and intrauterine growth. Environ Health Perspect 2009;117:1313–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Laubereau B, Brockow I, Zirngibl A, Koletzko S, Gruebl A, von Berg A et al. Effect of breast‐feeding on the development of atopic dermatitis during the first 3 years of life–results from the GINI‐birth cohort study. J Pediatr 2004;144:602–607. [DOI] [PubMed] [Google Scholar]
- 22. Berg AV, Kramer U, Link E, Bollrath C, Heinrich J, Brockow I et al. Impact of early feeding on childhood eczema: development after nutritional intervention compared with the natural course–the GINIplus study up to the age of 6 years. Clin Exp Allergy 2010;40:627–636. [DOI] [PubMed] [Google Scholar]
- 23. Aguilera I, Guxens M, Garcia‐Esteban R, Corbella T, Nieuwenhuijsen MJ, Foradada CM et al. Association between GIS‐based exposure to urban air pollution during pregnancy and birth weight in the INMA Sabadell cohort. Environ Health Perspect 2009;117:1322–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Heinrich J, Bolte G, Holscher B, Douwes J, Lehmann I, Fahlbusch B et al. Allergens and endotoxin on mothers’ mattresses and total immunoglobulin E in cord blood of neonates. Eur Respir J 2002;20:617–623. [DOI] [PubMed] [Google Scholar]
- 25. Nickel R, Niggemann B, Gruber C, Kulig M, Wahn U, Lau S. How should a birth cohort study be organised? Experience from the German MAS cohort study. Paediatr Respir Rev 2002;3:169–176. [DOI] [PubMed] [Google Scholar]
- 26. Illi S, von Mutius E, Lau S, Nickel R, Gruber C, Niggemann B et al. The natural course of atopic dermatitis from birth to age 7 years and the association with asthma. J Allergy Clin Immunol 2004;113:925–931. [DOI] [PubMed] [Google Scholar]
- 27. Nickel R, Illi S, Lau S, Sommerfeld C, Bergmann R, Kamin W et al. Variability of total serum immunoglobulin E levels from birth to the age of 10 years. A prospective evaluation in a large birth cohort (German multicenter allergy study). Clin Exp Allergy 2005;35:619–623. [DOI] [PubMed] [Google Scholar]
- 28. Illi S, von Mutius E, Lau S, Niggemann B, Gruber C, Wahn U. Perennial allergen sensitisation early in life and chronic asthma in children: a birth cohort study. Lancet 2006;368:763–770. [DOI] [PubMed] [Google Scholar]
- 29. Clarisse B, Demattei C, Nikasinovic L, Just J, Daures JP, Momas I. Bronchial obstructive phenotypes in the first year of life among Paris birth cohort infants. Pediatr Allergy Immunol 2009;20:126–133. [DOI] [PubMed] [Google Scholar]
- 30. Wijga A, Smit HA, Brunekreef B, Gerritsen J, Kerkhof M, Koopman LP et al. Are children at high familial risk of developing allergy born into a low risk environment? The PIAMA birth cohort study. Prevention and incidence of asthma and mite allergy. Clin Exp Allergy 2001;31:576–581. [DOI] [PubMed] [Google Scholar]
- 31. Chatzi L, Plana E, Daraki V, Karakosta P, Alegkakis D, Tsatsanis C et al. Metabolic syndrome in early pregnancy and risk of preterm birth. Am J Epidemiol 2009;170:829–836. [DOI] [PubMed] [Google Scholar]
- 32. Porta D, Forastiere F, Di Lallo D, Perucci CA. Enrolment and follow‐up of a birth cohort in Rome. Epidemiol Prev 2007;31:303–308. [PubMed] [Google Scholar]
- 33. Maier D, Kalus W, Wolff M, Kalko SG, Roca J, Marin de Mas I et al. Knowledge management for systems biology a general and visually driven framework applied to translational medicine. BMC Syst Biol 2011;5:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lupinek C, Wollmann E, Baar A, Banerjee S, Breiteneder H, Broecker BM et al. Advances in allergen‐microarray technology for diagnosis and monitoring of allergy: the MeDALL allergen‐chip. Methods 2014;66:106–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Skrindo I, Lupinek C, Valenta R, Hovland V, Pahr S, Baar A et al. The use of the MeDALL‐chip to assess IgE sensitization: a new diagnostic tool for allergic disease? Pediatr Allergy Immunol 2015;26:239–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Westman M, Lupinek C, Bousquet J, Andersson N, Pahr S, Baar A et al. Early childhood IgE reactivity to pathogenesis‐related class 10 proteins predicts allergic rhinitis in adolescence. J Allergy Clin Immunol 2015;135:1199–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Asarnoj A, Hamsten C, Waden K, Lupinek C, Andersson N, Kull I et al. Sensitization to cat and dog allergen molecules in childhood and prediction of symptoms of cat and dog allergy in adolescence: a BAMSE/MeDALL study. J Allergy Clin Immunol 2016;137:813–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pinart M, Albang R, Maier D, Duran‐Tauleria E, Mena G, Gimeno‐Santos E et al. Systematic review on the definition of allergic diseases in children: the MeDALL study. Int Arch Allergy Immunol 2015;168:110–121. [DOI] [PubMed] [Google Scholar]
- 39. Belgrave DC, Buchan I, Bishop C, Lowe L, Simpson A, Custovic A. Trajectories of lung function during childhood. Am J Respir Crit Care Med 2014;189:1101–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bousquet J, Van Cauwenberge P, Khaltaev N. Allergic rhinitis and its impact on asthma. J Allergy Clin Immunol 2001;108(5 Suppl):S147–S334. [DOI] [PubMed] [Google Scholar]
- 41. Pinart M, Benet M, Annesi‐Maesano I, von Berg A, Berdel D, Carlsen KC et al. Comorbidity of eczema, rhinitis, and asthma in IgE‐sensitised and non‐IgE‐sensitised children in MeDALL: a population‐based cohort study. Lancet Respir Med 2014;2:131–140. [DOI] [PubMed] [Google Scholar]
- 42. Garcia‐Aymerich J, Benet M, Saeys Y, Pinart M, Basagana X, Smit HA et al. Phenotyping asthma, rhinitis and eczema in MeDALL population‐based birth cohorts: an allergic comorbidity cluster. Allergy 2015;70:973–984. [DOI] [PubMed] [Google Scholar]
- 43. Bousquet J, Becker WM, Hejjaoui A, Chanal I, Lebel B, Dhivert H et al. Differences in clinical and immunologic reactivity of patients allergic to grass pollens and to multiple‐pollen species. II. Efficacy of a double‐blind, placebo‐controlled, specific immunotherapy with standardized extracts. J Allergy Clin Immunol 1991;88:43–53. [DOI] [PubMed] [Google Scholar]
- 44. Bousquet J, Hejjaoui A, Becker WM, Cour P, Chanal I, Lebel B et al. Clinical and immunologic reactivity of patients allergic to grass pollens and to multiple pollen species. I. Clinical and immunologic characteristics. J Allergy Clin Immunol 1991;87:737–746. [DOI] [PubMed] [Google Scholar]
- 45. Pene J, Rivier A, Lagier B, Becker WM, Michel FB, Bousquet J. Differences in IL‐4 release by PBMC are related with heterogeneity of atopy. Immunology 1994;81:58–64. [PMC free article] [PubMed] [Google Scholar]
- 46. Ballardini N, Bergstrom A, Wahlgren CF, van Hage M, Hallner E, Kull I et al. IgE antibodies in relation to prevalence and multimorbidity of eczema, asthma, and rhinitis from birth to adolescence. Allergy 2016;71:342–349. [DOI] [PubMed] [Google Scholar]
- 47. Just J, Deslandes‐Boutmy E, Amat F, Desseaux K, Nemni A, Bourrat E et al. Natural history of allergic sensitization in infants with early‐onset atopic dermatitis: results from ORCA study. Pediatr Allergy Immunol 2014;25:668–673. [DOI] [PubMed] [Google Scholar]
- 48. Burte E, Bousquet J, Varraso R, Gormand F, Just J, Matran R et al. Characterization of rhinitis according to the asthma status in adults using an unsupervised approach in the EGEA study. PLoS One 2015;10:e0136191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gehring UW, Hoek A, Bellander G, Berdel T, Brüske D, Fuertes I et al. Exposure to air pollution and development of asthma and rhinoconjunctivitis throughout childhood and adolescence: a population‐based birth cohort study. Lancet Respir Med 2015;3:933–942. [DOI] [PubMed] [Google Scholar]
- 50. Guerra S, Halonen M, Vasquez MM, Spangenberg A, Stern DA, Morgan WJ et al. Relation between circulating CC16 concentrations, lung function, and development of chronic obstructive pulmonary disease across the lifespan: a prospective study. Lancet Respir Med 2015;3:613–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Coquet JM, Schuijs MJ, Smyth MJ, Deswarte K, Beyaert R, Braun H et al. Interleukin‐21‐producing CD4(+) T cells promote type 2 immunity to house dust mites. Immunity 2015;43:318–330. [DOI] [PubMed] [Google Scholar]
- 52. Schuijs MJ, Willart MA, Vergote K, Gras D, Deswarte K, Ege MJ et al. Farm dust and endotoxin protect against allergy through A20 induction in lung epithelial cells. Science 2015;349:1106–1110. [DOI] [PubMed] [Google Scholar]
- 53. Palomares O, Ruckert B, Jartti T, Kucuksezer UC, Puhakka T, Gomez E et al. Induction and maintenance of allergen‐specific FOXP3+ Treg cells in human tonsils as potential first‐line organs of oral tolerance. J Allergy Clin Immunol 2012;129:510–520. [DOI] [PubMed] [Google Scholar]
- 54. Palomares O, Martin‐Fontecha M, Lauener R, Traidl‐Hoffmann C, Cavkaytar O, Akdis M et al. Regulatory T cells and immune regulation of allergic diseases: roles of IL‐10 and TGF‐beta. Genes Immun 2014;15:511–520. [DOI] [PubMed] [Google Scholar]
- 55. Kucuksezer UC, Palomares O, Ruckert B, Jartti T, Puhakka T, Nandy A et al. Triggering of specific Toll‐like receptors and proinflammatory cytokines breaks allergen‐specific T‐cell tolerance in human tonsils and peripheral blood. J Allergy Clin Immunol 2013;131:875–885. [DOI] [PubMed] [Google Scholar]
- 56. Stanic B, van de Veen W, Wirz OF, Ruckert B, Morita H, Sollner S et al. IL‐10‐overexpressing B cells regulate innate and adaptive immune responses. J Allergy Clin Immunol 2015;135:771–780. [DOI] [PubMed] [Google Scholar]
- 57. van de Veen W, Stanic B, Yaman G, Wawrzyniak M, Sollner S, Akdis DG et al. IgG4 production is confined to human IL‐10‐producing regulatory B cells that suppress antigen‐specific immune responses. J Allergy Clin Immunol 2013;131:1204–1212. [DOI] [PubMed] [Google Scholar]
- 58. Wegrzyn AS, Jakiela B, Ruckert B, Jutel M, Akdis M, Sanak M et al. T‐cell regulation during viral and nonviral asthma exacerbations. J Allergy Clin Immunol 2015;136:194–197. [DOI] [PubMed] [Google Scholar]
- 59. Wawrzyniak M, Ochsner U, Wirz O, Wawrzyniak P, van de Veen W, Akdis CA et al. A novel, dual cytokine‐secretion assay for the purification of human Th22 cells that do not co‐produce IL‐17A. Allergy 2016;71:47–57. [DOI] [PubMed] [Google Scholar]
- 60. Kast JI, Wanke K, Soyka MB, Wawrzyniak P, Akdis D, Kingo K et al. The broad spectrum of interepithelial junctions in skin and lung. J Allergy Clin Immunol 2012;130:544–547. [DOI] [PubMed] [Google Scholar]
- 61. Soyka MB, Holzmann D, Akdis CA. Regulatory cells in allergen‐specific immunotherapy. Immunotherapy 2012;4:389–396. [DOI] [PubMed] [Google Scholar]
- 62. Kubo T, Wawrzyniak P, Morita H, Sugita K, Wanke K, Kast JI et al. CpG‐DNA enhances the tight junction integrity of the bronchial epithelial cell barrier. J Allergy Clin Immunol 2015;136:1413–1416. [DOI] [PubMed] [Google Scholar]
- 63. Smit HA, Pinart M, Anto JM, Keil T, Bousquet J, Carlsen KH et al. Childhood asthma prediction models: a systematic review. Lancet Respir Med 2015;3:973–984. [DOI] [PubMed] [Google Scholar]
- 64. Bousquet PJ, Devillier P, Tadmouri A, Mesbah K, Demoly P, Bousquet J. Clinical relevance of cluster analysis in phenotyping allergic rhinitis in a real‐life study. Int Arch Allergy Immunol 2015;166:231–240. [DOI] [PubMed] [Google Scholar]
- 65. Canonica GW, Bachert C, Hellings P, Ryan D, Valovirta E, Wickman M et al. Allergen immunotherapy (AIT): a prototype of precision medicine. World Allergy Organ J 2015;8:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bousquet J, Burney PG, Zuberbier T, Cauwenberge PV, Akdis CA, Bindslev‐Jensen C et al. GA2 LEN (Global Allergy and Asthma European Network) addresses the allergy and asthma ‘epidemic’. Allergy 2009;64:969–977. [DOI] [PubMed] [Google Scholar]
- 67. Kauffmann F, Cambon‐Thomsen A. Tracing biological collections: between books and clinical trials. JAMA 2008;299:2316–2318. [DOI] [PubMed] [Google Scholar]
- 68. Anastasova V, Mahalatchimy A, Rial‐Sebbag E, Anto Boque JM, Keil T, Sunyer J et al. Communication of results and disclosure of incidental findings in longitudinal paediatric research. Pediatr Allergy Immunol 2013;24:389–394. [DOI] [PubMed] [Google Scholar]
- 69. Bousquet J, Kiley J, Bateman ED, Viegi G, Cruz AA, Khaltaev N et al. Prioritised research agenda for prevention and control of chronic respiratory diseases. Eur Respir J 2010;36:995–1001. [DOI] [PubMed] [Google Scholar]
- 70. Bousquet J, Dahl R, Khaltaev N. Global alliance against chronic respiratory diseases. Allergy 2007;62:216–223. [DOI] [PubMed] [Google Scholar]
- 71. Bousquet J, Khaltaev N. Global Surveillance, Prevention and Control of Chronic Respiratory Diseases. A Comprehensive Approach. Global Alliance Against Chronic Respiratory Diseases. Geneva, Switzerland: World Health Organization, 2007: 148 pages. ISBN 978 92 4 156346 8. [Google Scholar]
- 72. Bousquet J, Mantzouranis E, Cruz AA, Ait‐Khaled N, Baena‐Cagnani CE, Bleecker ER et al. Uniform definition of asthma severity, control, and exacerbations: document presented for the World Health Organization consultation on severe asthma. J Allergy Clin Immunol 2010;126:926–938. [DOI] [PubMed] [Google Scholar]
- 73. Bousquet J, Addis A, Adcock I, Agache I, Agusti A, Alonso A et al. Integrated care pathways for airway diseases (AIRWAYS‐ICPs). Eur Respir J 2014;44:304–323. [DOI] [PubMed] [Google Scholar]
- 74. Bousquet J, Barbara C, Bateman E, Bel E, Bewick M, Chavannes N et al. AIRWAYS ICPs (European Innovation Partnership on Active and Healthy Ageing) from concept to implementation. Eur Respir J 2016;47:1028–1033. [DOI] [PubMed] [Google Scholar]
- 75. Bousquet J, Schunemann HJ, Fonseca J, Samolinski B, Bachert C, Canonica GW et al. MACVIA‐ARIA Sentinel NetworK for allergic rhinitis (MASK‐rhinitis): the new generation guideline implementation. Allergy 2015;70:1372–1392. [DOI] [PubMed] [Google Scholar]
- 76. Bousquet J, Bourquin C, Augé P, Domy P, Bringer J, Camuzat T et al. MACVIA‐LR reference site of the European innovation partnership on active and healthy ageing. Eur GeriatrMed 2014;5:406–415. [Google Scholar]
- 77. Bousquet J, Michel J, Standberg T, Crooks G, Iakovidis I, Gomez M. The European innovation partnership on active and healthy ageing: the European geriatric medicine introduces the EIP on AHA column. Eur Geriatr Med 2014;5:361–362. [Google Scholar]
- 78. Bousquet J, Mercier J, Avignon A, Bourret R, Camuzat T. MACVIA‐LR (France) case study. Report EUR 27150 EN In: Abadie F, editor. Strategic Intelligence Monitor on Personal Health Systems Phase 3 (SIMPHS3). JRC94487. Luxembourg: Publications Office of the European Union: JRC (Joint Research Centre) Science and Policy Report, 2015: https://ec.europa.eu/jrc. [Google Scholar]
- 79. Samolinski B, Fronczak A, Kuna P, Akdis CA, Anto JM, Bialoszewski AZ et al. Prevention and control of childhood asthma and allergy in the EU from the public health point of view: Polish Presidency of the European Union. Allergy 2012;67:726–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Samolinski B, Fronczak A, Wlodarczyk A, Bousquet J. Council of the European Union conclusions on chronic respiratory diseases in children. Lancet 2012;379:e45–e46. [DOI] [PubMed] [Google Scholar]
- 81. Bousquet J, Tanasescu CC, Camuzat T, Anto JM, Blasi F, Neou A et al. Impact of early diagnosis and control of chronic respiratory diseases on active and healthy ageing. A debate at the European Union parliament. Allergy 2013;68:555–561. [DOI] [PubMed] [Google Scholar]
- 82. Bousquet J, Anto JM, Berkouk K, Gergen P, Antunes JP, Auge P et al. Developmental determinants in non‐communicable chronic diseases and ageing. Thorax 2015;70:595–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Samolinski B, Raciborski F, Bousquet J, Kosiniak‐Kamysz W, Radziewicz‐Winnicki I, Kłak A et al. Development of senioral policy in Poland. Eur Geriatr Med 2015;6:389–395. [Google Scholar]
- 84. Haahtela T, von Hertzen L, Makela M, Hannuksela M. Finnish allergy programme 2008–2018–time to act and change the course. Allergy 2008;63:634–645. [DOI] [PubMed] [Google Scholar]
- 85. Lodrup Carlsen KC, Haahtela T, Carlsen KH, Smith A, Bjerke M, Wickman M et al. Integrated allergy and asthma prevention and care: report of the MeDALL/AIRWAYS ICPs meeting at the Ministry of Health and Care Services, Oslo, Norway. Int Arch Allergy Immunol 2015;167:57–64. [DOI] [PubMed] [Google Scholar]
- 86. Just J, Gouvis‐Echraghi R, Couderc R, Guillemot‐Lambert N, Saint‐Pierre P. Novel severe wheezy young children phenotypes: boys atopic multiple‐trigger and girls nonatopic uncontrolled wheeze. J Allergy Clin Immunol 2012;130:103–110. [DOI] [PubMed] [Google Scholar]
- 87. Just J, Gouvis‐Echraghi R, Rouve S, Wanin S, Moreau D, Annesi‐Maesano I. Two novel, severe asthma phenotypes identified during childhood using a clustering approach. Eur Respir J 2012;40:55–60. [DOI] [PubMed] [Google Scholar]
- 88. Price DB, Rigazio A, Campbell JD, Bleecker ER, Corrigan CJ, Thomas M et al. Blood eosinophil count and prospective annual asthma disease burden: a UK cohort study. Lancet Respir Med 2015;3:849–858. [DOI] [PubMed] [Google Scholar]
- 89. Angus DC. Fusing randomized trials with big data: the key to self‐learning health care systems? JAMA 2015;314:767–768. [DOI] [PubMed] [Google Scholar]
- 90. Herr M, Just J, Nikasinovic L, Foucault C, Le Marec AM, Giordanella JP et al. Risk factors and characteristics of respiratory and allergic phenotypes in early childhood. J Allergy Clin Immunol 2012;130:389–396. [DOI] [PubMed] [Google Scholar]
- 91. Bousquet J, Jorgensen C, Dauzat M, Cesario A, Camuzat T, Bourret R et al. Systems medicine approaches for the definition of complex phenotypes in chronic diseases and ageing. From concept to implementation and policies. Curr Pharm Des 2014;20:5928–5944. [DOI] [PubMed] [Google Scholar]
- 92. Gough H, Grabenhenrich L, Reich A, Eckers N, Nitsche O, Schramm D et al. Allergic multimorbidity of asthma, rhinitis, and eczema over 20 years in the German birth cohort MAS. Pediatr Allergy Immunol 2015;26:431–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Bousquet J, Anto JM, Bachert C, Bousquet PJ, Colombo P, Crameri R et al. Factors responsible for differences between asymptomatic subjects and patients presenting an IgE sensitization to allergens. A GALEN project. Allergy 2006;61:671–680. [DOI] [PubMed] [Google Scholar]
- 94. Amaral AF, Minelli C, Guerra S, Wjst M, Probst‐Hensch N, Pin I et al. The locus C11orf30 increases susceptibility to poly‐sensitization. Allergy 2015;70:328–333. [DOI] [PubMed] [Google Scholar]
- 95. Ferreira MA, Matheson MC, Tang CS, Granell R, Ang W, Hui J et al. Genome‐wide association analysis identifies 11 risk variants associated with the asthma with hay fever phenotype. J Allergy Clin Immunol 2014;133:1564–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]


