Abstract
Shewanella oneidensis is an important model organism for bioremediation studies because of its diverse respiratory capabilities. However, the genetic basis and regulatory mechanisms underlying the ability of S. oneidensis to survive and adapt to various environmentally relevant stresses is poorly understood. To define this organism's molecular response to elevated growth temperatures, temporal gene expression profiles were examined in cells subjected to heat stress by using whole-genome DNA microarrays for S. oneidensis. Approximately 15% (n = 711) of the total predicted S. oneidensis genes (n = 4,648) represented on the microarray were significantly up- or downregulated (P < 0.05) over a 25-min period after shift to the heat shock temperature. As expected, the majority of the genes that showed homology to known chaperones and heat shock proteins in other organisms were highly induced. In addition, a number of predicted genes, including those encoding enzymes in glycolysis and the pentose cycle, serine proteases, transcriptional regulators (MerR, LysR, and TetR families), histidine kinases, and hypothetical proteins were induced. Genes encoding membrane proteins were differentially expressed, suggesting that cells possibly alter their membrane composition or structure in response to variations in growth temperature. A substantial number of the genes encoding ribosomal proteins displayed downregulated coexpression patterns in response to heat stress, as did genes encoding prophage and flagellar proteins. Finally, a putative regulatory site with high conservation to the Escherichia coli σ32-binding consensus sequence was identified upstream of a number of heat-inducible genes.
Shewanella oneidensis MR-1 (formerly Shewanella putrefaciens strain MR-1), a facultatively anaerobic γ-proteobacterium, possesses remarkably diverse respiratory capacities and is widely distributed in nature, with aquatic environments and sediments as its primary habitats (39). In addition to utilizing oxygen as a terminal electron acceptor, S. oneidensis can anaerobically respire various organic and inorganic substrates, including fumarate, nitrate, thiosulfate, trimethylamine N-oxide (TMAO), Fe(III), Mn(III), and (IV), Cr(VI), and U(VI). The metal ion-reducing capabilities of this bacterium may possibly be exploited for the remediation of metal contaminants in the environment. However, little is known about the molecular basis underlying the microorganism's perturbation response behavior or the impact of environmental stresses (e.g., temperature upshift, pH fluctuations, and nutrient limitation) on its ability to reduce metals and radionuclides.
Variation in growth temperature is a common stress encountered in nature. The heat shock response, which is elicited by a sudden increase in growth temperature, has been widely used as a model system for studying the impact of stress on biological systems (27). The hallmark of this adaptive cellular response is the induction of a limited set of proteins, called heat shock proteins (Hsps) or, more generally, molecular chaperones. The spectrum of Hsps synthesized in different organisms after a stress challenge displays notable similarities (25, 28, 47). Several Hsp families can be distinguished and are designated according to their average apparent molecular mass, e.g., Hsp100, Hsp90, Hsp70 (DnaK), Hsp60 (GroEL), and small Hsps (2, 3, 10, 12, 23, 28, 40). In addition, ATP-dependent proteases such as ClpP and Lon are known to be Hsps (11). In general, Hsps play important roles in protein folding, protein degradation, assembly of protein complexes, and transport of proteins across membranes.
DNA microarrays have already been used to characterize the transcriptomes of bacteria responding to different growth conditions and environmental stresses (6, 8, 16, 17, 22, 29, 32, 33, 35). In the present study, we used DNA microarrays covering ∼99% of the total predicted protein-encoding open reading frames (ORFs) in S. oneidensis to investigate the dynamics of global gene expression profiles in response to a temperature upshift from 30 to 42°C over a period of 25 min. Microarray hybridization results revealed that S. oneidensis homologues of known Hsps, together with genes not previously demonstrated to be affected by heat stress and those unique to this metal-reducing bacterium, showed significant differential expression in response to the temperature upshift. Based on the gene expression data and computational analysis, a putative consensus sequence for S. oneidensis heat shock gene promoters was derived that closely resembles the E. coli σ32 recognition site.
DNA microarray construction.
PCR primers for 4,648 of 4931 predicted ORFs in the S. oneidensis genome (excluding 43 unique and 240 multicopy genes) were designed by using PRIMEGENS (44) and then synthesized by MWG Biotech (MWG Biotech., Inc., Highpoint, N.C.). The following criteria were used to identify optimal forward and reverse primers to generate PCR products specific to each of the selected ORFs: (i) the entire ORF was used as a probe if it was <75% similar to all other genes in the genome; (ii) for homologous genes, the maximal portion of the genes showing <75% similarity were selected as specific probes; (iii) for homologous genes where no specific fragments can be identified, one of the genes was selected as a probe to represent the entire gene group; and (iv) each oligonucleotide primer contained 20 to 25 bases. To simplify PCR amplification, most of the primer sets were designed to have annealing temperatures of ∼65°C.
ORF-specific fragments were amplified by using the following cycling conditions: 30-s denaturation at 95°C, 1-min annealing at 60°C, and 1.5-min extension at 72°C, along with an initial 5-min denaturation at 95°C and a final extension reaction at 72°C for 7 min. All PCR products were purified by using the QIAquick 96-well purification kit (Qiagen, Valencia, Calif.). The quality of the amplified products was checked by 1.5% agarose gel electrophoresis and ethidium bromide staining. Amplified DNA fragments were considered correct if PCRs contained a single product of the expected size. PCRs for 451 genes consistently failed to yield satisfactory products (e.g., no product, product of the wrong size, multiple bands, or faint bands), even after redesigned primers were used in the amplification reaction. Of the 4,648 total predicted genes, 4,197 ORFs were correctly amplified, representing ca. 90% of the genome. We used specific 50-mer oligonucleotides to represent the 451 ORFs not successfully amplified by PCR. In total, the PCR amplicons and oligonucleotide probes represented 99% of the total predicted gene content of S. oneidensis. Probes were printed in duplicate onto Telechem Superamine slides (Telechem, Inc., Sunnyvale, Calif.) as described previously (36).
Microarray experiments and data analysis.
S. oneidensis DSP10, a spontaneous rifampin-resistant derivative of S. oneidensis MR-1, was used in the present study because it has been used for mutant construction in our laboratory and thus maintains the consistency in the genotype background among all our microarray studies (36). For all experiments, a single colony of DSP10 was used to inoculate 1 ml of Luria-Bertani (LB) medium (Difco, Detroit, Mich.) in 12-ml plastic tubes and grown overnight at 30°C (optimal growth temperature) on a rotary platform (200 rpm). This culture was then used to inoculate 50 ml of medium prewarmed to 30°C at an optical density at 600 nm (OD600) of 0.01. The flask was shaken on a rotary platform (250 rpm) until a mid-log OD600 of 0.60 was attained. Samples (zero time) were taken from the 50-ml culture, and a 25-ml aliquot was transferred to a 250-ml flask prewarmed to 42°C and then incubated in a 42°C water bath shaker. A parallel identical experiment was performed with a prewarmed 250-ml flask at 30°C. Preliminary experiments were carried out to determine proper heat shock conditions with samples at 5, 15, 25, 40, and 60 min at either 37 or 42°C. Samples were removed from cultures grown at 30 and 42°C at 5, 15, and 25 min and centrifuged for 10 s at the maximum speed in a 5415R centrifuge (Eppendorf, Westbury, N.Y.). The culture supernatant was removed instantly and the tubes containing the cell pellet were placed in liquid nitrogen. Cell density changed only slightly after the 25-min heat shock period (OD600 = 0.60 to 0.66 [on average]). RNA isolation, cDNA labeling, hybridization, and microarray scanning were performed as described previously (36).
To determine signal fluorescence intensities for each spot, 16-bit TIFF scanned images were analyzed by using the software ImaGene version 5.5 (Biodiscovery, Marina Del Rey, Calif.). Any spot that had <75% of pixels and >2 standard deviations above the local background in both channels was rejected. The resulting data files were normalized and further analyzed by using GeneSpring version 5.1 (Silicon Genetics, Redwood City, Calif.).
Assessment of array data quality.
The reliability of the microarray data was assessed by cohybridization of two cDNA samples prepared from the same total cellular RNA. The pattern of hybridization revealed a linear correlation with no more than a twofold change in the relative expression level of 99.7% of the genes (data not shown). This control experiment suggested that genes with an expression ratio beyond this range were either down- or upregulated. Therefore, only genes identified as being down- or upregulated by an expression ratio of at least 2-fold were chosen for further analysis, even though a 1.5-fold cutoff has recently been reported as being biologically significant (18, 32, 33).
To validate the microarray results, eight ORFs were selected for real-time quantitative reverse transcription-PCR (RT-PCR) analysis with the same RNA samples used in the array hybridizations based on the level and reproducibility of changes observed in the microarray experiments. Primers were designed by using Omiga software (Oxford Molecular, Ltd., San Diego, Calif.) and were synthesized by Applied Biosystems. PCR products amplified from these ORFs were single-band fragments of 99 to 101 bp, as confirmed by agarose gel electrophoresis. A 100-bp fragment of the dnaK gene, which was amplified by PCR with genomic DNA as the template, was used to construct the standard curve. The reaction was performed with 50 cycles of 30 s at 94°C, 30 s at 55°C, and 1 min at 72°C and monitored in an iCycler iQ real-time PCR detection system (Bio-Rad, Hercules, Calif.). A standard curve was derived from PCR products representing each ORF with genomic DNA as the template and used to convert threshold crossings to log copy numbers. The expression of each gene was determined from three replicates on a single real-time RT-PCR experiment. The expression ratio was recorded as the fold difference in quantity of real-time RT-PCR product from samples grown at the treatment versus control temperature. A high level of concordance was observed between the microarray and real-time RT-PCR data despite quantitative differences in the level of change (see Fig. S1 in the supplemental material). Overall, real-time RT-PCR and DNA microarray data differed by an average of 2.5-fold, suggesting that microarray hybridization may underestimate the change in expression. This underestimation of fold changes by DNA microarray analysis has been reported previously (16, 33, 43).
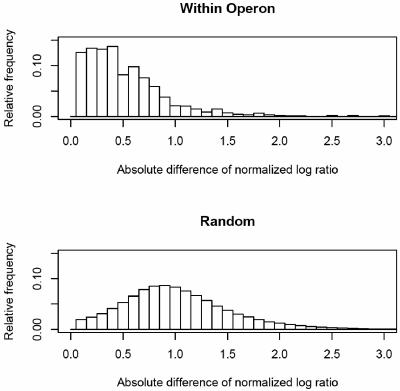
In addition to real-time RT-PCR analysis, expression differences for gene pairs within the same predicted operon or gene pairs selected at random were compared to determine whether changes in gene expression were experimentally significant (Fig. 1). All adjacent gene pairs in S. oneidensis were classified into same-operon and nonoperon pairs by using a Bayesian prediction method based on the distance between the genes, the similarity of the COG functional classes assigned to each gene, the similarity of codon usage, and the degree to which gene adjacency is conserved in 129 publicly available genome sequences (20) (details of the methods are available online[http://vimssftp.lbl.gov/UnsupervisedOperons/home.html]).Deviations from the Gaussian distribution were determined by using the Kolmogorov-Smirnov test (21). Consistent with our expectation, we observed that genes within the same operon responded more similarly to heat shock conditions than did genes randomly selected from the genome. As shown in Fig. 1, the within-operon pairs showed much smaller log-ratio differences than did gene pairs chosen at random, thus confirming the high quality of the expression data particularly for the significantly over- or underexpressed genes. To demonstrate that the underlying distributions of log-ratio differences for the samples of gene pairs within operons and those selected at random are significantly different, the Kolmogorov-Smirnov test was performed for data from the three time points (5 min, D = 0.4128, P < 2.2e−16; 15 min, D = 0.398, P < 2.2e−16; 25 min, D = 0.348, P < 2.2e−16). Highly significant P values were obtained even without the requirement that one gene be significantly over- or underexpressed.
FIG. 1.
Histogram of log ratio expression difference of gene pairs within the same operon versus gene pairs selected at random. The normalized frequency was plotted against the ratio expression difference between the treatments and control. Genes within the same operon responded more similarly than genes randomly selected from the genome under heat shock.
Genomic response of S. oneidensis to heat stress. Whole-genome DNA microarrays were used to obtain a comprehensive, general description of the molecular response mounted by S. oneidensis when challenged by heat stress (a complete microarray data set is available in Table S1 in the supplemental material). In total, 609 genes (323 induced and 286 repressed) at 5 min, 711 genes (358 and 353) at 15 min, and 466 genes (240 and 226) at 25 min exhibited significant (P < 0.05) differential expression at a ≥2-fold level in at least four of the six replicates in response to a temperature upshift from 30 to 42°C. These total gene numbers represent ca. 13% (5 min), 16% (15 min), and 10% (25 min) of the 4,648 ORFs represented on the array. Figure 2 summarizes the overall genomic response of S. oneidensis to the temperature upshift by grouping the differentially expressed genes into their functional role categories, as assigned based on The Institute for Genomic Research 's annotation of the MR-1 genome sequence (14; http://www.tigr.org/).
FIG. 2.
Differentially expressed genes grouped by functional classification according to the TIGR S. oneidensis genome database (www.tigr.org). Columns: 1, amino acid biosynthesis; 2, biosynthesis of cofactors, prosthetic groups, and carriers; 3, cell envelope; 4, cellular processes; 5, central intermediary metabolism; 6, DNA metabolism; 7, energy metabolism; 8, fatty acid and phospholipid metabolism; 9, other categories; 10, protein fate; 11, protein synthesis; 12, purines, pyrimidines, nucleosides, and nucleotides; 13, regulatory functions; 14, signal transduction; 15, transcription; 16, transport and binding proteins; 17, unknown function; 18, hypothetical proteins.
The wide distribution of putative functional roles attributed to the differentially expressed genes indicates the extent of the molecular response that enables S. oneidensis cells to survive and eventually adapt to thermal stress. As shown in Fig. 2, a large number of the genes that were downregulated in response to heat shock had annotated functions in energy metabolism (bar 7), whereas most of the differentially expressed genes related to protein fate (bar 10) were induced upon the temperature upshift. For genes of known function, those encoding proteins involved in cellular processes (bar 4), energy metabolism (bar 7), protein fate (bar 10), regulatory functions (bar 13), and substrate transport (bar 16) were among the most upregulated genes in response to heat stress. Most notably, many of the genes whose expression was altered by the temperature increase encoded proteins of unknown function (bar 18), thus suggesting a more extensive heat shock stimulon than what can be deduced based solely on the sequence annotation. Along with genes involved in energy metabolism, functionally undefined genes (bar 18) were among the most downregulated genes in response to heat shock.
In contrast to steady-state or single-time-point studies, time course experiments are particularly valuable in providing insight into the mechanism regulating a bacterial response to stress and provide useful data for generating computational models of stress response pathways. Our temporal gene expression analysis indicated that the global changes in mRNA levels upon the temperature increase were largely transient. S. oneidensis cells responded with large changes in the expression level of selected genes (≥2-fold) during the first 15 min, and then the percentage of such genes decreased ∼20% at 25 min. The data from our preliminary experiments showed that <20% of these genes remained differentially expressed at 1 h (data not shown). These gene expression profiles suggest that S. oneidensis quickly readjusts its transcript levels to a new steady state at the heat shock temperature (42°C), thereby allowing the bacterium to survive the stress. This is in agreement with previous findings reported for both Escherichia coli and Campylobacter jejuni (29, 33).
Hierarchical clustering of temporal gene expression data.
To identify coregulated patterns of gene expression, we classified all differentially expressed genes in response to heat shock into seven hierarchical clusters based on their expression log ratio (Fig. 3). Cluster A contains the majority of downregulated genes. Significantly downregulated genes (P < 0.05 and ≥2-fold change in expression) primarily included those encoding hypothetical proteins (30%), proteins involved in metabolic pathways, translation or DNA replication (60%), and proteins involved in regulation (6%). The most notable subgroup of genes displaying transcriptional repression included 37 of 52 ribosomal structural genes (rpl, rpm, and rps operons). In several gene expression studies with Saccharomyces cerevisiae and C. jejuni, it has been observed that the expression level of ribosomal genes is similarly affected by heat stress, as well as alkylating agents (9, 19, 33). This observation may provide insight into the mechanism of ribosomes as sensors of heat shock (38), although several lines of evidence support the proposal that the free pool of DnaK and DnaJ may serve as a cellular thermometer, monitoring changes in cellular concentration of unfolded or denatured proteins (26, 37). Expression of ribosomal genes in S. oneidensis was repressed mostly at the early stage of heat shock and nearly returned to a basal level 25 min after the temperature upshift. Such a pattern suggests a brief initial growth arrest, during which the cell redirects and/or rechannels its energy usage to the increased expression of genes encoding proteins involved in the protective response to heat stress. An interesting finding was the observation that a number of genes in cluster A showing decreased expression encode proteins belonging to prophage families. S. oneidensis has three prophages, two of which are phylogenetically distinct phages related to the E. coli Mu (MuSo1 and MuSo2) and a lambda-like phage (LambdaSo). Most of genes (67% [12 induced/18 repressed]) encoding LambdaSo proteins were downregulated more than twofold, whereas ca. 33% (3 of 9) and 43% (6 of 14) of the genes for MuSo1 and MuSo2 proteins, respectively, displayed at least a twofold decrease in expression. This phenomenon has been observed under various stress conditions (unpublished results), suggesting that it may be just a natural decrease in expression of these superfluous genes under unfavorable growth conditions. Finally, another subset of functionally defined genes from cluster A encoded flagellar proteins and shared a similar expression pattern as that of the prophage genes. The nature of these prophage and flagellar proteins in the heat shock response is currently unknown.
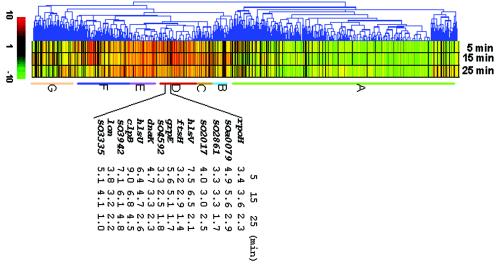
FIG. 3.
Hierarchical clustering of selected genes that varied significantly (P < 0.05 and a fold change of >2 at least at one time point) in their expression profiles in response to a temperature change from 30 to 42°C. The red color indicates the levels of induction, while the green color represents repression. Each row represents the expression of a single gene, and each column represents an individual time point after the temperature increase.
Cluster B was composed of genes whose expression fluctuated with time, whereas cluster C contained genes whose expression was constantly induced but to only a slight degree. Few genes in these clusters were significantly affected by heat stress.
Cluster D consisted of genes whose expression increased immediately after the temperature upshift and then decreased with time. Genes encoding hypothetical proteins dominate this cluster at the level of 50%. The major group of known proteins encoded by genes in this cluster have predicted functions related to protein fate and included mainly chaperones, chaperonins, and Hsps. Sequence annotation of the S. oneidensis MR-1 genome revealed at least 22 homologues of chaperones or chaperonins and Hsps, several of which have been well characterized in other bacteria and include DnaK, DnaJ, GroEL, GroES, GrpE, HtpG, and Lon/La proteases. Also present in the MR-1 genome are genes predicted to encode known regulators of the heat shock response, namely, σ32 and σE. In E. coli, the induction of the majority of Hsps results from a rapid and transient increase in the cellular level of the alternative 32-kDa sigma subunit (σ32), encoded by rpoH, which complexes with the core RNA polymerase (RNAP) and directs the RNAP holoenzymes to transcribe specifically from heat-regulated promoters (13, 25, 42, 46, 47), thus permitting both steady-state and stress-induced levels of Hsp expression (5, 48). The increase in the intracellular concentration of the σ32 transcription factor is due to a concomitant increase in both the stability and synthesis of σ32. In addition, alternative σ factors, σE (σ24) and σ54, encoded by rpoE and rpoN, respectively, are involved in the regulation of certain subsets of Hsps. σE is essential for transcription from one (rpoHp3) of the promoters of rpoH and the promoter of htrA encoding a periplasmic endopeptidase essential for growth at high temperature, whereas σ54 plays roles in regulation of α-Hsps (24). The embedded table in Fig. 3 shows the temporal expression levels for some of the S. oneidensis Hsps. Although the magnitude of induction for each of these genes varied, they displayed a maximal fold change at an early stage in the response, with a decrease in induction over time. This is consistent with the notion that the rapid cellular accumulation of Hsps upon a temperature upshift is followed by an adaptation period, during which the levels of Hsps are readjusted to the new steady-state growth conditions at the higher temperature (29). In our study, σ32, encoded by rpoH, had a relatively lower induction at the early stage and kept the same level for the first 15 min after the temperature upshift. This finding differs somewhat from the observation in E. coli that the increase in rpoH mRNA synthesis (<2-fold) after heat shock appears to contribute negligibly to the overall σ32 induction (7, 34). Other genes grouped in cluster D encode proteins involved in energy metabolism such as Ni/Fe hydrogenase (hyaB and hydC), formate dehydrogenase (fdhB and SO4509), and anaerobic reductase (dmaA-2, dmsB-1, nrdD, and nrdG). The hyaB and hydC genes encode a classic [NiFe] hydrogenase, an important component in anaerobic respiratory electron transport systems. It is unclear why these genes, whose products are involved in anaerobic respiration, would be induced in response to heat stress.
Cluster E comprises genes that showed a constant level of induction over the 25-min heat shock period. Genes encoding hypothetical proteins (SO0886, 1443, 2542, 2911, 3274, 3377, 3381, 3512, 3585, 3682, 3765, and 4593) account for 45% of this cluster. The majority of the remaining genes code for proteins involved in energy metabolism (aceA, aceB, cfa, edd, gapA-2, pgl, SO1471, 3683, and zwf). The mechanism underlying the involvement of isocitrate lyase and malate synthase A, encoded by aceA and aceB, respectively, in the heat shock response of S. oneidensis is currently unknown. These proteins work together to short circuit the tricarboxylic acid cycle, thereby rendering most of the tricarboxylic acid components unnecessary. One possible explanation is that S. oneidensis cells challenged with heat stress, unlike those growing under normal physiological conditions, diverted the usage of the protein synthesis machinery from rapid growth to producing proteins important or critical for survival and cell maintenance only. Genome sequence analysis suggests that the bacterium appears to bypass the missing phosphofructokinase step in glycolysis via the pentose phosphate pathway. Consistent with this supposition is the observation that the expression of zwf, pgl, and edd, whose products catalyze the key steps connecting the glycolytic pathway to the pentose phosphate pathway, were highly induced in response to heat shock (41). Interestingly, steps catalyzed by most of these metabolic enzymes produce NADPH or ATP, which provides both reducing power and energy for cells to overcome the stress.
Cluster F included genes that exhibited variable expression levels during the 25-min heat shock period. The cluster was dominated by genes encoding hypothetical proteins. The remaining genes included a subset of genes encoding proteins known or presumed to be involved in chemotaxis and ion transport. This subset of genes included cheA, cheB, cheR-1, cheW, cheY, motA, and motB genes (encoding chemotaxis proteins) and ktrA, ktrB, nhaA, nosF, nosy, pstB-2, pstC, SO0534, 2045, 2865, 3333, 3690, 3768, 3801, 3802, and 4598 genes (encoding ion transport proteins). This observation is in agreement with the findings of Richmond et al. (29), who demonstrated that the expression of chemotaxis and ion transport genes in E. coli was induced in response to heat shock. Cluster F also included genes encoding transcriptional regulators belonging to MerR, LysR, and TetR families. Expression of these genes was increased slightly early in the heat shock response and exhibited higher expression levels at 25 min. S. oneidensis has a relatively small repertoire of regulatory genes compared to Vibrio cholerae, phylogenetically closely related to S. oneidensis. This repertoire includes 57 response regulators and 88 two-component regulatory system proteins, which could allow rapid detection and response to environmental changes (14, 15). Our microarray data revealed that 46 of these regulatory genes were upregulated, and an additional 17 members grouped in other clusters showed a similar expression pattern. It is not clear how these regulators are involved in the heat shock response, and further investigation is needed.
Cluster G comprised genes that showed fluctuating expression after the temperature upshift but neither up- nor downregulation was significant.
Hypothetical proteins.
More than 41% of the S. oneidensis ORFs encode hypothetical proteins. In our study, genes for hypothetical proteins make up to 38% of twofold above changes in both up- and downregulated gene expression. As for possible operons containing multiple genes encoding hypothetical proteins, we observed co-upregulation of all members from four such operons: SO1442/SO1443, SO2861/SO2862/SO2863, SO3764/SO3765/SO3766, and SO4260/SO4261 (see Table S1 in the supplemental material). The consistency of expression of these genes under the heat shock condition provides basic evidence to support such operon structure. Unfortunately, little is known about these proteins, although the protein encoded by SO3765 may be a member of PspA that suppresses sigma54-dependent transcription (www.tigr.org). Two genes (SO1094 and SO3386) displayed the highest induction through the whole period of heat shock. The SO1094 protein may be a GreA-like transcription elongation factor enabling continuation of RNA transcription past template-encoded arresting sites, whereas the SO3386 protein belongs to a family of uncharacterized bacterial proteins. Interestingly, SO1264 and SO1274, undoubtedly from different operons, encode possible members of a beta subunit family of sarcosine oxidase, suggesting that sarcosine oxidase may have a role in heat shock response.
Computational prediction of σ32-binding consensus motif in S. oneidensis.
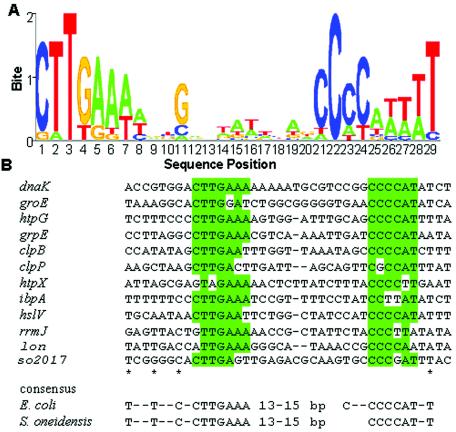
To predict genes regulated by the σ32 transcription factor, we used AlignACE (30) to search for potential regulatory motifs upstream of heat-inducible genes, as identified by microarray analysis, and compared the derived S. oneidensis consensus sequence to that of the E. coli σ32-binding motif. In the upstream regions of these genes, there exists a sequence that is nearly identical to the consensus sequence derived for E. coli heat shock promoters. Based on a comparison of these promoters from a couple of bacteria, including E. coli and V. cholerae, we propose an S. oneidensis consensus sequence for σ32-controlled promoters having T-n-n-T-n-n-C-n-C-T-T-G-A-A-A in the −35 region and C-C-C-C-A-T-n-T-a in the −10 region with 13 to 15 bp separating the two elements (Fig. 4). Together with the induction of annotated heat shock genes at the elevated temperature, these observations suggest that the heat shock response in S. oneidensis is mediated by a mechanism similar to that occurring in E. coli.
FIG. 4.
Consensus sequence for σ32 promoters. Upstream sequences of heat-induced genes in both S. oneidensis and E. coli were analyzed with AlignACE to find potential regulatory motifs. (A) A motif conserved in the upstream regions of upregulated S. oneidensis genes is nearly identical to the E. coli σ32 binding site. The sequence logo was prepared by using public software at http://ep.ebi.ac.uk/EP/SEQLOGO (31). (B) Listed are promoters that display at least 7 of 12 matches in highlighted base pairs and 13- to 15-bp spacing. Asterisks indicate the positions with at least 50% matches.
Sequence BLAST analysis revealed an extremely high similarity (85% positives) between rrmJ of S. oneidensis, encoding the rRNA large subunit methyltransferase J, and ftsJ of E. coli, indicating that the rrmJ operon, as in E. coli (4), is a heat shock operon. A hypothetical protein, encoded by SO2017, is very likely under the control of σ32. The protein contains a domain closely related to thioredoxin, suggesting that it may participate in redox reactions. Although protein alignment analysis confirmed it as a thioredoxin domain-containing protein that may be involved in posttranslational modification and protein turnover, little information has been obtained by using Prospect protein structure prediction software (1, 45).
In addition to σ32, σ24, encoded by rpoE, has been reported to be involved in the regulation of the heat shock response in E. coli. The promoter sequence recognized by the S. oneidensis σ24 is not known because of insufficient knowledge. Our data showed that changes in expression of rpoE were in the range of natural experimental variation, suggesting that σ24 may only play a minor role in the heat shock response of S. oneidensis or its gene may be induced at a higher temperature as in E. coli.
Conclusion.
The primary objective of the present study was to characterize the transcriptome of S. oneidensis in response to heat stress by using whole-genome DNA microarrays to monitor temporal gene expression. More than 10% of transcriptionally active genes displayed at least a twofold induction upon the temperature upshift. The expression patterns of eight of these genes were independently confirmed by using real-time RT-PCR. In general, the heat shock response in S. oneidensis is similar to that in E. coli. Moreover, the identified consensus sequences of both bacteria for heat shock gene promoters are virtually the same. However, it is noteworthy that the hypothetical protein SO2017, likely under the control of σ32, shows no similarity to any protein known thus far. Our analysis illustrated the value and utility of microarray expression profiling in defining bacterial stimulons and regulons. Our future work will focus on inactivating sigma factors and other transcriptional regulators that might control critical stress responses.
Supplementary Material
Acknowledgments
This research was supported by The U.S. Department of Energy under the Genomics:GTL and Microbial Genome Programs of the Office of Biological and Environmental Research, Office of Science. Oak Ridge National Laboratory is managed by University of Tennessee-Battelle LLC for the Department of Energy under contract DE-AC05-00OR22725.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Presented in part at the 11th International Conference on Microbial Genomes, Durham, N.C., 28 September to 2 October 2003.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buchner, J. 1999. Hsp90 & Co.—a holding for folding. Trends Biochem. Sci. 24:136-141. [DOI] [PubMed] [Google Scholar]
- 3.Bukau, B., E. Deuerling, C. Pfund, and E. A. Craig. 2000. Getting newly synthesized proteins into shape. Cell 101:119-122. [DOI] [PubMed] [Google Scholar]
- 4.Caldas, T., E. Binet, P. Bouloc, A. Costa, J. Desgres, and G. Richarme. 2000. The FtsJ/RrmJ heat shock protein of Escherichia coli is a 23S ribosomal RNA methyltransferase. J. Biol. Chem. 275:16414-16419. [DOI] [PubMed] [Google Scholar]
- 5.Cowing, D. W., J. C. Bardwell, E. A. Craig, C. Woolford, R. W. Hendrix, and C. A. Gross. 1985. Consensus sequence for Escherichia coli heat-shock promoters. Proc. Natl. Sci. Sci. USA 82:2679-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eisen, M. B., and P. O. Brown. 1999. DNA arrays for analysis of gene expression. Methods Enzymol. 303:179-205. [DOI] [PubMed] [Google Scholar]
- 7.Erickson, J., V. Vaughn, W. A. Walter, F. C. Neidhardt, and C. A. Gross. 1987. Regulation of the promoters and transcripts of rpoH, the Escherichia coli heat shock regulatory gene. Genes Dev. 1:419-432. [DOI] [PubMed] [Google Scholar]
- 8.Fisher, M. A., B. B. Plikaytis, and T. M. Shinnick. 2002. Microarray analysis of the Mycobacterium tuberculosis transcriptional response to the acidic conditions found in phagosomes. J. Bacteriol. 184:4025-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goloubinoff, P., A. Mogk, A. Peres Ben Zvi, T. Tomoyasu, and B. Bukau. 1999. Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc. Natl. Acad. Sci. USA 96:13732-13737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gottesman, S. 1996. Proteases and their targets in Escherichia coli. Annu. Rev. Genet. 30:465-506. [DOI] [PubMed] [Google Scholar]
- 12.Grimaud, R., M. Kessel, F. Beuron, A. C. Steven, and M. R. Maurizi. 1998. Enzymatic and structural similarities between the Escherichia coli ATP-dependent proteases, ClpXP and ClpAP. J. Biol. Chem. 273:12476-12481. [DOI] [PubMed] [Google Scholar]
- 13.Gross, C. A. 1996. Function and regulation of the heat shock proteins, p. 1382-1399. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 14.Heidelberg, J., I. Paulsen, K. Nealson, E. Gaidos, W. Nelson, T. Read, et al. 2002. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat. Biotechnol. 20:1118-1123. [DOI] [PubMed] [Google Scholar]
- 15.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, et al. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helmann, J. D., M. F. Wu, P. A. Kobel, F. J. Gamo, M. Wilson, M. M. Morshedi, M. Navre, and C. Paddon. 2001. Global transcriptional response of Bacillus subtilis to heat shock. J. Bacteriol. 183:7318-7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hihara, Y., A. Kamei, M. Kanehisa, A. Kaplan, and M. Ikeuchi. 2001. DNA micro-array analysis of cyanobacterial gene expression during acclimation to high light. Plant Cell 13:793-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hughes, T. R., M. J. Marton, A. R. Jones, C. J. Roberts, R. Stoughton, C. D. Armour, H. A. Bennett, E. Coffey, H. Dai, Y. D. He, M. J. Kidd, A. M. King, M. R. Meyer, D. Slade, P. Y. Lum, S. B. Stepaniants, D. D. Shoemaker, D. Gachotte, K. Chakraburtty, J. Simon, M. Bard, and S. H. Friend. 2000. Functional discovery via a compendium of expression profiles. Cell 102:109-126. [DOI] [PubMed] [Google Scholar]
- 19.Jelinsky, S. A., and L. D. Samson. 1999. Global response of Saccharomyces cerevisiae to an alkylating agent. Proc. Natl. Acad. Sci. USA 96:1486-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karp, P. D., M. Riley, M. Saier, I. T. Paulsen, J. Collado-Vides, S. M. Paley, A. Pellegrini-Toole, C. Bonavides, and S. Gama-Castro. 2002. The EcoCyc database. Nucleic Acids Res. 30:56-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khamis, H. J. 2000. The two-stage delta-corrected Kolmogorov-Smirnov test. J. Appl. Stat. 27:439-450. [Google Scholar]
- 22.Khodursky, A., B. Peter, N. Cozzerelli, D. Botstein, P. Brown, and C. Yanofsky. 2000. DNA microarray analysis of gene expression in response to physiological and genetic changes that affect tryptophan metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:12170-12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, K. I., G. W. Cheong, S. C. Park, J. S. Ha, K. M. Woo, S. J. Choi, and C. H. Chung. 2000. Heptameric ring structure of the heat-shock protein ClpB, a protein-activated ATPase in Escherichia coli. J. Mol. Biol. 303:655-666. [DOI] [PubMed] [Google Scholar]
- 24.Kuczynska-Wisnik, D., E. Laskowska, and A. Taylor. 2001. Transcription of ibpB heat shock gene is under control of σ32- and σ54-promoters, a third regulon of heat-shock response. Biochem. Biophys. Res. Commun. 284:57-64. [DOI] [PubMed] [Google Scholar]
- 25.Lindquist, S., and E. A. Craig. 1988. The heat-shock proteins. Annu. Rev. Genet. 22:631-677. [DOI] [PubMed] [Google Scholar]
- 26.Macarty, J. S., and G. C. Walker. 1991. DnaK as a thermometer: threonine 199 is site of aurophosphorylation and is critical for ATPase activity. Proc. Natl. Acad. Sci. USA 88:9513-9517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mager, W. H., and A. J. J. De Kruijff. 1995. Stress-induced transcriptional activation. Microbiol. Rev. 59:506-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Narberhaus, F. 2002. α-Crystallin-type heat shock proteins: socializing minichaperones in the context of a multichaperone network. Microbiol. Mol. Biol. Rev. 66:64-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richmond, C. S., J. D. Glasner, R. Mau, H. Jin, and F. R. Blattner. 1999. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 27:3821-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roth, F. P., J. D. Hughes, P. W. Estep, and G. M. Church. 1998. Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nat. Biotechnol. 16:939-945. [DOI] [PubMed] [Google Scholar]
- 31.Schneider, T. D., and R. M. Stephens. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18:6097-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smoot, L. M., J. C. Smoot, M. R. Graham, G. A. Somerville, D. E. Sturdevant, C. A. Migliaccio, G. L. Sylva, and J. M. Musser. 2001. Global differential gene expression in response to growth temperature alteration in group A Streptococcus. Proc. Natl. Acad. Sci. USA 98:10416-10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stintzi, A. 2003. Gene expression profile of Campylobacter jejuni in response to growth temperature variation. J. Bacteriol. 185:2009-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Straus, D. B., W. A. Walter, and C. A. Gross. 1987. The heat shock response of Escherichia coli is regulated by changes in the concentration of sigma32. Nature 329:348-351. [DOI] [PubMed] [Google Scholar]
- 35.Tao, H., C. Bausch, C. Richmond, F. Blattner, and T. Conway. 1999. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J. Bacteriol. 181:6425-6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson, D. K., A. Beliaev, C. S. Giometti, D. P. Lies, K. H. Nealson, H. Lim, J. Yates III, J. Tiedje, and J. Zhou. 2002. Transcription and proteomic analysis of a ferric uptake regulator (Fur) mutant of Shewanella oneidensis: possible involvement of Fur in energy metabolism, regulation, and oxidative stress. Appl. Environ. Microbiol. 68:881-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tomoyasu, T., T. Ogura, T. Tatsuta, and B. Bukau. 1998. Levels of DnaK and DnaJ provide tight control of heat shock gene expression and protein repair in Escherichia coli. Mol. Microbiol. 30:567-581. [DOI] [PubMed] [Google Scholar]
- 38.VanBogelen, R. A., and F. C. Neidhardt. 1990. Ribosomes as sensors of heat and cold shock in Escherichia coli. Proc. Natl. Acad. Sci. USA 87:5589-5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Venkateswaran, K., D. P. Moser, M. E. Dollhopf, D. P. Lies, D. A. Saffarini, B. J. MacGregor, D. B. Ringelberg, D. C. White, M. Nishijima, H. Sano, J. Burghardt, E. Stackebrandt, and K. H. Nealson. 1999. Polyphasic taxonomy of the genus Shewanella and description of Shewanella oneidensis sp. nov. Int. J. Syst. Bacteriol. 49:705-724. [DOI] [PubMed] [Google Scholar]
- 40.Weber-Ban, E. U., B. G. Reid, A. D. Miranker, and A. L. Horwich. 1999. Global unfolding of a substrate protein by the Hsp100 chaperone ClpA. Nature 401:90-93. [DOI] [PubMed] [Google Scholar]
- 41.Wei, B., S. Shin, D. LaPorte, A. J. Wolfe, and T. Romeo. 2000. Global regulatory mutations in csrA and rpoS cause severe central carbon stress in Escherichia coli in the presence of acetate. J. Bacteriol. 182:1632-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wösten, M. M. S. M. 1998. Eubacterial sigma-factors. FEMS Microbiol. Rev. 22:127-150. [DOI] [PubMed] [Google Scholar]
- 43.Wurmbach, E., T. Yuen, B. J. Ebersole, and S. C. Sealfon. 2001. Gonadotropin-releasing hormone receptor-coupled gene network organization. J. Biol. Chem. 276:47195-47201. [DOI] [PubMed] [Google Scholar]
- 44.Xu, D., G. Li, L. Wu, J. Zhou, and Y. Xu. 2002. PRIMEGENS: robust and efficient design of gene-specific probes for microarray analysis. Bioinformatics 18:1432-1437. [DOI] [PubMed] [Google Scholar]
- 45.Xu, Y., and Xu, D. 2000. Protein threading using PROSPECT: design and evaluation. Proteins 40:343-354. [PubMed] [Google Scholar]
- 46.Yura, T., M. Kanemori, and M. T. Morita. 2000. The heat shock response: regulation and function, p. 3-18. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 47.Yura, T., and K. Nakahigashi. 1999. Regulation of the heat-shock response. Curr. Opin. Microbiol. 2:159-165. [DOI] [PubMed] [Google Scholar]
- 48.Zhou, Y., N. Kusakawa, J. W. Erickson, C. A. Gross, and T. Yura. 1988. Isolation and characterization of Escherichia coli mutants that lack the heat shock sigma factor σ32. J. Bacteriol. 170:3640-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.