TO THE EDITOR
Recurrent mutations in the promoter of the telomerase reverse transcriptase (TERT) gene were first discovered in melanoma (Horn et al., 2013; Huang et al., 2013) and subsequently in several other cancer types (Killela et al., 2013). These mutations occur mainly at positions −124 bp (chr5, 1,295,228 C>T hg19 coordinate) and −146 bp (chr5, 1,295,250 C>T hg19 coordinate) from the ATG translation start site (Heidenreich et al., 2014; Horn et al., 2013; Huang et al., 2013), and are hereafter termed as −124C>T and −146C>T, respectively (Figure 1a). Mutually exclusive −124C>T and −146C>T mutations have been detected in more than 65% of melanomas (Network, 2015). These mutations contribute to TERT transcriptional upregulation by recruiting the GABPA/B1 transcription factor (Bell et al., 2015). The association of TERT promoter mutations with increased TERT expression has been demonstrated in various tumor types such as bladder cancer and glioblastoma (Borah et al., 2015; Heidenreich et al., 2015), but the functional consequences of these mutations have not yet been clarified in melanoma.
Figure 1.
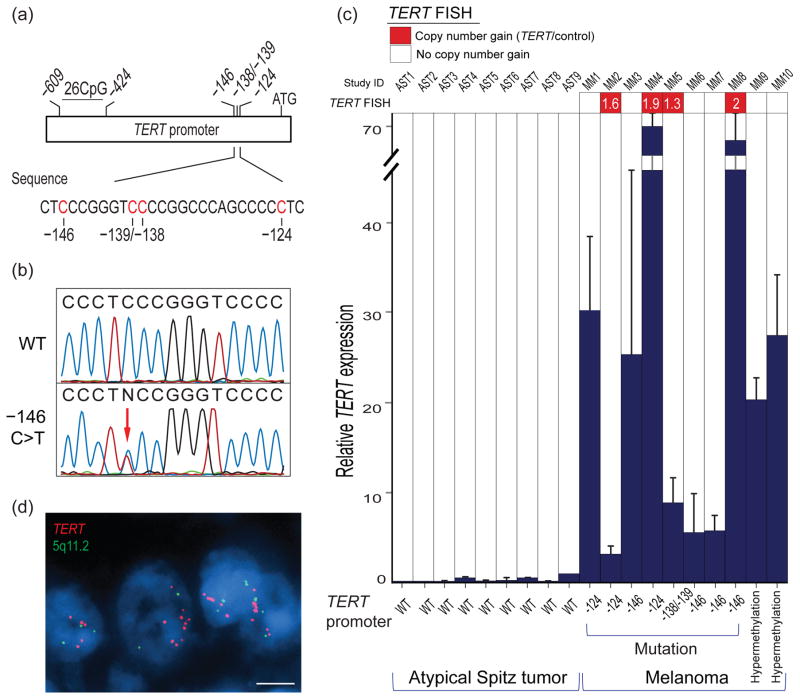
TERT mRNA expression by RT-qPCR in 10 melanomas with TERT promoter alterations and 9 atypical Spitz tumors with the wild-type (WT) TERT promoter. a) Schematic of the TERT promoter showing the position of hotspot mutations. b) Sequence chromatogram of a wild-type (top) and a mutated (bottom) TERT promoter sequence. The bottom sequence is heterozygous at the chr5, 1,295,250 residue, indicated as an N in the printed sequence (red arrow), representing 2 overlapping peaks, a C (WT allele) and a T (mutant allele). c) TERT copy number and relative TERT mRNA expression data in melanomas with TERT promoter −146C>T, −124C>T, or −138/−139CC>TT mutations or TERT promoter hypermethylation versus atypical Spitz tumors with the WT TERT promoter. The TERT expression value in sample AST9 was used as the reference. The numbers in red boxes represent the TERT to PLK2 ratio. d) Representative image of interphase fluorescence in situ hybridization (DAPI counterstain) in a melanoma sample (MM4) showing TERT copy gains in tumor nuclei (scale bar = 10 μm).
In a study of 39 primary melanoma samples, Heidenreich et al. (2014) reported higher TERT expression levels in aggregate in 10 melanomas with various types of mutations compared to 29 samples with the wild-type TERT promoter (Heidenreich et al., 2014). Their study provided circumstantial evidence that all cancer-associated TERT promoter mutations upregulate TERT expression in melanoma. However, an mRNA expression analysis by The Cancer Genome Atlas (TCGA) network, using RNA sequencing data from a large set of melanoma samples, found that only the −124C>T mutation was associated with increased TERT expression (Network, 2015). Therefore, the effect of the −146C>T mutation or the −138/−139CC>TT (chr5, 1,295,242–243 CC>TT) tandem mutation on telomerase expression in melanoma is still unclear.
To assess TERT mRNA expression levels in melanoma samples harboring −146C>T or −138/−139CC>TT mutations, in this study we used real-time quantitative reverse transcriptase polymerase chain reaction (RT-qPCR) in a sample set of metastatic pediatric melanocytic tumors for which the methylation status and the mutational profile of the TERT promoter were known. We previously showed that in a subset of melanomas, TERT expression is mediated epigenetically by promoter hypermethylation rather than by promoter point mutations (Fan et al., 2016). For RT-qPCR, we used formalin-fixed paraffin-embedded (FFPE) tissues from 10 melanomas (7 conventional; 1 fatal spitzoid; 2 arising in giant congenital nevi) and included 9 atypical Spitz tumors with a wild-type unmethylated TERT promoter for comparison. The TERT promoter mutation and methylation data on these samples have been previously published (Fan et al., 2016; Lu et al., 2015). The conventional and spitzoid melanomas each harbored a hotspot TERT promoter mutation (−146C>T in 4; −124C>T in 3; −138/−139CC>TT in 1), and the two melanomas arising in giant congenital nevi (samples MM9, MM10) carried a wild-type hypermethylated TERT promoter.
For RT-qPCR, total RNA was isolated from FFPE tumor tissues by using the Maxwell® 16 LEV RNA FFPE Purification Kit (Promega, AS1260) according to the manufacturer’s protocol. To quantify full-length TERT mRNA expression levels, 2 μg of total RNA from each sample was converted to cDNA by using the SuperScript® VILO cDNA Synthesis Kit (Invitrogen, 11754-010). RT-qPCR was performed in triplicate by using the TaqMan® Gene Expression Assays with gene-specific primers (Life Technologies) for TERT (Hs00972656_m1), and normalized with GAPDH (Hs02758991_g1) as the endogenous control. Relative expression levels were determined by using transcript levels of an atypical Spitz tumor as the reference.
TERT mRNA was undetectable or expressed at low levels by RT-qPCR in the atypical Spitz tumor samples (Figure 1c). Thus, we selected an atypical Spitz tumor with the highest TERT mRNA expression (sample # AST9) as the reference sample (Figure 1c). TERT mRNA in all melanomas, including the four samples with the −146C>T TERT promoter mutation (Figure 1b), was expressed at higher levels than those in atypical Spitz tumors with a 3- to 71-fold (average, 26-fold) increase relative to the control value (Figure 1c). RT-qPCR showed a wide range of expression across samples with similar TERT promoter mutations, whereas the range of expression overlapped between −124C>T (3- to 68-fold increase, average 33-fold) and −146C>T (5- to 71-fold, average 27-fold) mutant melanomas (Figure 1c). The two melanomas arising in congenital nevi showed a 20- and 27-fold increase relative to the control value.
To investigate whether gene copy number gains or amplification contributed to the observed variations in mRNA expression levels, we evaluated the 10 melanoma samples for TERT copy number alterations by interphase fluorescence in situ hybridization (FISH), using a probe specific to the TERT gene together with a control probe containing a specific sequence at 5q11.2. The probe sets were developed using bacterial artificial chromosome clones (BACPAC Resources, Oakland, CA) CTD-3080P12 covering the TERT sequence and RP11-350C7 covering the sequence of the PLK2 control gene at 5q11.2. FISH analysis revealed that the TERT to PLK2 ratio was >1 (1.6 to 2) in at least 40% of cells in four melanomas and there was a normal ratio of 1 in the remaining six melanomas. Interestingly, FISH showed a TERT/PLK2 ratio of ≥1.9 in each of the two samples that had the highest TERT expression levels (MM4 and MM8) (Figure 1c and 1d). Although the findings suggested that gene copy gains might have contributed to the observed variations in telomerase expression, these assays could not determine whether the extra copies of TERT had a normal or a mutant promoter. It is conceivable that additional unaccounted biological mechanisms other than promoter mutations, copy number variations, or promoter hypermethylation contributed to TERT transcriptional upregulation. It is also possible that confounding factors secondary to tissue processing and formalin fixation affected the TERT transcript levels. One major shortcoming of our study is that our comparison is based on telomerase expression rather than the enzymatic activity of TERT mRNA. The direct enzymatic activity of TERT could not be evaluated in these tumors for which the only available tissue was FFPE material. Also, the small sample size (n=10) precluded statistical analysis and therefore the results need to interpreted with caution. Further, because all the melanoma samples were from pediatric patients only, it is necessary to reproduce these findings in a larger number of melanoma samples from adults.
The lack of an association between −146C>T mutations and elevated TERT mRNA expression in the melanoma TCGA data might be because the expression levels were compared relative to melanomas with wild-type promoters, of which some could be telomerase positive due to a different mechanism, such as promoter hypermethylation, rather than point mutations. There are several possible reasons for higher TERT expression in the −124C>T mutant melanomas than in other melanomas in the TCGA study. It has been shown that the −124C>T and −146C>T mutations activate TERT by distinct mechanisms (Li et al., 2015). The −146C>T promoter, unlike the −124C>T promoter, requires non-canonical NF-κB signaling in cooperation with ETS factors to induce TERT reactivation (Li et al., 2015). Although once differentiated into somatic cells both−146C>T and −124C>T mutations increased telomerase activity, a study in which embryonic stem cells were engineered to harbor a TERT promoter mutation reported that only the −124C>T mutation increased TERT transcription in pluripotent stem cells (Chiba et al., 2015). Also, an mRNA expression analysis in glioma samples showed that tumors with the −124C>T mutation had higher TERT expression levels than those with the −146C>T mutations (Heidenreich et al., 2015). Altogether, these findings suggest that the −124C>T genotype is more potent than the −146C>T genotype in inducing TERT expression, which could explain the observed difference seen in the TCGA data for TERT expression levels.
In conclusion, our RT-qPCR findings demonstrate that in melanoma, TERT promoter hotspot −146C>T and −138/−139CC>TT mutations, similar to the −124C>T mutation, correlate with TERT overexpression. Our findings also support that the −146C>T and −138/−139CC>TT mutations contribute biologically to tumorigenesis in melanoma.
Acknowledgments
Funding: This research was supported in part by the National Cancer Institute of the National Institutes of Health under Award Number P30CA021765 and by ALSAC.
Footnotes
Competing Interests: The authors declare that they have no competing interests.
References
- Bell RJ, Rube HT, Kreig A, Mancini A, Fouse SD, Nagarajan RP, Choi S, Hong C, He D, Pekmezci M, et al. Cancer The transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer. Science. 2015;348:1036–9. doi: 10.1126/science.aab0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borah S, Xi L, Zaug AJ, Powell NM, Dancik GM, Cohen SB, Costello JC, Theodorescu D, Cech TR. Cancer TERT promoter mutations and telomerase reactivation in urothelial cancer. Science. 2015;347:1006–10. doi: 10.1126/science.1260200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba K, Johnson JZ, Vogan JM, Wagner T, Boyle JM, Hockemeyer D. Cancer-associated TERT promoter mutations abrogate telomerase silencing. eLife. 2015;4 doi: 10.7554/eLife.07918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Lee S, Wu G, Easton J, Yergeau D, Dummer R, Vogel P, Kirkwood JM, Barnhill RL, Pappo A, et al. Telomerase Expression by Aberrant Methylation of the TERT Promoter in Melanoma Arising in Giant Congenital Nevi. J Invest Dermatol. 2016;136:339–42. doi: 10.1038/JID.2015.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidenreich B, Nagore E, Rachakonda PS, Garcia-Casado Z, Requena C, Traves V, Becker J, Soufir N, Hemminki K, Kumar R. Telomerase reverse transcriptase promoter mutations in primary cutaneous melanoma. Nature communications. 2014;5:3401. doi: 10.1038/ncomms4401. [DOI] [PubMed] [Google Scholar]
- Heidenreich B, Rachakonda PS, Hosen I, Volz F, Hemminki K, Weyerbrock A, Kumar R. TERT promoter mutations and telomere length in adult malignant gliomas and recurrences. Oncotarget. 2015;6:10617–33. doi: 10.18632/oncotarget.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, Kadel S, Moll I, Nagore E, Hemminki K, et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339:959–61. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339:957–9. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killela PJ, Reitman ZJ, Jiao Y, Bettegowda C, Agrawal N, Diaz LA, Jr, Friedman AH, Friedman H, Gallia GL, Giovanella BC, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A. 2013;110:6021–6. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhou QL, Sun W, Chandrasekharan P, Cheng HS, Ying Z, Lakshmanan M, Raju A, Tenen DG, Cheng SY, et al. Non-canonical NF-kappaB signalling and ETS1/2 cooperatively drive C250T mutant TERT promoter activation. Nat Cell Biol. 2015;17:1327–38. doi: 10.1038/ncb3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Zhang J, Nagahawatte P, Easton J, Lee S, Liu Z, Ding L, Wyczalkowski MA, Valentine M, Navid F, et al. The genomic landscape of childhood and adolescent melanoma. J Invest Dermatol. 2015;135:816–23. doi: 10.1038/jid.2014.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Network TCGA. Genomic Classification of Cutaneous Melanoma. Cell. 2015;161:1681–96. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]