Abstract
Z-prenyl diphosphate synthases catalyze the sequential condensation of isopentenyl diphosphate with allylic diphosphates to synthesize polyprenyl diphosphates. In mycobacteria, these are precursors of decaprenyl phosphate, a molecule which plays a central role in the biosynthesis of essential mycobacterial cell wall components, such as the mycolyl-arabinogalactan-peptidoglycan complex and lipoarabinomannan. Recently, it was demonstrated that open reading frame Rv2361c of the Mycobacterium tuberculosis H37Rv genome encodes a unique prenyl diphosphate synthase (M. C. Schulbach, P. J. Brennan, and D. C. Crick, J. Biol. Chem. 275:22876-22881, 2000). We have now purified the enzyme to near homogeneity by using an Escherichia coli expression system and have shown that the product of this enzyme is decaprenyl diphosphate. Rv2361c has an absolute requirement for divalent cations and an optimal pH range of 7.5 to 8.5, and the activity is stimulated by both detergent and dithiothreitol. The enzyme catalyzes the addition of isopentenyl diphosphate to geranyl diphosphate, neryl diphosphate, ω,E,E-farnesyl diphosphate, ω,E,Z-farnesyl diphosphate, or ω,E,E,E-geranylgeranyl diphosphate, with Km values for the allylic substrates of 490, 29, 84, 290, and 40 μM, respectively. The Km value for isopentenyl diphosphate is 89 μM. The catalytic efficiency is greatest when ω,E,Z-farnesyl diphosphate is used as the allylic acceptor, suggesting that this is the natural substrate in vivo, a conclusion that is supported by previous structural studies of decaprenyl phosphoryl mannose isolated from M. tuberculosis. This is the first report of a bacterial Z-prenyl diphosphate synthase that preferentially utilizes an allylic diphosphate primer having the α-isoprene unit in the Z configuration, indicating that Rv1086 (ω,E,Z-farnesyl diphosphate synthase) and Rv2361c act sequentially in the biosynthetic pathway that leads to the formation of decaprenyl phosphate in M. tuberculosis.
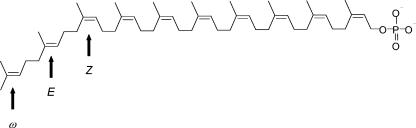
A common feature of many bacterial cell wall polymer biosynthetic pathways is the requirement for polyprenyl phosphate (Pol-P) (2, 20, 23), which acts as a carrier of activated saccharides. Pol-P is a low-abundance lipid, and its availability may be rate limiting for cell wall synthesis (14). It has been suggested that the rate of formation of polyprenyl diphosphoryl muramyl pentapeptide (lipid I), the first lipid-linked intermediate in peptidoglycan synthesis, in Escherichia coli may be dependent on the pool levels of Pol-P (29), and mutant cells deficient in Pol-P synthesis do show reduced levels of cell wall biosynthesis (17). In bacteria, the predominant Pol-P molecule is typically ω,diE,polyZ-undecaprenyl phosphate (6, 15); exceptions have been reported for Paracoccus denitrificans, Mycobacterium smegmatis, and Mycobacterium tuberculosis (16, 28, 30, 31). In M. tuberculosis the predominant Pol-P molecule is decaprenyl phosphate (28), which occurs in the ω,E,polyZ configuration (Fig. 1).
FIG. 1.
Mycobacterial decaprenyl phosphate. The structure is drawn as described by Wolucka et al. (30). Arrows indicate the ω isoprene unit and representative double bonds in the E and Z stereoconfigurations.
Pol-P molecules are synthesized via sequential condensation of isopentenyl diphosphate (IPP) with allylic diphosphates, reactions which are catalyzed by prenyl diphosphate synthases. These enzymes are specific for the type of stereochemistry (E or Z) of the introduced allylic double bond, and amino acid alignments have shown that there is no amino acid sequence homology between the E- and Z-prenyl families of diphosphate synthases (12). The first amino acid sequence of a Z-prenyl diphosphate synthase was reported in 1998, when undecaprenyl diphosphate synthase (UPS) was cloned from Micrococcus luteus and characterized (27). Since then, UPS homologs have been cloned from E. coli, Streptococcus pneumoniae, and Haemophilus influenzae (3). In previously studied bacteria, these Z-prenyl diphosphate synthases utilize ω,E,E-FPP as the allylic acceptor and synthesize long-chain products with the typical ω,diE,polyZ stereoconformation (1, 3, 4, 21). M. tuberculosis is unusual in that it has two genes (Rv1086 and Rv2361c) that encode proteins with amino acid sequence similarity to that of UPS. Researchers have previously shown that Rv1086 encodes an E,Z-FPP synthase and implicated Rv2361c in the synthesis of decaprenyl diphosphate in crude cell extracts (25). Based on the reported structure of mycobacterial decaprenyl phosphate, we hypothesized that both enzymes were involved in the synthesis of this molecule. We now report the cloning, purification, and characterization of Rv2361c, which adds seven isoprene units to ω,E,Z-farnesyl diphosphate to form decaprenyl diphosphate, indicating that Rv1086 and Rv2361c act sequentially in the synthesis of decaprenyl diphosphate, a precursor of decaprenyl phosphate, which is utilized in many aspects of cell wall biosynthesis in M. tuberculosis (7, 8, 20).
MATERIALS AND METHODS
Materials.
[1-14C]IPP (58 mCi/mmol) was purchased from Amersham Pharmacia Biotech. Kanamycin, farnesol (mixed stereoisomers), ω,E,E-farnesol, geraniol, ω,E,E,E-geranylgeraniol, and dimethylallyl diphosphate (DMAPP) were purchased from Sigma. ω,E,E-farnesyl diphosphate (E,E-FPP) and geranyl diphosphate (GPP) were synthesized as described by Davisson et al. (10). Neryl diphosphate (NPP) was a gift from C. J. Waechter and J. S. Rush (University of Kentucky). Various authentic long-chain prenols and isoprenyl phosphates were obtained from the Institute of Biochemistry and Biophysics, Polish Academy of Sciences (Warsaw, Poland).
Radiolabeled ω,E,Z-FPP was prepared enzymatically using purified, recombinant ω,E,Z-FPP synthase (Rv1086) (26). The product of this enzyme was quantitated by liquid scintillation spectrometry and was analyzed for chain length and stereochemistry by thin-layer chromatography (TLC), confirming that the material was exclusively ω,E,Z-[14C]FPP.
LKC18F reverse-phase TLC plates were from Whatman; potato acid phosphatase (grade 2) was purchased from Roche Molecular Biochemicals; Luria-Bertani (LB) agar and LB broth were from Invitrogen; and restriction enzymes NdeI and XhoI, deoxynucleoside triphosphates, and T4 DNA ligase were purchased from New England Biolabs. E. coli-competent cells (DH5α), pET28a vector, and E. coli BL21(DE3)pLysS were purchased from Novagen (Madison, Wis.). Qiaprep Spin Miniprep kits were purchased from QIAGEN, Inc. Monoclonal anti-polyhistidine unconjugated antibody from mouse ascites fluid, anti-mouse immunoglobulin G conjugated to alkaline phosphatase, and 5-bromo-4-chloro-3-indoylphosphate tablets were purchased from Sigma. Oligonucleotide synthesis and sequencing were done by Macromolecular Resources (Colorado State University).
Cloning, expression, and purification of Rv2361c.
The M. tuberculosis gene, Rv2361c, was amplified by PCR using the primers 5′-AGGCTCGAGACTAGGCGCTCCCGAACC-3′ and 5′-TATAACATATGGCTAGGGATGCACGGA-3′, which were designed to create XhoI and NdeI restriction endonuclease sites (underlined). The PCR product was digested with NdeI and XhoI and was subjected to agarose (0.8%) gel electrophoresis. The desired band was purified and ligated into pET28a(+) vector, yielding an expression plasmid for M. tuberculosis Rv2361c which was used to transform DH5α cells. The construct was then isolated, sequenced, and used to transform E. coli BL21(DE3)pLysS cells.
Transformed cells were grown overnight in 1 ml of LB broth containing 50 μg of kanamycin/ml and 40 μg of chloramphenicol/ml and was transferred to 1 liter of the same medium. The cells were grown at 37°C to an A600 of 0.6, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM, and incubation was continued for an additional 7 h at 15°C. Expression of the protein was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot using the monoclonal anti-polyhistidine antibody.
E. coli cells expressing recombinant enzyme were harvested by centrifugation and were resuspended in buffer containing 50 mM morpholinepropanesulfonic acid (MOPS) (pH 7.9), 10 mM MgCl2, 5 mM 2-mercaptoethanol, 10% glycerol, and 0.1% Triton X-100. The cells were disrupted by probe sonication on ice with a Sanyo Soniprep 150 (6 cycles of 20 s on and 90 s off). The resulting suspension was agitated at 4°C for 1 h and was centrifuged at 27,000 × g for 20 min. The pellet was discarded, and the resulting supernatant containing the soluble His-tagged fusion protein was loaded on a cobalt-based TALON (BD Biosciences Clontech) immobilized metal affinity column (IMAC) which was washed with a solution of 50 mM MOPS (pH 8), 0.1% Triton X-100, 7 mM imidazole, and 500 mM NaCl. The His-tagged proteins were eluted using washing buffer containing a linear gradient of 25 to 200 mM imidazole. Protein-containing fractions were analyzed by SDS-PAGE and Western blot. Fractions that were estimated to be at least 90% pure by Coomassie brilliant blue 250R staining were pooled, desalted, and used for further characterization. Protein concentrations were estimated using a bicinchoninic acid protein assay kit (Pierce).
Prenyl diphosphate synthase assay.
Typically, enzyme activity was measured in a 50-μl reaction mixture containing 50 mM MOPS (pH 7.9), 2.5 mM dithiothreitol, 10 mM sodium orthovanadate, 1 mM MgCl2, 0.3% Triton X-100, 100 μM allylic diphosphate, and 30 μM [14C]IPP. Reactions were initiated by the addition of 0.25 μg (6.9 pmol) of recombinant enzyme. The mixture was incubated for 10 min at 37°C, and the reaction was stopped with 1 ml of water saturated with NaCl. The products were then extracted with n-butanol saturated with water, and an aliquot was taken for liquid scintillation spectrometry. All assays were conducted under conditions where product formation was linear for both time and protein concentration.
To analyze the chain length of the radioactive prenyl diphosphate products, the enzymatically synthesized radiolabeled compounds were hydrolyzed to the corresponding alcohols with potato acid phosphatase as described previously (25). The alcohols were extracted from the reaction mixture with hexane and were analyzed on reverse-phase TLC plates developed in methanol/water (8/2). Authentic standards were visualized with an anisaldehyde spray, and the radioactive compounds were detected with a Bioscan System 200 Imaging System and/or autoradiography.
To study the effect of different divalent metal ions on the activity of Rv2361c, the enzyme was incubated with Bio-Rex70 (sodium form, 200 to 400 mesh; Bio-Rad) on ice for 20 min to remove the endogenous divalent cations. Resin was removed, and MgCl2, MnCl2, CaCl2, ZnCl2, or EDTA was added to the reaction mixtures at the indicated concentrations. In the Triton X-100 dependence studies, Triton X-100 was added to assay mixtures at the indicated concentrations. The effect of pH on the activity of Rv2361c was studied by using a broad-range buffer (50 mM Tris-HCl, 25 mM MOPS, and 25 mM acetic acid) adjusted to the indicated pH with tetraethylammonium hydroxide. All values reported are averages of duplicate measurements from representative experiments. Km and Vmax values were calculated by nonlinear regression analysis (SigmaPlot; SPSS).
RESULTS
Overexpression of recombinant Rv2361c in E. coli.
In order to characterize Rv2361c from M. tuberculosis H37Rv, the enzyme was overexpressed in E. coli by using the pET-28a(+) expression system. After inducing expression in E. coli with IPTG, the cells were harvested and disrupted by sonication. Cytosolic and membrane fractions from recombinant and wild-type E. coli were tested for activity using ω,E,E-FPP as the allylic substrate. Membranes from recombinant bacteria showed a 10-fold increase in [14C]IPP incorporation into butanol-extractable material compared to incorporation in wild-type membrane preparations. Enzymatic activity in the membranes was solubilized with 0.1% Triton X-100, affinity purified using a TALON Co2+ column, and analyzed by SDS-PAGE (Fig. 2). A major band was observed at the expected size of 36 kDa, and the identity of the band as the Rv2361c-His tag fusion protein was confirmed by Western blot.
FIG. 2.
Expression and purification of Rv2361c. An expression vector was constructed in which Rv2361c is fused to a histidine repeat at the N terminus and used to transform E. coli BL21(DE3)pLysS. Expression was induced by the addition of 1 mM IPTG, and the protein was purified by IMAC chromatography. Fractions eluted from the column were subjected to SDS-PAGE, and proteins were visualized by staining with Coomassie brilliant blue 250R. Lane 1, uninduced soluble cell extract. Lane 2, soluble cell extract after IPTG induction. Lane 3, purified Rv2361c.
Reaction requirements.
To confirm the activity of recombinant Rv2361c, a standard activity assay was performed using radiolabeled IPP and ω,E,E-FPP as substrates. [14C]IPP incorporation into butanol-soluble material increased linearly with both time and recombinant protein concentration (data not shown). The purified enzyme was tested for its ability to use six different allylic compounds (DMAPP, GPP, NPP, ω,E,E-FPP, ω,E,Z-FPP, and ω,E,E,E-GGPP) as substrates. Of these, only DMAPP did not support enzymatic activity. The enzyme was absolutely dependent on the presence of divalent cation, as the activity was abolished by addition of 10 mM EDTA, and enzyme activity was optimal in the presence of 1 mM Mg2+ (Table 1). MnCl2 at 0.1 mM also supported the activity; however, CaCl2 and ZnCl2 were much less effective at the concentrations tested. The enzyme was maximally active in the presence of 0.1% Triton X-100 and over a range of pH between 7.5 and 8.5 (data not shown).
TABLE 1.
Effect of divalent cation concentration on the activity of Rv2361ca
| Concn (mM) | Divalent cation activity (pmol/mg/min)
|
|||
|---|---|---|---|---|
| MgCl2 | MnCl2 | ZnCl2 | CaCl2 | |
| 0 | 580 | 250 | 260 | 260 |
| 0.01 | 480 | 830 | 220 | 320 |
| 0.1 | 5,000 | 3,800 | 470 | 660 |
| 1.0 | 11,000 | 2,100 | 520 | 150 |
| 5.0 | 2,900 | 270 | 190 | 51 |
The recombinant enzyme was preincubated with Bio-Rex 70 (sodium form) on ice for 20 min. Resin was removed, and MgCl2, MnCl2, CaCl2, or ZnCl2 was added at the indicated concentrations. Assays were performed as described in Materials and Methods. Reactions were incubated at 37°C for 10 min, stopped by the addition of water saturated with NaCl, and extracted with butanol-saturated with water. An aliquot was then taken for liquid scintillation spectrometry.
Kinetic characterization.
The kinetic parameters of Rv2361c were investigated with different amounts of allylic diphosphate as cosubstrates with IPP (Fig. 3). Data from Fig. 3 were used to calculate Km and Vmax values for the different substrates by nonlinear regression analysis. The Michaelis constants calculated for GPP, NPP, ω,E,E-FPP, ω,E,Z-FPP, ω,E,E,E-GGPP, and IPP were 490, 29, 84, 290, 40, and 89 μM, respectively. When ω,E,Z-FPP was used as the allylic acceptor, Vmax, Kcat, and Kcat/Km were all found to be greater than those calculated for the other allylic substrates (Table 2).
FIG.3.
Effect of substrate concentration on the rate of product formation by Rv2361c. Reaction mixtures contained 50 mM MOPS (pH 7.9), 2.5 mM dithiothreitol, 10 mM sodium orthovanadate, 5 mM MgCl2, 0.3% Triton X-100, 0.25 μg (6.9 pmol) of protein, and the indicated amount of substrate in a final volume of 50 μl. (A) The concentration of ω,E-FPP was fixed at 100 μM, and the concentration of IPP was varied as indicated. The other reactions contained 30 μM IPP and various amounts of ω,E,Z-FPP (B), GPP (C), NPP (D), ω,E,E-FPP (E), and ω,E,E,E,GGPP (F). The reactions were incubated at 37°C for 10 min and were stopped with 1 ml of water saturated with NaCl and extracted with 1 ml of butanol saturated with water. An aliquot was then taken for liquid scintillation spectrometry. Structures of the substrates that were varied in concentration are inset in the appropriate panels.
TABLE 2.
Calculated kinetic parameters of Rv2361ca
| Parameter | Substrate
|
|||||
|---|---|---|---|---|---|---|
| IPP | GPP | NPP | E,E-FPP | E,Z-FPP | GGPP | |
| Km (μM) | 89 | 490 | 29 | 84 | 290 | 40 |
| Vmax (pmol/min) | 34 | 25 | 21 | 30 | 4,800 | 12 |
| Kcat (min−1) | 4.8 | 3.6 | 3.1 | 4.3 | 690 | 1.8 |
| Kcat/Km | 5.4 × 104 | 7.4 × 103 | 1.1 × 105 | 5.1 × 104 | 2.4 × 106 | 4.3 × 105 |
To determine the kinetic parameters, assays were done in the presence of 30 μM [14C]IPP and various concentrations of an allylic primer or of 100 μM ω,E,E-FPP and various concentrations of [14C]IPP. Reaction conditions were as described in Materials and Methods, and 6.9 pmols of Rv2361c was used to initiate the reaction. The data were subjected to nonlinear regression analysis using SigmaPlot for Windows.
Analysis of the reaction products.
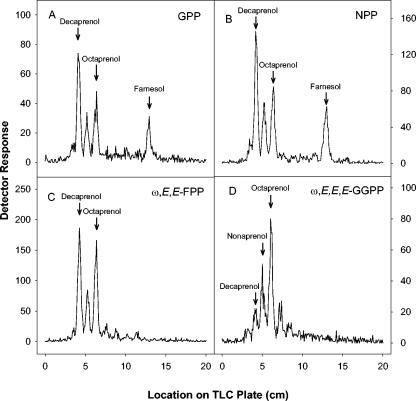
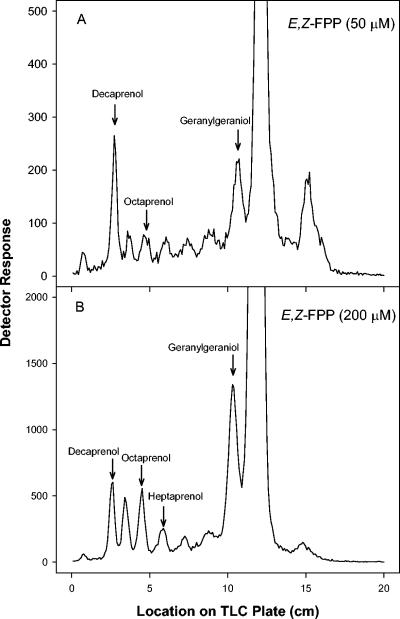
The products of the assays using various allylic diphosphate primers were subjected to enzymatic dephosphorylation for analysis of chain length. When GPP, NPP, or ω,E,E-FPP was used as the substrate, decaprenyl diphosphate, nonaprenyl diphosphate, and octaprenyl diphosphate were synthesized (Fig. 4). Interestingly, the relative amounts of both decaprenyl diphosphate and octaprenyl diphosphate were greater than that of nonaprenyl diphosphate. When either GPP or NPP was used as the allylic primer, FPP (probably in the E,Z or Z,Z conformation, respectively) was also synthesized (Fig. 4A and B). When GGPP was used, the major product synthesized was octaprenyl diphosphate, followed by nonaprenyl diphosphate, decaprenyl diphosphate, and heptaprenyl diphosphate (Fig. 4D). When ω,E,Z-FPP was used as the allylic substrate at 50 μM, decaprenyl diphosphate was the predominant product synthesized (Fig. 5A), with smaller amounts of intermediates of shorter chain lengths also being produced. However, as the concentration of ω,E,Z-FPP was increased in the assays, there was an increase in the amount of the shorter products synthesized relative to decaprenyl diphosphate. In addition, at 200 μM ω,E,Z-FPP there is significantly more GGPP synthesized than decaprenyl diphosphate (Fig. 5B).
FIG. 4.
TLC analysis of enzymatically radiolabeled products. The alcohols obtained by enzymatic dephosphorylation of the products formed by incubation of [1-14C]IPP and GPP (A), NPP (B), ω,E,E-FPP (C), and ω,E,E,E,GGPP (D) were analyzed by reverse-phase TLC as described in Materials and Methods. Arrows indicate the migration of authentic standards.
FIG. 5.
Effect of increasing substrate concentration on product chain length. TLC analysis of the radiolabeled products synthesized by Rv2361c in the presence of 30 μM [1-14C]IPP and 50 μM ω,E,Z-FPP (A) or 200 μM ω,E Z-FPP (B). The alcohols obtained by enzymatic dephosphorylation of the products formed were analyzed by reverse-phase TLC as described in Materials and Methods. Products labeled with [1-14C]IPP were visualized by a Bioscan System 200 imaging Scanner (Bioscan Inc). Standard polyprenols were located with an anisaldehyde spray reagent. The large peak that goes off the scale in both panels corresponds to the allylic substrate, E,Z-FPP.
DISCUSSION
The enzyme encoded by Rv2361c in M. tuberculosis is a decaprenyl diphosphate synthase. Purification of recombinant, histidine-tagged, active enzyme to near homogeneity was achieved by affinity chromatography on an IMAC. The enzyme is capable of catalyzing the addition of [14C]IPP to GPP, NPP, ω,E,E-FPP, ω,E,Z-FPP, or ω,E,E,E,GGPP. This observation indicates that this enzyme is not strictly specific for either the chain length or stereoconfiguration of the acceptor substrate. A number of other prenyl diphosphate synthases, which are capable of utilizing allylic primers of various chain lengths and configurations as substrates, have previously been reported (5, 9, 16, 21). However, this property contrasts with that of the other Z-prenyl diphosphate synthase found in M. tuberculosis (encoded by Rv1086), which has strict specificity for an allylic substrate containing 10 carbon atoms (26). Of the six allylic diphosphates tested as substrates for Rv2361c, only DMAPP did not serve as an acceptor. Interestingly, DMAPP also is not a substrate for the Z-prenyl diphosphate encoded by Rv1086, which synthesizes ω,E,Z-FPP, a molecule containing only 15 carbon atoms. This suggests that the M. tuberculosis genome encodes an unidentified GPP synthase to provide the required substrate for the cis-prenyl diphosphate synthases in M. tuberculosis.
Rv2361c has a strict requirement for divalent cations, as do all other prenyl-diphosphate synthases reported to date. These enzymes use the divalent cation to coordinate the diphosphate moieties of the substrates in the active site (18). Of the divalent cations tested, Mg2+ supports maximal activity of Rv2361c. Mn2+ also supported enzymatic activity, although at a significantly reduced rate. Zn2+ and Ca2+ supported little or no activity, and the addition of EDTA to the reaction mixtures completely abolished activity. The activity of the enzyme was slightly enhanced by the addition of Triton X-100 to the reaction mixtures, likely by increasing the solubility of the reaction products. However, the protein is predicted to be membrane bound with a single membrane-spanning region based on amino acid sequence, a prediction which is supported by experimental evidence (25). Thus, it is possible that the observed stimulation by the detergent is at least partially due to the physical nature of the protein.
Of the five allylic diphosphates that could be used as substrates, NPP, ω,E,E-FPP, and ω,E,E,E-GGPP all had Km values that were an order of magnitude lower than that for GPP or ω,E,Z-FPP. However, only the products generated by using these latter two compounds would have structures that are consistent with the known structure of decaprenyl phosphate (Fig. 1). Thus, these compounds are the only potential natural substrates in vivo. The 15-carbon molecule, ω,E,Z-FPP, appears to be the preferred allylic substrate for Rv2361c due to the fact that its catalytic efficiency is three orders of magnitude greater than that seen for GPP, thus confirming the hypothesis that the enzymes encoded by Rv1086 and Rv2361c are both part of the biosynthetic pathway leading to the formation of decaprenyl phosphate in M. tuberculosis (25).
When either GPP or NPP was used as the allylic acceptor, significant amounts of FPP (likely E,Z-FPP and Z,Z-PP, respectively) accumulated. It is not obvious why this should occur, but the accumulation may be due to a combination of high Km values (290 μM for E,Z-FPP) and the relatively greater solubility of these intermediates in water relative to the longer intermediates, which presumably are more tightly bound to the enzyme. Interestingly, citronellyl diphosphate, a C10 prenyl diphosphate in which the α-isoprene unit is saturated, did not inhibit Rv2361c as it did Rv1086 (26), even when tested at concentrations as high as 1 mM.
The distribution of chain lengths of the polyprenyl diphosphates synthesized by Rv2361c changes depending on the concentration of ω,E,Z-FPP used in the reaction; the amount of 40-carbon and 45-carbon products relative to 50-carbon products increases as the ω,E,Z-FPP concentration is increased, suggesting that the enzyme loses specificity as the substrate concentration increases. It has previously been reported that in some cases prenyl diphosphate synthases produce small amounts of intermediates and that the relative amounts of these intermediates change with the reaction conditions (11, 19, 22). This appears to be due to the fact that the allylic substrate added to the reaction can compete with newly synthesized allylic intermediates for binding to the active site and that the enzymatically synthesized intermediates can be displaced from the enzyme by the allylic substrate added to the reaction mixture (13).
In the case of Rv2361c, the chain length of the product synthesized also appears to be dependent on the stereoconfiguration of the allylic diphosphate used to prime the reaction. That is, when Rv2361c is incubated with ω,E,E,E-GGPP the enzyme produces octaprenyl diphosphate as the major product followed by nonaprenyl and decaprenyl diphosphate (Fig. 4D), suggesting that the ω,E,E,E stereoconformation of the omega end of longer allylic products either cannot be accommodated by the enzyme or that the substrate conformation affects the conformation of the protein itself, resulting in the formation of products with shorter chain lengths.
High-density transposon mutagenesis experiments have shown that Rv2361c is likely essential in M. tuberculosis H37Rv (24). Surprisingly, the enzyme encoded by Rv1086 is not essential (24), even though our data indicate that Rv1086 synthesizes the precursor used by Rv2361c for synthesis of decaprenyl diphosphate. However, the kinetic data presented in this report indicates that Rv2361c is also capable of synthesizing ω,E,Z-FPP and subsequently decaprenyl diphosphate from GPP, although at a much reduced catalytic efficiency, suggesting that Rv2361c can compensate for the deletion of Rv1086. Therefore, Rv1086 may not be a good drug target in M. tuberculosis as speculated earlier (26), even though synthesis of decaprenyl phosphate appears to be essential.
Acknowledgments
This work was supported by grants (AI18357 and AI49151) from the National Institute of Allergy and Infectious Disease, NIH.
REFERENCES
- 1.Allen, C. M., M. V. Kennan, and J. Sack. 1976. Lactobacillus plantarum undecaprenyl pyrophosphate synthetase: purification and reaction requirements. Arch. Biochem. Biophys. 175:236-248. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, R. G., H. Hussey, and J. Baddiley. 1972. The mechanism of wall synthesis in bacteria. The organization of enzymes and isoprenoid phosphates in the membrane. Biochem. J. 127:11-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apfel, C. M., B. Takacs, M. Fountoulakis, M. Stieger, and W. Keck. 1999. Use of genomics to identify bacterial undecaprenyl pyrophosphate synthetase: cloning, expression, and characterization of the essential uppS gene. J. Bacteriol. 181:483-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baba, T., and C. M. Allen. 1978. Substrate specificity of undecaprenyl pyrophosphate synthetase from Lactobacillus plantarum. Biochemistry 17:5598-5604. [DOI] [PubMed] [Google Scholar]
- 5.Baba, T., and C. M. Allen. 1980. Prenyl transferases from Micrococcus luteus: characterization of undecaprenyl pyrophosphate synthetase. Arch. Biochem. Biophys. 200:474-484. [DOI] [PubMed] [Google Scholar]
- 6.Baddiley, J. 1972. Teichoic acids in cell walls and membranes of bacteria. Essays Biochem. 8:35-77. [PubMed] [Google Scholar]
- 7.Besra, G. S., T. Sievert, R. E. Lee, R. A. Slayden, P. J. Brennan, and K. Takayama. 1994. Identification of the apparent carrier in mycolic acid synthesis. Proc. Natl. Acad. Sci. USA 91:12735-12739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Besra, G. S., C. B. Morehouse, C. M. Rittner, C. J. Waechter, and P. J. Brennan. 1997. Biosynthesis of mycobacterial lipoarabinomannan. J. Biol. Chem. 272:18460-18466. [DOI] [PubMed] [Google Scholar]
- 9.Crick, D. C., J. S. Rush, and C. J. Waechter. 1991. Characterization and localization of a long-chain isoprenyltransferase activity in porcine brain: proposed role in the biosynthesis of dolichyl phosphate. J. Neurochem. 57:1354-1362. [DOI] [PubMed] [Google Scholar]
- 10.Davisson, V. J., A. B. Woodside, and C. D. Poulter. 1985. Synthesis of allylic and homoallylic isoprenoid pyrophosphates. Methods Enzymol. 110:130-144. [DOI] [PubMed] [Google Scholar]
- 11.Ericsson, J., A. Thelin, T. Chojnacki, and G. Dallner. 1992. Substrate specificity of cis-prenyltransferase in rat liver microsomes. J. Biol. Chem. 267:19730-19735. [PubMed] [Google Scholar]
- 12.Fujihashi, M., N. Shimizu, Y. W. Zhang, T. Koyama, and K. Miki. 1999. Crystallization and preliminary X-ray diffraction studies of undecaprenyl diphosphate synthase from Micrococcus luteus B-P 26. Acta Crystallogr. D Biol. Crystallogr. 55:1606-1607. [DOI] [PubMed] [Google Scholar]
- 13.Fujii, H., H. Sagami, T. Koyama, K. Ogura, S. Seto, T. Baba, and C. M. Allen. 1980. Variable product specificity of solanesyl pyrophosphate synthetase. Biochem. Biophys. Res. Commun. 96:1648-1653. [DOI] [PubMed] [Google Scholar]
- 14.Higashi, Y., G. Siewert, and J. L. Strominger. 1970. Biosynthesis of the peptidoglycan of bacterial cell walls. XIX. Isoprenoid alcohol phosphokinase. J. Biol. Chem. 245:3683-3690. [PubMed] [Google Scholar]
- 15.Higashi, Y., J. L. Strominger, and C. C. Sweeley. 1967. Structure of a lipid intermediate in cell wall peptidoglycan synthesis: a derivative of a C55 isoprenoid alcohol. Proc. Natl. Acad. Sci. USA 57:1878-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishii, K., H. Sagami, and K. Ogura. 1986. A novel prenyltransferase from Paracoccus denitrificans. Biochem. J. 233:773-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kato, J.-I., S. Fujisaki, K.-I. Nakajima, Y. Nishimura, M. Sato, and A. Nakano. 1999. The Escherichia coli homologue of yeast RER2, a key enzyme of dolichol synthesis, is essential for carrier lipid formation in bacterial cell wall synthesis. J. Bacteriol. 181:2733-2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koyama, T. 1999. Molecular analysis of prenyl chain elongating enzymes. Biosci. Biotechnol. Biochemistry 63:1671-1676. [DOI] [PubMed] [Google Scholar]
- 19.Matsuoka, S., H. Sagami, A. Kurisaki, and K. Ogura. 1991. Formation of Z,E,E-geranylgeranyl diphosphate by rat liver microsomes J. Biol. Chem. 266:3458-3463. [PubMed] [Google Scholar]
- 20.Mikusova, K., M. Mikus, G. S. Besra, I. Hancock, and P. J. Brennan. 1996. Biosynthesis of the linkage region of the mycobacterial cell wall. J. Biol. Chem. 271:7820-7828. [DOI] [PubMed] [Google Scholar]
- 21.Muth, J. D., and C. M. Allen. 1984. Undecaprenyl pyrophosphate synthetase from Lactobacillus plantarum: a dimeric protein. Arch. Biochem. Biophys. 230:49-60. [DOI] [PubMed] [Google Scholar]
- 22.Ohnuma, S., T. Koyama, and K. Ogura. 1992. Chain length distribution of the products formed in solanesyl diphosphate synthase reaction. J. Biochem. 112:743-749. [DOI] [PubMed] [Google Scholar]
- 23.Raetz, C. R. 1996. Bacterial lipopolysaccharides: a remarkable family of bioactive macroamphiphiles, p. 1035-1063. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 24.Sassetti, C. M., D. H. Boyd, and E. Rubin. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. J. Mol. Microbiol. 48:77-84. [DOI] [PubMed] [Google Scholar]
- 25.Schulbach, M. C., P. J. Brennan, and D. C. Crick. 2000. Identification of a short (C15) chain Z-isoprenyl diphosphate synthase and a homologous long (C50) chain isoprenyl diphosphate synthase in Mycobacterium tuberculosis. J. Biol. Chem. 275:22876-22881. [DOI] [PubMed] [Google Scholar]
- 26.Schulbach, M. C., S. Mahapatra, M. Macchia, C. Papi. Barontini, F. Minutolo, S. Bertini, P. J. Brennan, and D. C. Crick. 2001. Purification, enzymatic characterization, and inhibition of the Z-farnesyl diphosphate synthase from Mycobacterium tuberculosis. J. Biol. Chem. 276:11624-11630. [DOI] [PubMed] [Google Scholar]
- 27.Shimizu, N., T. Koyama, and K. Ogura. 1998. Molecular cloning, expression, and purification of undecaprenyl diphosphate synthase. No sequence similarity between E- and Z-prenyl diphosphate synthases. J. Biol. Chem. 273:9476-19481. [DOI] [PubMed] [Google Scholar]
- 28.Takayama, K., and D. S. Goldman. 1970. Enzymatic synthesis of mannosyl-1-phosphoryl-decaprenol by a cell-free system of Mycobacterium tuberculosis. J. Biol. Chem. 245:6251-6257. [PubMed] [Google Scholar]
- 29.van Heijenoort, J. 1996. Murein synthesis, p. 1025-1034. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 30.Wolucka, B. A., M. R. McNeil, E. de Hoffman, T. Chojnacki, and P. J. Brennan. 1994. Recognition of the lipid intermediate for arabinogalactan/arabinomannan biosynthesis and its relation to the mode of action of ethambutol on mycobacteria. J. Biol. Chem. 269:23328-23335. [PubMed] [Google Scholar]
- 31.Wolucka, B. A., and E. de Hoffmann. 1998. Isolation and characterization of the major form of polyprenyl-phospho-mannose from Mycobacterium smegmatis. Glycobiology 8:955-962. [DOI] [PubMed] [Google Scholar]