Abstract
Bacillus cereus strains ATCC 10987 and ATCC 14579 harbor a ∼155-bp repeated element, bcr1, which is conserved in B. cereus, B. anthracis, B. thuringiensis, and B. mycoides but not in B. subtilis and B. licheniformis. In this study, we show by Southern blot hybridizations that bcr1 is present in all 54 B. cereus group strains tested but absent in 11 Bacillus strains outside the group, suggesting that bcr1 may be specific and ubiquitous to the B. cereus group. By comparative analysis of the complete genome sequences of B. cereus ATCC 10987, B. cereus ATCC 14579, and B. anthracis Ames, we show that bcr1 is exclusively present in the chromosome but absent from large plasmids carried by these strains and that the numbers of full-length bcr1 repeats for these strains are 79, 54, and 12, respectively. Numerous copies of partial bcr1 elements are also present in the three genomes (91, 128, and 53, respectively). Furthermore, the genomic localization of bcr1 is not conserved between strains with respect to chromosomal position or organization of gene neighbors, as only six full-length bcr1 loci are common to at least two of the three strains. However, the intergenic sequence surrounding a specific bcr1 repeat in one of the three strains is generally strongly conserved in the other two, even in loci where bcr1 is found exclusively in one strain. This finding indicates that bcr1 either has evolved by differential deletion from a very high number of repeats in a common ancestor to the B. cereus group or is moving around the chromosome. The identification of bcr1 repeats interrupting genes in B. cereus ATCC 10987 and ATCC 14579 and the presence of a flanking TTTAT motif in each end show that bcr1 exhibits features characteristic of a mobile element.
Bacillus cereus and Bacillus anthracis are both members of the B. cereus group of bacteria but are widely different with respect to pathogenicity (reviewed by Jensen et al. [17]); whereas B. cereus is an opportunistic pathogen frequently linked to food-borne disease in humans, B. anthracis is the etiological agent of anthrax, a highly infectious and fatal human and animal disease, and was used in biological attacks through the U.S. Postal Service during the fall of 2001 (15).
Bacterial genomes frequently contain interspersed, non-protein-encoding repetitive sequences of diverse length, type, and copy number. Repetitive DNA may often account for a substantial portion of the genomes, sometimes even higher than 10% (14, 37). For many of these repeats, no function is known, although the number of repeats with assigned roles is increasing. Such functions include promoter activity, regulation of mRNA stability, transcription termination, maintenance of chromosome structure and function, DNA uptake or recombination signals, methylation sites, hotspots for insertion of insertion sequence elements, phage integration signals, constituents of integrons, and substrates for intrachromosomal recombination events contributing to chromosome rearrangements and genome plasticity (reviewed in references 14 and 37). Interspersed sequence repeats have previously been identified in bacilli; the determination of the complete genome sequence of B. subtilis 168 revealed a 190-bp sequence repeated 10 times in the chromosome, with 5 copies located on each side of oriC and which in all cases but one were cooriented with the direction of replication (21). The repeat, named Bs-rep, was suggested to form a structural RNA molecule, and a highly conserved repeat was identified in the close relative B. licheniformis (21, 27, 28).
During analyses of piecemeal sequences from B. cereus ATCC 10987 and ATCC 14579 (type strain) genomes (25), we identified a 155-bp chromosomal intergenic repeat, bcr1, which was unrelated in sequence to the 190-bp repeat from B. subtilis. The bcr1 element was shown by Southern blotting to be present in all 17 B. thuringiensis and B. cereus strains tested but was not found in B. subtilis 168 (25). Sequence comparison of two corresponding genetic loci in the two B. cereus strains revealed that bcr1 could be present at a given locus in one strain but absent at the corresponding locus in the other, indicating a heterogenous chromosomal distribution. In this paper, we extend the results from our previous study and show that the bcr1 element is ubiquitous in and specific to the members of the B. cereus group, including B. cereus, B. thuringiensis, B. anthracis, B. mycoides, and B. weihenstephanensis. Furthermore, we perform an extensive three-way comparative analysis of bcr1 elements in the complete genome sequences of B. cereus ATCC 10987 (29), B. cereus ATCC 14579 (16), and B. anthracis Ames (31), examining the distribution of full-length and partial repeats, organization of their neighboring genes, and conservation of the flanking regions. Taken together, the results show that bcr1 exhibits several features characteristic of a mobile element.
MATERIALS AND METHODS
PCR amplification and cloning of bcr1.
Oligonucleotide primers for amplification of bcr1 elements were designed from the bcr1 element bcr1_trp1 (Bc14579_19R [Fig. 4]) located in the upstream region of the transcriptional regulator trp1 in B. cereus ATCC 14579 (26) (left primer, bcr1forBam [5′-CCC GGA TCC GGC AGT AAG ACC TCC ACC TC-3′]; right primer, bcr1revBam [5′-GCG GGA TCC ATA AAG TGA AAC TTT AAT CAG TGG G-3′]). Both primers contained BamHI restriction sites at the ends (underlined) to facilitate cloning of PCR products. Following an initial denaturation step at 95°C for 6 min, PCR was run for 35 cycles, each consisting of a 1-min denaturation step at 95°C, a 1-min annealing step at 58°C, and a 10-s polymerization step at 72°C, using a solution containing 100 ng of template DNA (11 kb of chromosomal DNA fragment bc301 carrying bcr1_trp1, cloned in the pUC19 vector) (26), 0.4 μM each primer, 2.5 mM MgCl2, 0.2 mM each deoxynucleoside triphosphate, 1× Dynazyme reaction buffer, and 1 U of Dynazyme (Finnzymes Oy, Espoo, Finland) in a total volume of 50 μl. After PCR cycling, a polymerization step was performed (7 min at 72°C) to complete end polymerization of all DNA fragments.
FIG. 4.
Multiple alignment of 145 full-length bcr1 elements (≥120 bp) from B. anthracis Ames, B. cereus ATCC 10987, and B. cereus ATCC 14579. Sequences were aligned by using CLUSTALW (35) followed by manual editing with SEAVIEW (11). Consensus sequences computed with thresholds of 51, 60, 70, 80, and 90% conservation are included at the bottom of the alignment. When a threshold T is used, a residue appears in the consensus if it is present in ≥T% of the 145 bcr1 elements; otherwise, the character N is assigned. Furthermore, a plot of the conservation of each site was generated by CLUSTALX (36) and is shown below the alignment. The asterisk above the alignment indicates the single site where all 145 sequences have an identical nucleotide. Sequence lengths are given to the left of the alignment. Indels are represented by dashes. Bc denotes B. cereus.
bcr1 PCR products were purified with the QIAquick PCR purification kit (QIAGEN GmbH, Hilden, Germany) and cloned into a pUC19 vector by using electrocompetent Escherichia coli XL1-Blue MRF′ cells (Stratagene, Cedar Falls, Tex.). Transformants were checked for the presence of the bcr1 insert by restriction digests and DNA sequencing.
Southern blotting and hybridization.
The cloned bcr1 repeat (141-bp PCR-amplified fragment) from the trp1 locus in B. cereus ATCC 14579 (bcr1_trp1) (26) was used as probe in Southern hybridizations after agarose gel purification of the cloned fragment with a QIAquick gel purification kit. The probe was randomly labeled with [α32P]dATP and [α32P]dCTP (Amersham Biosciences, Little Chalfont, United Kingdom) by using the Klenow fragment of E. coli DNA polymerase I (New England Biolabs, Beverly, Mass.) as previously described in the work of Økstad et al. (25). Genomic DNA (approximately 1 to 10 μg) from 65 Bacillus strains was digested to completion with HincII and run on a 0.8% agarose gel. After electrophoresis, DNA was transferred to nylon filters (MagnaCharge; Micron Separations Inc., Westboro, Mass.) by capillary blotting overnight, essentially as described previously by Kolstø et al. (20). DNA hybridization was performed at 68°C overnight with a rotating oven as previously described (20).
Sequence analysis. (i) Iterative BLAST searches.
bcr1 repeats in the complete chromosome sequences of B. anthracis Ames (31) (GenBank accession number NC_003997), B. cereus ATCC 14579 (16) (accession number NC_004722), and B. cereus ATCC 10987 (29) (accession number NC_003909) were identified by iterative runs of gapped BLASTN searches (1) using lowered gap penalties (opening cost, G = 2; extension cost, E = 1), no filtering of low-complexity regions, and a maximum E value of 1.0. Default values were employed for all other BLASTN parameters. A 141-bp sequence from the bcr1 element bcr1_trp1 (26) was originally used as the seed for the process. However, a comparison of the flanking regions of the sequences obtained after the first BLASTN run revealed that the bcr1 repeat could be redefined as a ∼160-bp sequence flanked by TTTAT motifs. The extended bcr1_trp1 sequence (Bc14579_19R; Fig. 4) was then used to restart the iterative searches. After each BLASTN round, full-length hits, defined as matches of 120 bp or more, from all three chromosome sequences were used as seeds in a new round of searches against the three chromosomes. The process was repeated until no further full-length sequences could be found. The whole procedure stopped after three iterations, giving a final data set of 145 bcr1 elements of 120 bp in length or longer. We defined all remaining hits of at least 30 bp as partial bcr1 repeats. Sequences of the following plasmids harbored by the three bacteria were also searched: B. anthracis pXO1 and pXO2 (GenBank accession numbers NC_003980 and NC_003981) (24, 30), pBc10987 from B. cereus ATCC 10987 (accession number NC_005707) (29), and pBClin15 from B. cereus ATCC 14579 (accession number NC_004721) (16).
(ii) Phylogenetic analysis of bcr1 sequences.
Full-length and partial bcr1 elements were aligned with CLUSTALW (35), followed by manual editing using SEAVIEW (11), and submitted to the EMBL-ALIGN database (accession numbers ALIGN_000716 and ALIGN_000717 for alignments of full-length and full-length plus partial sequences, respectively). Based on the multiple alignment, a phylogenetic tree of full-length bcr1 repeats was built by using the neighbor-joining method (32) applied to a matrix of pairwise distances between bcr1 sequences. Distances were computed according to Kimura's (19) two-parameter substitution model, which takes into account multiple substitutions at a given site and the bias between transition and transversion rates. Sites with gaps (representing insertions and deletions) were excluded from distance computations; to avoid exclusion of a prohibitively large number of sites, gaps were removed specifically for each pair of sequences compared instead of globally for all sequences.
(iii) Comparative analysis of flanking regions and neighboring genes.
A comparative analysis of the bcr1 loci in the genomes of B. anthracis Ames, B. cereus ATCC 10987, and B. cereus ATCC 14579 was performed by examining three neighboring genes upstream and downstream of all bcr1 elements. For a given bcr1 locus in a given genome, the organization of the neighboring genes was compared to that of their orthologues in the other two genomes. Open reading frames (ORFs) were considered as putative orthologues in two bacterial strains when they satisfied a reciprocal best-hit relationship based on TBLASTX (1) searches of the complete gene sets of the two organisms. Hits were judged as significant if the E value was lower than 10−5 and the match covered at least 70% of the length of both ORFs. Furthermore, for all bcr1 loci in a genome, the nucleotide sequence covering the bcr1 element, the flanking 5′ and 3′ intergenic regions and the first upstream and downstream gene, was aligned with the homologous sequences (if any) in the other two genomes by means of CLUSTALW. Multiple alignments were then manually corrected with the aid of dot plots using SEAVIEW.
RESULTS
Mapping of bcr1 in Bacillus species.
The bcr1 element bcr1_trp1, located upstream of the transcriptional regulator trp1 in B. cereus ATCC 14579 (26) (see Materials and Methods), was amplified by PCR, cloned, and used as a probe for hybridization in Southern blots to test for the presence of bcr1 repeats in various strains of bacilli (Fig. 1). Altogether, 65 strains covering all five 16S rRNA groups in the Bacillus genus (2), 54 of which were from the B. cereus group, were examined. The B. cereus group isolates were widely distributed in a phylogenetic tree constructed by multilocus enzyme electrophoresis analysis (13; E. Helgason, personal communication) and were all found to harbor the bcr1 repeat, while no repeats were identified in the 11 Bacillus strains outside the group (Table 1). This finding suggests that bcr1 may be ubiquitous and specific to members of the B. cereus group of bacteria and could be employed as a molecular marker sequence to rapidly identify bacterial isolates belonging to the group.
FIG. 1.
Southern blot of genomic DNA from a sample of B. cereus group strains hybridized with the bcr1 probe (see Table 1 for strain numbers). Species designations are as follows: Bc, B. cereus; Bt, B. thuringiensis; Bw, B. weihenstephanensis; Bct, B. cereus/B. thuringiensis (not investigated for crystal toxin formation).
TABLE 1.
Hybridization of bcr1 probe to genomic DNA from various Bacillus spp. belonging to all five rRNA groups
| Strain | Origin | Hybridizationc | rRNA group |
|---|---|---|---|
| AH 75 | B. cereus ATCC 10987 | + | 1 |
| AH 181 | B. cereus ATCC 10876 | + | 1 |
| AH 182 | B. cereus ATCC 11778 | + | 1 |
| AH 188 | B. cereus F0837/76 | + | 1 |
| AH 226 | B. cereus ATCC 4342 | + | 1 |
| AH 233 | B. cereus ATCC 33018 | + | 1 |
| AH 234 | B. cereus ATCC 33019 | + | 1 |
| AH 248 | B. thuringiensis subsp. kurstaki 4D1 | + | 1 |
| AH 249 | B. thuringiensis subsp. kurstaki 4D4 | + | 1 |
| AH 250 | B. thuringiensis subsp. sotto/dendrolimus 4E2 | + | 1 |
| AH 252 | B. thuringiensis subsp. canadensis 4H2 | + | 1 |
| AH 253 | B. thuringiensis subsp. entomocidus/subtoxicus 4I4 | + | 1 |
| AH 255 | B. thuringiensis subsp. morrisoni 4K1 | + | 1 |
| AH 256 | B. thuringiensis subsp. darmstadiensis 4M1 | + | 1 |
| AH 257 | B. thuringiensis subsp. israelensis 4Q1 | + | 1 |
| AH 259 | B. cereus 6A1 | + | 1 |
| AH 260 | B. cereus 6A3 | + | 1 |
| AH 263 | B. cereus 6E1 | + | 1 |
| AH 264 | B. cereus 6E2 | + | 1 |
| AH 265 | B. thuringiensis 4A3 | + | 1 |
| AH 338 | B. mycoides | + | 1 |
| AH 401 | B. cereus 181 (Sweden) | + | 1 |
| AH 408 | B. cereus 3102 (Finland) | + | 1 |
| AH 418 | B. thuringiensis subsp. berliner 1715 | + | 1 |
| AH 424 | B. thuringiensis 4Q7 | + | 1 |
| AH 442 | B. thuringiensis HD-73 | + | 1 |
| AH 535 | B. cereus/B. thuringiensis 2-21 (soil, moss, Norway) | + | 1 |
| AH 607 | B. cereus vet 59 (cow milk)a | + | 1 |
| AH 608 | B. cereus vet 61 (cow milk)a | + | 1 |
| AH 612 | B. cereus vet 131 (cow milk)a | + | 1 |
| AH 613 | B. cereus vet 132 (cow milk)a | + | 1 |
| AH 681 | B. cereus/B. thuringiensis K64 (soil, Tromsø, Norway) | + | 1 |
| AH 696 | B. thuringiensis subsp. alesti | + | 1 |
| AH 697 | B. thuringiensis subsp. gelechiae | + | 1 |
| AH 724 | B. cereus/B. thuringiensis 41859/86 (urine, patient, Skien, Norway) | + | 1 |
| AH 727 | B. cereus/B. thuringiensis 27999/87 (burn, patient, Skien, Norway) | + | 1 |
| AH 775 | B. thuringiensis subsp. israelensis 4Q2 | + | 1 |
| AH 812 | B. cereus 599-1-95 (periodontitis patient)b | + | 1 |
| AH 819 | B. cereus 600-3-95 (periodontitis patient)b | + | 1 |
| AH 831 | B. cereus 202-1-96 (periodontitis patient)b | + | 1 |
| AH 840 | B. thuringiensis subsp. thuringiensis 4A3 | + | 1 |
| AH 874 | B. cereus SIC (United States) | + | 1 |
| AH 923 | B. cereus ATCC 14579 | + | 1 |
| AH 1031 | B. thuringiensis 407 (Paris, France) | + | 1 |
| AH 1123 | B. cereus 9823 (clinical isolate, France) | + | 1 |
| AH 1143 | B. weihenstephanensis WSBC 10201 | + | 1 |
| AH 1144 | B. weihenstephanensis WSBC 10202 | + | 1 |
| AH 1146 | B. weihenstephanensis WSBC 10205 | + | 1 |
| AH 1270 | B. cereus 0001 + 31175 (cervix, patient, Landsspitali, Reykjavik, Iceland) | + | 1 |
| AH 1271 | B. cereus 9903 + 02049 (secretary lamp, Landsspitali, Reykjavik, Iceland) | + | 1 |
| AH 1272 | B. cereus 9901 + 17036 (amniotic fluid, patient, Landsspitali, Reykjavik, Iceland) | + | 1 |
| AH 1273 | B. cereus 9708 + 03060 (blood, patient, Landsspitali, Reykjavik, Iceland) | + | 1 |
| AH 1293 | B. cereus group sp. 493/02 (blood, patient, Norwegian Institute of Public Health, Oslo, Norway) | + | 1 |
| AH 1294 | B. cereus group sp. 440/02 (blood, patient, Norwegian Institute of Public Health, Oslo, Norway) | + | 1 |
| AH 212 | B. licheniformis | − | 1 |
| AH 399 | B. firmus | − | 1 |
| AH 699 | B. subtilis 168 | − | 1 |
| AH 1275 | B. circulans DSMZ 11 | − | 1 |
| AH 1277 | B. pumilus DSMZ 27 | − | 1 |
| AH 1278 | B. megaterium DSMZ 32 | − | 1 |
| AH 1274 | B. sphaericus DSMZ 28 | − | 2 |
| AH 1281 | B. polymyxa DSMZ 36 | − | 3 |
| AH 1283 | B. alvei DSMZ 29 | − | 3 |
| AH 1280 | B. laterosporus DSMZ 25 | − | 4 |
| AH 1276 | B. alcalophilus DSMZ 485 | − | 5 |
Strains isolated at the Norwegian College of Veterinary Medicine, Oslo, Norway.
Strains isolated at Institute of Oral Biology, Faculty of Dentistry, University of Oslo, Oslo, Norway.
+, bcr1 repeats were identified; −, no bcr1 repeats were identified.
Pattern of distribution of bcr1 repeats in B. cereus ATCC 10987, B. cereus ATCC 14579, and B. anthracis Ames.
By employing iterative BLASTN searches of the complete genome sequences of B. cereus ATCC 10987 (5.2 Mb) (29), B. cereus ATCC 14579 (5.4 Mb) (16), and B. anthracis Ames (5.2 Mb) (31) (see Materials and Methods), 145 full-length bcr1 repeats were identified in the chromosomes of the three strains, 79 in B. cereus ATCC 10987, 54 in B. cereus ATCC 14579, and 12 in B. anthracis Ames (see http://www.salmongenome.no/htdocs/Suppl_info_for_web_Okstadetal2004.html). As was shown previously (25, 26), bcr1 could be found as a full-length repeat of ∼155 bp or as shorter versions containing parts of the full-length sequence. Here, our sequence searches detected 272 partial bcr1 elements (of a minimal defined length of 30 bp) in the three chromosomes altogether, and the ratio of full-length to partial copies was highly divergent among the strains (0.87, 0.42, and 0.23, for B. cereus ATCC 10987, B. cereus ATCC 14579, and B. anthracis Ames, respectively). In contrast, no bcr1 repeats, full length or partial, were identified in any of the plasmids in B. cereus ATCC 10987 (pBc10987 [208 kb]), B. cereus ATCC 14579 (pBClin15 [15-kb linear plasmid]), or B. anthracis (pXO1 [182 kb] and pXO2 [96 kb]), indicating that bcr1 could be a feature limited to the chromosomes of these bacteria.
bcr1 chromosomal orientation and localization.
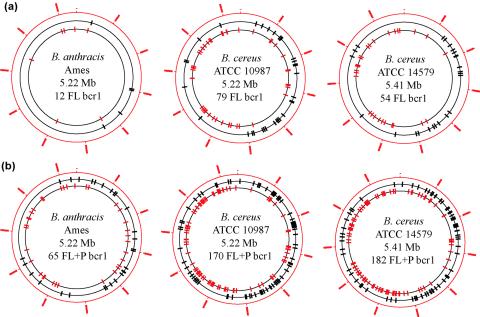
B. cereus, B. anthracis, and other gram-positive bacteria are known to exhibit a strong gene transcription orientation bias (∼75%) with respect to movement of the replication fork (16, 21, 31). Interestingly, in the three B. cereus group strains examined in this study, we observed an analogous coorientation bias of full-length bcr1 repeats with the direction of replication in the two halves of the chromosome bordered by the oriC and ter regions (77.2, 81.5, and 83.3%, respectively) (Fig. 2a). That is, the chromosome half richest in genes encoded on the forward strand also contained a higher proportion of forward-oriented bcr1 repeats, and conversely for the other chromosome half. When the partial bcr1 repeats were included in the analysis, the bias was less prominent but still significant (71.2, 73.6, and 64.6%) (Fig. 2b). Considering both DNA strands together, bcr1 repeats did not exhibit a high degree of clustering and were in general evenly distributed over the chromosomes. Among the chromosomal regions devoid of bcr1 (full length and partial), 18 (B. anthracis Ames), 11 (B. cereus ATCC 10987), and 8 (B. cereus ATCC 14579) regions spanned more than 100 kb, the largest region in each chromosome being 523, 198, and 181 kb in length, respectively. Only three of these regions (in the range of 100 to 130 kb) corresponded (in terms of homologous gene content) in all three strains (Fig. 2b). Each of the three regions contained 100 to 120 genes of diverse functions, with one region containing a cluster of transcription- and translation-related proteins (ribosomal proteins, translation factors, and RNA polymerase subunits) covering ∼40 kb.
FIG. 2.
Chromosomal distribution of bcr1 repeats in B. anthracis Ames, B. cereus ATCC 10987, and B. cereus ATCC 14579. Full-length (FL) (121 to 163 bp) elements are shown only in (a), while full-length and partial (P) (30 to 119 bp) elements are shown in (b). Each tick mark corresponds to a repeat in the circular representation of the chromosome of a strain; black, repeat located on the forward DNA strand (outer circle); red, repeat located on the reverse DNA strand (inner circle). Position 1 on the ruler indicates the origin of replication.
Comparative sequence analysis of bcr1 repeats.
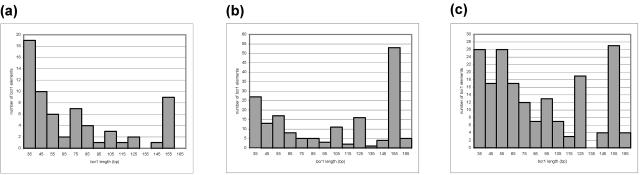
The 145 full-length repeats ranged from 121 to 163 bp in length (Fig. 3), exhibited a GC content (average, 44.2%; range, 36.8 to 50.4%) higher than the average for the complete chromosome (∼35% in each bacterium) (16, 29, 31), and shared between 62.7 and 99.4% (average, 84.5%) pairwise nucleotide sequence identity. A multiple alignment of the sequences showed that certain subregions of the repeat were more conserved than others, in particular, the defined 3′ end (Fig. 4). Whole-genome comparisons have demonstrated that B. cereus ATCC 10987 is phylogenetically more closely related to B. anthracis Ames than to B. cereus ATCC 14579 (29). However, by constructing a phylogenetic tree of all 145 aligned full-length repeats from the three organisms, we did not observe any specific clustering of bcr1 elements with respect to host strain (Fig. 5) or chromosomal locus.
FIG. 3.
Size distribution of bcr1 elements (≥30 bp) from (a) B. anthracis Ames, (b) B. cereus ATCC 10987, and (c) B. cereus ATCC 14579.
FIG. 5.
Phylogenetic tree of 145 full-length bcr1 sequences (≥120 bp) from B. anthracis Ames, B. cereus ATCC 10987, and B. cereus ATCC 14579. The tree was based on the multiple alignment shown in Fig. 4 and was built by using the neighbor-joining method (32) applied to a matrix of pairwise distances between sequences. Distances were computed following Kimura's (19) two-parameter substitution model. Gaps (indels) were removed specifically for each pair of sequences compared. The scale bar shows the average number of nucleotide substitutions per site. bcr1 repeats are color coded based on the strain of origin: blue, B. cereus ATCC 14579; green, B. cereus ATCC 10987; red, B. anthracis Ames.
A remarkable feature of bcr1 is the presence of a 5-bp direct repeat sequence, TTTAT, at its ends. The TTTAT consensus was conserved in the 3′ end of bcr1 in 138 out of the 145 full-length copies while showing a somewhat larger variation at the 5′ end (conserved in 109 out of 145 sequences). As described previously, the bcr1 sequence also contains two short internal inverted repeat motifs (25), which may have structural implications (consensus, 5′-GGGCAG3′↔5′-CTGCCC-3′ and 5′-TAATCAGTGGG-3′↔5′-CCCACTGATTA-3′) (Fig. 4). Although the first motif was present in all full-length bcr1 sequences, the region containing the second motif was deleted in 33 sequences (Fig. 4). The 272 partial bcr1 elements identified in the three genomes examined corresponded to various combinations of fragments of the full-length bcr1 and had a diverse size distribution (Fig. 3). A given element could lack either or both of the TTTAT motifs and/or the internal repeat motifs (data not shown).
Comparative analysis of bcr1 loci.
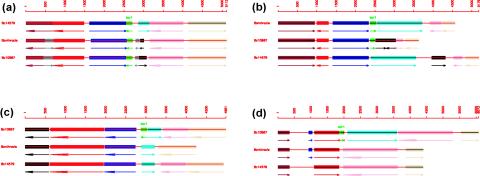
Strikingly, when the local chromosomal contexts for all 145 cases of full-length bcr1 were compared, only one locus was conserved in all three strains with respect to the presence of homologous flanking regions and neighboring genes, and only five loci were common to two genomes exclusively. The closest gene neighbor on each side of bcr1 was used in the computations (Fig. 6a and b). There were also nine homologous regions for which a full-length bcr1 matched a partial repeat(s) in one or two of the other genomes, 10 loci where partial elements were conserved in all three strains, and 18 cases where partial bcr1 repeats were conserved in two strains only. For 123 loci, however, a full-length bcr1 repeat present in any one strain was not found (either as a full-length or partial element) in the corresponding locus in the two other bacteria (Fig. 6c), although the organization of the gene neighbors per se was largely conserved (Fig. 7), as would be expected from the overall conservation in gene organization among B. cereus ATCC 10987, B. cereus ATCC 14579, and B. anthracis Ames (16, 29, 31). The same pattern emerged from the partial repeats; out of a total of 272 partial repeats, 194 were unique to any one strain. These findings clearly show that the global distribution of bcr1 in the chromosome is not fixed. Interestingly, in 14 cases, a partial bcr1 repeat was identified within another partial or full-length element but in the reverse orientation. When the region covering three genes upstream and downstream of bcr1 was investigated, 85 out of the 145 loci contained additional genes (usually one or two) inserted between orthologues of bcr1 neighbors in one strain relative to the other. Conversely, in 98 occurrences, one or more (usually one or two) orthologues of bcr1 neighbors were missing from the locus and either were found at another genomic location or were absent from the genome altogether. By investigating the annotations for all genes surrounding bcr1 repeats in the three strains, we were not able to detect any pattern of conserved or related functions (see http://www.salmongenome.no/htdocs/Suppl_info_for_web_Okstadetal2004.html).
FIG. 6.
Examples of genetic organization around chromosomal bcr1 loci in B. anthracis Ames, B. cereus ATCC 10987, and B. cereus ATCC 14579. Green blocks represent full-length bcr1 elements, while all other blocks represent genes. (a) bcr1 locus conserved in all three strains (repeats Bc14579_1R, Banthracis_1R, and Bc10987_59R [see http://www.salmongenome.no/htdocs/Suppl_info_for_web_Okstadetal2004.html]); (b) bcr1 locus shared by B. anthracis Ames and B. cereus ATCC 10987 only (Banthracis_4F and Bc10987_64F, respectively); (c) bcr1 locus unique to B. cereus ATCC 10987 (Bc10987_28R); (d) bcr1 element (Bc10987_60F) inserted within a gene encoding a putative DNase of the TatD family (BCE0237) in B. cereus ATCC 10987. In each panel, the top strain is the reference. Additional genes in the bottom two strains that are not orthologous (as defined in Materials and Methods) to genes in the reference strain are coded in grey (orthologous in the bottom two strains) or black (not orthologous to any gene in the corresponding region of the other strains). Genes orthologous in all three strains are drawn in the same color. Right- and left-oriented arrows indicate that the gene or repeat is on the forward or reverse strand, respectively. Rulers are in base pairs.
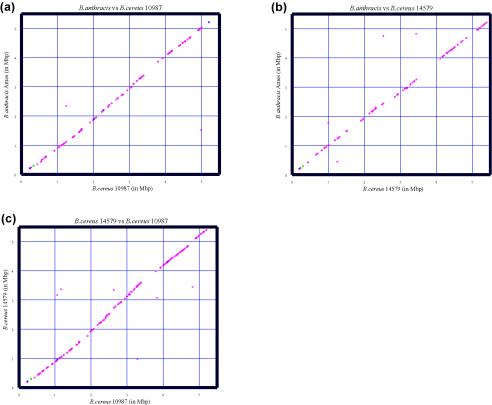
FIG. 7.
Chromosomal organization of bcr1 gene neighbors and their orthologues in B. anthracis Ames, B. cereus ATCC 10987, and B. cereus ATCC 14579. Each panel shows the comparison of two strains. Red circles represent a protein-encoding ORF neighboring a full-length bcr1 element in one strain and its orthologue (as defined in Materials and Methods) in the other. Green crosses represent rRNA genes neighboring bcr1 and their orthologues. Blue triangles correspond to shared bcr1 elements in the two organisms. All genes and repeats are plotted according to their positions in the chromosomes. For any given bcr1 repeat, three upstream and three downstream genes were examined; 411, 327, and 573 orthologous pairs are displayed in (a), (b), and (c), respectively.
bcr1 was overwhelmingly overrepresented in intergenic regions, with only 21 full-length or partial repeats positioned within genes (Table 2). A pairwise comparison of corresponding intergenic regions in separate strains showed that the distance between bcr1 and a neighbor gene, either upstream or downstream, was highly variable, ranging from 0 bp to 1 kbp (in most cases 0 to 300 bp), with no dominant length (see http://www.salmongenome.no/htdocs/Suppl_info_for_web_Okstadetal2004.html). The DNA flanking bcr1 was usually well conserved, with an average sequence identity of ∼85%, although in 65 loci, the upstream and/or downstream DNA was partially or entirely nonconserved in one of the strains (average identity of ∼91% considering the 78 loci with conserved intergenic regions only). When individual intergenic regions upstream or downstream from bcr1 were examined, for 38 out of 46 nonhomologous regions, the gene preceding or following the nonconserved region was also nonhomologous. Conversely, when the intergenic region was conserved (partly or entirely), the neighboring gene was also conserved (in 194 out of 201 cases). Furthermore, for loci where bcr1 was present in one genome and absent in another, the difference in distance between neighbor genes was centered around 150 bp, equivalent to the full length of bcr1 (Fig. 8). The outliers may be explained by the few cases in which gene neighbors have been rearranged in one strain relative to the other, by the occasional insertion of genes in between bcr1 neighbors in one of the strains (Fig. 6b), or by the localization of a partial repeat in one strain corresponding to a full-length repeat in another. Interestingly, in 169 out of 182 loci where bcr1 was missing in one strain relative to another and intergenic regions were homologous, one copy of the TTTAT repeat (or a variant with one mismatch) was still present at the corresponding position.
TABLE 2.
bcr1 elements inserted within genes in B. anthracis Ames, B. cereus ATCC 10987, and B. cereus ATCC 14579
| Strain and interrupted ORF | ORF description | Orthologue(s) in other strainsa | bcr1 repeat namei | bcr1 repeat coordinates | Location of insertion in ORF |
|---|---|---|---|---|---|
| B. anthracis Ames | |||||
| BA1809 | Conserved hypothetical protein, degenerate, putative (authentic frameshift) | BCE1881c | Banthracis_20R_Pg | 1693777-1693697 | 5′ end (opposite strand) |
| BA3940 | Hypothetical protein | BCE3841c | Banthracis_41F_Pf,g,h | 3616545-3616589 | 5′ end/middle |
| B. cereus ATCC 10987 | |||||
| BCE0237 | Deoxyribonuclease, TatD family, putative | BA0222b | Bc10987_60F | 240329-240472 | 3′ end |
| BC0234d | |||||
| BCE0562 | Glycosyl transferase, group 2 family protein | BA0508b | Bc10987_5F_P | 575508-575561 | 3′ end |
| BC0489d | |||||
| BCE2164 | Conserved hypothetical protein | BA2080b | Bc10987_23F | 2076392-2076548 | 5′ end (opposite strand) |
| BCE2974 | Conserved hypothetical protein | BA2937b | Bc10987_29R | 2783476-2783320 | 3′ end |
| BCE3520 | Transcriptional regulator, ArsR family | BA3565b | Bc10987_35F | 3275290-3275437 | 3′ end |
| BC3497d | |||||
| BCE3537e | Transposase, degenerate | Bc10987_37Rg | 3288664-3288509 | 5′ end | |
| BCE3571 | Hypothetical protein | BA3611b | Bc10987_39R | 3326177-3326020 | 5′ end (opposite strand) |
| BCE3841 | Hypothetical protein | BA3940b | Bc10987_62F_Pf,g,h | 3589331-3589369 | 5′ end/middle |
| BCE4739e | Hypothetical protein | Bc10987_72F_Pf,g | 4395530-4395564 | 3′ end (opposite strand) | |
| Bc10987_73F_Pf,g | 4395574-4395603 | 3′ end (opposite strand) | |||
| B. cereus ATCC 14579 | |||||
| BC0401 | Cystine transport system permease protein | BA0367b | Bc14579_7R_P | 382621-382592 | 5′ end |
| BCE0467c | |||||
| BC0556 | Microbial collagenase (EC 3.4.24.3) | BA0555b | Bc14579_15R_P | 538399-538344 | 3′ end (opposite strand) |
| BCE0616c | |||||
| BC0673 | Flavin-dependent dehydrogenase | BA0680b | Bc14579_20F_P | 673072-673127 | 3′ end |
| BCE0747c | |||||
| BC2622 | Macrolide glycosyltransferase (EC 2.4.1.-) | BA2638b | Bc14579_60R_P | 2587064-2587012 | 3′ end (opposite strand) |
| BCE2664c | |||||
| BC2949 | Hypothetical membrane-spanning protein | BA2965b | Bc14579_70F_P | 2909064-2909094 | 5′ end (opposite strand) |
| BCE3003c | Bc14579_18R | 2909154-2908997 | 5′ end | ||
| BC3375 | Similar to glucose-6-phosphate-1-dehydrogenase (fragment) (EC 1.1.1.49) | BA3434b | Bc14579_22R | 3340147-3339994 | 5′ end |
| BC4095 | Hypothetical protein | BA4317b | Bc14579_100R_P | 4061796-4061757 | 3′ end (opposite strand) |
| BCE4164c | |||||
| BC4349 | Hypothetical protein | BA4581b | Bc14579_25R | 4288074-4287918 | 5′ end (opposite strand) |
| BCE4434c |
ORFs were considered orthologues based on reciprocal best hits using TBLASTX (see Materials and Methods).
ORF from B. anthracis Ames.
ORF from B. cereus ATCC 10987.
ORF from B. cereus ATCC 14579.
There is no orthologue for these ORFs, but the entire bcr1 element is included in the predicted gene.
In these cases, the entire bcr1 element is part of the translated protein (complete readthrough).
In these six cases, the entire bcr1 element is included within a predicted gene.
These two bcr1 elements are shared by B. anthracis Ames and B. cereus ATCC 10987.
Names ending with “P” refer to partial bcr1 elements.
FIG. 8.
Histogram of the difference in distance between bcr1 gene neighbors and their orthologues in B. anthracis Ames, B. cereus ATCC 10987, and B. cereus ATCC 14579. The graph shows the distribution of the difference between two distances, the distance between the upstream and downstream genes neighboring a bcr1 element in a strain and the distance between the orthologues of these genes in another strain lacking bcr1 in the corresponding locus. The difference in distance was computed for all loci where bcr1 is present in one strain and absent in another and for all pairwise comparisons of the three strains (178 comparisons in total).
bcr1 gene disruption and overlap.
For 40 out of the 145 full-length bcr1 repeats identified, the element overlapped a neighboring gene, either the upstream ORF (9 cases), the downstream ORF (30 cases), or both (1 case) (see http://www.salmongenome.no/htdocs/Suppl_info_for_web_Okstadetal2004.html). Additionally, one full-length bcr1 copy was located within a predicted gene (BCE3537 from B. cereus ATCC 10987, annotated as a degenerate transposase), introducing a premature stop codon (Table 2 and see http://www.salmongenome.no/htdocs/Suppl_info_for_web_Okstadetal2004.html). Among the 41 overlapped genes, 36 were terminated at a stop codon within the overlapping region, while the remaining five ORFs were initiated from a start codon within the overlapping region. In 21 cases, the overlapping downstream gene was encoded on the opposite strand relative to bcr1 and ended within the reverse complement of the TTTAT motif, creating a TAA stop codon (underlined). In the corresponding region from a strain lacking bcr1, the orthologous gene similarly stopped within the single TTTAT motif present in the sequence (see above). For the remaining 20 overlaps, the ORF either started or ended within bcr1. This leads to a part of bcr1 being expressed as the N- or C-terminal part (range, 2 to 47 amino acids) of the gene product, and adds extra amino acids to the termini of these proteins relative to their orthologues in strains lacking bcr1 in the corresponding locus. From the functions of these proteins, the significance of this phenomenon is not clear. Also, among the 272 partial bcr1 elements found in B. anthracis Ames, B. cereus ATCC 10987, and B. cereus ATCC 14579, 50 elements overlapped either the upstream or the downstream gene, four elements overlapped both genes, and five elements were located within four predicted ORFs. Furthermore, for full-length or partial bcr1 elements unique to one strain and overlapping a gene on one side, we found 15 cases where the intergenic region on the other side of bcr1 matched a part of the orthologous gene (range, 2 to 122 bp) in one or both of the other strains (Fig. 6d and Table 2). Altogether, these findings could be taken as evidence of bcr1 insertion within protein-encoding genes, adding to the six cases of intragenic bcr1 described above (one full-length and five partial elements) and strongly indicating that bcr1 constitutes a novel mobile DNA element. In 17 of these 21 cases, the bcr1 insertions introduce premature translational stops in the synthesis of the resulting proteins. Either the interrupted genes may be nonessential to the bacterium or the bcr1 insertions may not have impaired the functions of the resulting proteins. Most remarkable were the remaining four cases, where a partial repeat of 30 to 45 bp was inserted within three predicted genes without causing interruption of the coding sequence (Table 2), again leading to bcr1 putatively being expressed as part of the protein. Two of the repeats were unique to B. cereus ATCC 10987 (inserted within BCE4739) and the other two were shared between B. cereus ATCC 10987 and B. anthracis Ames (BCE3841 and BA3940, respectively). All three bcr1-containing genes encode hypothetical proteins and have no homologues in B. cereus ATCC 14579 or any other organism.
DISCUSSION
The availability of complete genome sequences of three strains from the B. cereus group has allowed a global analysis of bcr1 repeat sequences, initially identified by Økstad et al. (25). The bcr1 element is similar in length and distribution to several DNA repeats identified previously in other bacteria, most prominently the Correia elements found in Neisseria species (4, 6, 7, 22). However, the composition of the elements is different; while the Correia elements have a fixed length, a highly modular substructure, and 25-bp inverted repeats followed by TA dinucleotide direct repeats flanking the ends (4, 22), bcr1 exhibits a mosaic substructure (Fig. 4) and a highly diverse length distribution which varies between strains (Fig. 3) and carries only a short 5-bp direct repeat flanking the termini.
Correia elements exhibit several features characteristic of transposable elements and have been suggested to constitute mobile DNA (4, 22, 23). Similarly, although lacking the classic terminal inverted repeat structures, several lines of evidence point to bcr1 as a novel mobile genetic element in the B. cereus group: the presence of bcr1 almost exclusively in noncorresponding genomic locations in different strains (see http://www.salmongenome.no/htdocs/Suppl_info_for_web_Okstadetal2004.html),the absence of phylogenetic clustering with respect to host strain and chromosomal locus (Fig. 5), and the cases of bcr1 localization inside protein-coding regions of genes (Fig. 6d and Table 2). Such insertion events are probably in most cases selected against due to decreased genome fitness of the insertion mutants. Alternatively, the heterogeneous distribution of bcr1 in the chromosome could have originated by differential deletion in different strains from an original ancestor organism. We find this unlikely, however, since the high genetic diversity observed in the B. cereus group (12, 13) together with the large heterogeneity in bcr1 organization observed from whole-genome analysis of three strains only (Fig. 2) implies that a common ancestor to the group must have been carrying an unreasonably high number of bcr1 repeats.
The occurrence of short terminal direct repeats (in the case of bcr1, TTTAT [Fig. 4]) is a classic feature of transposable elements. The direct repeats result from target site duplication during transposition, presumably due to a staggered cut in the host target DNA which is later filled (reviewed by Chandler [5]). The presence of only one TTTAT motif at corresponding loci lacking bcr1 suggests that TTTAT constitutes the bcr1 insertion site. Although bcr1 does not encode its own transposase, a possible moving mechanism could be supplied by a trans-acting enzyme, and the three B. cereus group genomes that were analyzed are indeed each known to contain a multitude of transposase-encoding genes when compared by using The Institute for Genomic Research annotation (http://www.tigr.org/tigr-scripts/CMR2/CMRGenomes.spl) (55 for B. cereus 14579, 17 for B. anthracis Ames, and 34 for B. cereus 10987) (16, 29, 31). Alternatively, both the bcr1 element and its chromosomal target could carry a copy of the TTTAT motif, allowing bcr1 to insert by site-specific recombination.
Our results from bcr1 hybridization in Southern blots indicate that bcr1 is ubiquitous in, and unique to, the B. cereus group and thus that its presence predates the divergence of B. cereus, B. anthracis, B. thuringiensis, B. weihenstephanensis, and B. mycoides. Most B. cereus group organisms carry various numbers of small and large plasmids which harbor many of the factors that take part in defining the species in the group (crystal toxin genes in B. thuringiensis and anthrax toxin genes in B. anthracis). Strikingly, the plasmids carried by the three strains studied here, as well as B. anthracis A2012 plasmids pXO1 and pXO2 (30) and pBtoxis from B. thuringiensis subsp. israelensis (3), appeared to be devoid of bcr1 elements (full length or partial). An examination of many plasmids from various isolates is required to determine if this is a general phenomenon. Still, it is tempting to speculate that bcr1 is an ancient element and that the origin of B. thuringiensis and B. anthracis is linked to the uptake of plasmid DNA in cells that were already carrying bcr1 in the chromosome. bcr1 may also provide a function important to the integrity of the chromosome but not the plasmids, which ensures its unique presence in chromosomal DNA.
B. anthracis Ames carries a considerably lower number of full-length and partial bcr1 elements than the two B. cereus strains analyzed (Fig. 2). This finding could be due to an increased deletion rate in B. anthracis, a lower evolutionary rate in B. anthracis caused by a lower number of replication cycles over time when the bacterium is stationary as dormant spores in the soil, or a genetic bottleneck caused by strong selective pressure applied to B. anthracis during the infectious cycle. A comparative analysis of the B. anthracis Ames strain and the unfinished genome sequence of B. anthracis Kruger B from The Institute for Genomic Research available at the National Center for Biotechnology Information (GenBank accession number NC_004126) showed that all full-length bcr1 loci were conserved, although the strains are from two different phylogenetic subgroups (A and B groups, respectively) (18, 33; data not shown). An examination of whole-genome sequences from additional B. cereus group strains will be necessary to determine if carrying low copy numbers of bcr1 is a specific feature of B. anthracis.
Although we have provided strong indications for the mobility of bcr1, a major question is what functional role this element might hold. It is interesting that the distribution of bcr1 elements is biased and correlated with the transcription orientation bias of the genes in each half of the chromosome (Fig. 2). A pattern search of the TTTAT motif in the chromosomes of the three sequenced B. cereus group strains revealed a random distribution of TTTAT sequences with respect to chromosome half and DNA strand and also when only intergenic regions were considered. If we assume that TTTAT is the bcr1 insertion site, this indicates that the observed bcr1 distribution is not the result of unequal distribution of target sites but may be due to functional or selective constraints.
Based on sequence analysis and experimental data, Correia elements have been shown to have the potential for creating functional promoters affecting transcription of nearby genes (4, 34). Furthermore, some Correia elements have been shown to be part of mRNA transcripts, possibly regulating mRNA stability by forming a stem-loop which is a substrate for RNase III processing (9). Whether bcr1 may play similar roles in transcriptional regulation is yet unclear. We were not able to detect bcr1-specific binding of whole-cell protein extracts from B. cereus ATCC 14579 (data not shown). Northern hybridizations have revealed that bcr1 may be part of RNA transcripts (data not shown), but whether this finding is of functional significance or is merely a result of a bcr1 element being positioned between a gene and its promoter or transcriptional terminator is at present not known. At this point, we cannot exclude the possibility that bcr1 may encode a small RNA molecule or act in transcription termination.
Natural competence for transformation in Neisseria and Haemophilus spp. requires the presence of short (9- to 10-bp), species-specific DNA uptake sequences in the transforming DNA (8, 10). The three sequenced B. cereus group genomes carry orthologues to most of the competence system genes from B. subtilis (29), and although bcr1 is longer than the previously characterized DNA uptake sequences from gram-negative bacteria, one could envisage a role for bcr1, or conserved part(s) of it, as a recognition sequence for the putative competence system. If bcr1 does play a key role in DNA uptake, it would essentially define the species in the classical reproductive isolation sense; if an organism lacks the repeats, it cannot effectively donate DNA to B. cereus group organisms. Indeed, previous studies have indicated that DNA exchange in the B. cereus group does occur and that the exchange seems to be limited to DNA from close relatives (31). This finding could also offer an alternative explanation for the lower number of bcr1 repeats in B. anthracis: the ecological isolation of B. anthracis compared to B. cereus and B. thuringiensis could mean that little uptake of new DNA is occurring in this bacterium. Future work on bcr1 elements in the B. cereus group should focus on functional analysis and could include investigations of other possible roles, such as methylation sites, hotspots for DNA recombination, or substrates for intrachromosomal recombination events contributing to chromosome rearrangements and genome plasticity.
Acknowledgments
We are most grateful to Erlendur Helgason for providing unpublished multilocus enzyme electrophoresis data.
This work was supported by a personal grant from the Norwegian Research Council to O.A.Ø. N.J.T. and F.B.S. were supported by grants to A.-B.K. from the Norwegian Research Council.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ash, C., J. A. Farrow, S. Wallbanks, and M. D. Collins. 1991. Phylogenetic heterogeneity of the genus Bacillus revealed by comparative analysis of small-subunit-ribosomal RNA sequences. Lett Appl. Microbiol. 13:202-206. [Google Scholar]
- 3.Berry, C., S. O'Neil, E. Ben-Dov, A. F. Jones, L. Murphy, M. A. Quail, M. T. Holden, D. Harris, A. Zaritsky, and J. Parkhill. 2002. Complete sequence and organization of pBtoxis, the toxin-coding plasmid of Bacillus thuringiensis subsp. israelensis. Appl. Environ. Microbiol. 68:5082-5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buisine, N., C. M. Tang, and R. Chalmers. 2002. Transposon-like Correia elements: structure, distribution and genetic exchange between pathogenic Neisseria sp. FEBS Lett. 522:52-58. [DOI] [PubMed] [Google Scholar]
- 5.Chandler, M. 1998. Insertion sequences and transposons, p. 30-37. In F. J. De Bruijn, J. R Lupski, and G. Weinstock (ed.), Bacterial genomes—physical structure and analysis. Chapman & Hall, New York, N.Y.
- 6.Correia, F. F., S. Inouye, and M. Inouye. 1986. A 26-base-pair repetitive sequence specific for Neisseria gonorrhoeae and Neisseria meningitidis genomic DNA. J. Bacteriol. 167:1009-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Correia, F. F., S. Inouye, and M. Inouye. 1988. A family of small repeated elements with some transposon-like properties in the genome of Neisseria gonorrhoeae. J. Biol. Chem. 263:12194-12198. [PubMed] [Google Scholar]
- 8.Danner, D. B., R. A. Deich, K. L. Sisco, and H. O. Smith. 1980. An eleven-base-pair sequence determines the specificity of DNA uptake in Haemophilus transformation. Gene 11:311-318. [DOI] [PubMed] [Google Scholar]
- 9.De Gregorio, E., C. Abrescia, M. S. Carlomagno, and P. P. Di Nocera. 2003. Ribonuclease III-mediated processing of specific Neisseria meningitidis mRNAs. Biochem. J. 374:799-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elkins, C., C. E. Thomas, H. S. Seifert, and P. F. Sparling. 1991. Species-specific uptake of DNA by gonococci is mediated by a 10-base-pair sequence. J. Bacteriol. 173:3911-3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galtier, N., M. Gouy, and C. Gautier. 1996. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 12:543-548. [DOI] [PubMed] [Google Scholar]
- 12.Helgason, E., N. J. Tourasse, R. Meisal, D. A. Caugant, and A.-B. Kolstø. 2004. Multilocus sequence typing scheme for bacteria of the Bacillus cereus group. Appl. Environ. Microbiol. 70:191-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Helgason, E., O. A. Økstad, D. A. Caugant, H. Johansen, A. Fouet, M. Mock, I. Hegna, and A.-B. Kolstø. 2000. Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis—one species on the basis of genetic evidence. Appl. Environ. Microbiol. 66:2627-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hofnung, M., and J. A. Shapiro. 1999. Introduction—special issue on repetitive DNA sequences in microbes. Res. Microbiol. 150:577-578. [Google Scholar]
- 15.Hughes, J. M., and J. L. Gerberding. 2002. Anthrax bioterrorism: lessons learned and future directions. Emerg. Infect. Dis. 8:1013-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ivanova, N., A. Sorokin, I. Anderson, N. Galleron, B. Candelon, V. Kapatral, A. Bhattacharyya, G. Reznik, N. Mikhailova, A. Lapidus, L. Chu, M. Mazur, E. Goltsman, N. Larsen, M. D'Souza, T. Walunas, Y. Grechkin, G. Pusch, R. Haselkorn, M. Fonstein, S. D. Ehrlich, R. Overbeek, and N. Kyrpides. 2003. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423:87-91. [DOI] [PubMed] [Google Scholar]
- 17.Jensen, G. B., B. M. Hansen, J. Eilenberg, and J. Mahillon. 2003. The hidden lifestyles of Bacillus cereus and relatives. Environ. Microbiol. 5:631-640. [DOI] [PubMed] [Google Scholar]
- 18.Keim, P., L. B. Price, A. M. Klevytska, K. L. Smith, J. M. Schupp, R. Okinaka, P. J. Jackson, and M. E. Hugh-Jones. 2000. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J. Bacteriol. 182:2928-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 20.Kolstø, A. B., A. Grønstad, and H. Oppegaard. 1990. Physical map of the Bacillus cereus chromosome. J. Bacteriol. 172:3821-3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, N. J. Cummings, R. A. Daniel, F. Denizot, K. M. Devine, A. Düsterhöft, S. D. Ehrlich, P. T. Emmerson, K. D. Entian, J. Errington, C. Fabret, E. Ferrari, D. Foulger, C. Fritz, M. Fujita, Y. Fujita, S. Fuma, A. Galizzi, N. Galleron, S.-Y. Ghim, P. Glaser, A. Goffeau, E. J. Golightly, G. Grandi, G. Guiseppi, B. J. Guy, K. Haga, J. Haiech, C. R. Harwood, A. Henaut, H. Hilbert, S. Holsappel, S. Hosono, M.-F. Hullo, M. Itaya, L. Jones, B. Joris, D. Karamata, Y. Kasahara, M. Klearr-Blanchard, C. Klein, Y. Kobayashi, P. Koetter, G. Koningstein, S. Krogh, M. Kumano, K. Kurita, A. Lapidus, S. Lardinois, J. Lauber, V. Lazarevic, S.-M. Lee, A. Levine, H. Liu, S. Masuda, C. Mauel, C. Medigue, N. Medina, R. P. Mellado, M. Mizuno, D. Moestl, S. Nakai, M. Noback, D. Noone, M. O'Reilly, K. Ogawa, A. Ogiwara, B. Oudega, S.-H. Park, V. Parro, T. M. Pohl, D. Portetelle, S. Porwollik, A. M. Prescott, E. Presecan, P. Pujic, B. Purnelle, G. Rapoport, M. Rey, S. Reynolds, M. Rieger, C. Rivolta, E. Rocha, B. Roche, M. Rose, Y. Sadaie, T. Sato, E. Scanlan, S. Schleich, R. Schroeter, F. Scoffone, J. Sekiguchi, A. Sekowska, S. J. Seror, P. Serror, B.-S. Shin, B. Soldo, A. Sorokin, E. Tacconi, T. Takagi, H. Takahashi, K. Takemaru, M. Takeuchi, A. Tamakoshi, T. Tanaka, P. Terpstra, A. Tognoni, V. Tosato, S. Uchiyama, M. Vandenbol, F. Vannier, A. Vassarotti, A. Viari, R. Wambutt, E. Wedler, H. Wedler, T. Weitzenegger, P. Winters, A. Wipat, H. Yamamoto, K. Yamane, K. Yasumoto, K. Yata, K. Yoshida, H.-F. Yoshikawa, E. Zumstein, H. Yoshikawa, and A. Danchin. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 22.Liu, S. V., N. J. Saunders, A. Jeffries, and R. F. Rest. 2002. Genome analysis and strain comparison of Correia repeats and Correia repeat-enclosed elements in pathogenic Neisseria. J. Bacteriol. 184:6163-6173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazzone, M., E. De Gregorio, A. Lavitola, C. Pagliarulo, P. Alifano, and P. P. Di Nocera. 2001. Whole-genome organization and functional properties of miniature DNA insertion sequences conserved in pathogenic Neisseriae. Gene 278:211-222. [DOI] [PubMed] [Google Scholar]
- 24.Okinaka, R. T., K. Cloud, O. Hampton, A. R. Hoffmaster, K. K. Hill, P. Keim, T. M. Koehler, G. Lamke, S. Kumano, J. Mahillon, D. Manter, Y. Martinez, D. Ricke, R. Svensson, and P. J. Jackson. 1999. Sequence and organization of pXO1, the large Bacillus anthracis plasmid harboring the anthrax toxin genes. J. Bacteriol. 181:6509-6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Økstad, O. A., I. Hegna, T. Lindbäck, A.-L. Rishovd, and A.-B. Kolstø. 1999. Genome organization is not conserved between Bacillus cereus and Bacillus subtilis. Microbiology 145:621-631. [DOI] [PubMed] [Google Scholar]
- 26.Økstad, O. A., M. Gominet, B. Purnelle, M. Rose, D. Lereclus, and A.-B. Kolstø. 1999. Sequence analysis of three Bacillus cereus loci carrying PIcR-regulated genes encoding degradative enzymes and enterotoxin. Microbiology 145:3129-3138. [DOI] [PubMed] [Google Scholar]
- 27.Popham, D. L., and P. Setlow. 1994. Cloning, nucleotide sequence, mutagenesis, and mapping of the Bacillus subtilis pbpD gene, which codes for penicillin-binding protein 4. J. Bacteriol. 176:7197-7205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Presecan, E., I. Moszer, L. Boursier, H. C. C. Ramos, V. de la Fuente, M. F. Hullo, C. Lelong, S. Schleich, A. Sekowska, B. H. Song, G. Villani, F. Kunst, A. Danchin, and P. Glaser. 1997. The Bacillus subtilis genome from gerBC (311 degrees) to licR (334 degrees). Microbiology 143:3313-3328. [DOI] [PubMed] [Google Scholar]
- 29.Rasko, D. A., J. Ravel, O. A. Økstad, E. Helgason, R. Z. Cer, L. Jiang, K. A. Shores, D. E. Fouts, N. J. Tourasse, S. V. Angiuoli, J. Kolonay, W. C. Nelson, A.-B Kolstø, C. M. Fraser, and T. D. Read. 2004. The genome sequence of Bacillus cereus ATCC 10987 reveals metabolic adaptations and a large plasmid related to Bacillus anthracis pXO1. Nucleic Acids Res. 32:977-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Read, T. D., S. L. Salzberg, M. Pop, M. Shumway, L. Umayam, L. Jiang, E. Holtzapple, J. D. Busch, K. L. Smith, J. M. Schupp, D. Solomon, P. Keim, and C. M. Fraser. 2002. Comparative genome sequencing for discovery of novel polymorphisms in Bacillus anthracis. Science 296:2028-2033. [DOI] [PubMed] [Google Scholar]
- 31.Read, T. D., S. N. Peterson, N. Tourasse, L. W. Baillie, I. T. Paulsen, K. E. Nelson, H. Tettelin, D. E. Fouts, J. A. Eisen, S. R. Gill, E. K. Holtzapple, O. A. Økstad, E. Helgason, J. Rilstone, M. Wu, J. F. Kolonay, M. J. Beanan, R. J. Dodson, L. M. Brinkac, M. Gwinn, R. T. DeBoy, R. Madpu, S. C. Daugherty, A. S. Durkin, D. H. Haft, W. C. Nelson, J. D. Peterson, M. Pop, H. M. Khouri, D. Radune, J. L. Benton, Y. Mahamoud, L. Jiang, I. R. Hance, J. F. Weidman, K. J. Berry, R. D. Plaut, A. M. Wolf, K. L. Watkins, W. C. Nierman, A. Hazen, R. Cline, C. Redmond, J. E. Thwaite, O. White, S. L. Salzberg, B. Thomason, A. M. Friedlander, T. M. Koehler, P. C. Hanna, A.-B. Kolstø, and C. M. Fraser. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 423:81-86. [DOI] [PubMed] [Google Scholar]
- 32.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 33.Smith, K. L., V. DeVos, H. Bryden, L. B. Price, M. E. Hugh-Jones, and P. Keim. 2000. Bacillus anthracis diversity in Kruger National Park. J. Clin. Microbiol. 38:3780-3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Snyder, L. A., W. M. Shafer, and N. J. Saunders. 2003. Divergence and transcriptional analysis of the division cell wall (dcw) gene cluster in Neisseria spp. Mol. Microbiol. 47:431-442. [DOI] [PubMed] [Google Scholar]
- 35.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Versalovic, J., and J. R. Lupski. 1998. Interspersed repetitive sequences in bacterial genomes, p. 38-48. In F. J. de Bruijn, J. R. Lupski, and G. M. Weinstock (ed.), Bacterial genomes—physical structure and analysis. Chapman & Hall, New York, N.Y.