Abstract
Chlamydiaphage Chp2 is a member of the family Microviridae, of which bacteriophage φX174 is the type species. Although grouped in the same family, the relationship between the Microviridae coliphages and the Chp2-like viruses, which infect obligate intracellular parasitic bacteria, is quite distant, with major differences in structural protein content and scaffolding protein dependence. To investigate the morphogenesis of Chp2, large particles were isolated from infected Chlamydophila abortus by equilibrium and rate zonal sedimentation. A monoclonal antibody that recognizes only assembled viral coat proteins was used in these detection assays. Thus, the detected particles represent virions and/or postcapsid formation assembly intermediates. Two distinct particle types were detected, differing in both protein and DNA content. Filled particles lacked VP3, the putative internal scaffolding protein, whereas empty particles contained this protein. These results indicate that VP3 is a scaffolding protein and that the isolated VP3-containing particles most likely represent Chp2 procapsids.
Although chlamydiae are widespread bacterial pathogens causing a wide range of illnesses, such as blindness, respiratory infections, and possibly coronary artery disease (23, 24, 26), detailed molecular mechanisms of pathogenesis will remain obscure as long as significant barriers hinder laboratory manipulation. One of these barriers is the lack of a stable gene transfer system. In recent years, five chlamydiaphages belonging to the family Microviridae, of which the prototype is bacteriophage φX174, have been isolated (11, 14, 22, 25). The mechanisms and techniques to package φX174 in vitro and in vivo are well defined (1, 12). Thus, the microviruses of chlamydiae have the potential of becoming the basis of a genetic transfer system.
Critical to this goal is the identification of a chlamydiaphage procapsid, the assembly intermediate into which DNA is packaged. Although the chlamydiaphages are within the same family as φX174, the relationship is distant. The family has a deep evolutionary divide between the φX174-like phages (coliphages) that infect free-living enterobacteria and the Chp-like phages infecting obligate intracellular parasites and mollicutes, such as the chlamydiae, Bdellovibrio bacteriovorus, and spiroplasma (3). The most striking differences are structural and morphogenetic (3, 5). Structurally, the φX174-like phages contain a large pentameric spike protein complex at each vertex of the T = 1 virion. These spikes and their requisite genes are not present in the Chp-like phages, which have elaborate viral coat protein protrusions on the three fold axes of symmetry not seen in the coliphages. Accordingly, structural differences have led to different morphogenetic requirements vis-à-vis scaffolding proteins. The φX174-like phages utilize two scaffolding proteins during assembly: an internal and external species, proteins B and D, respectively (10). While it is evident that the Chp-like phages do not encode an external scaffolding protein (3, 18), the exact identity or existence of the internal scaffolding protein remains obscure.
Amino acid sequence identity between the Chp-2-like VP3s and the internal scaffolding proteins of the φX174-like viruses is low. However, key amino acids known to make contacts between the φX174 internal scaffolding and coat proteins are conserved in Chp2-like particles (18). While VP3 is found in icosahedral particles, it is unclear whether these particles are virions or procapsids (the assembly intermediate into which DNA is packaged) or a mixture of the two. To define the assembly pathway of the Chp2 virions, post-coat protein assembly particles were isolated from Chp2-infected chlamydiae and characterized. Particles were analyzed for DNA and VP3 content, and the results indicated that VP3 is a scaffolding protein component of a putative procapsid species. The latter particle will be required for the development of a phage-based genetic transfer system for chlamydiae.
MATERIALS AND METHODS
Purification and separation of Chp2 virions and procapsids.
The Chp2-bearing strain of Chlamydophila abortus (strain MA) was propagated as previously described (8). Chp2-infected reticulate bodies were lysed by successive freezing and thawing. Cell debris was removed by low-speed centrifugation, and the supernatant was passed through 0.45- and 0.22-μm-pore filters. Virions and other post-coat assembly structures were isolated by two different protocols: pellet formation by rate zonal centrifugation (8) or equilibrium sedimentation in CsCl gradients (4). In the latter technique, material within the 1.25- to 1.35-g/cm3 density range was extracted for further analyses. Material was resuspended or dialyzed against the buffer used in the subsequent purification (4), layered over 5 to 30% sucrose gradients, and spun at 45,000 rpm for 75 min in a Beckman SW50.1Ti rotor. Gradients were divided into 50 0.1-ml fractions for analysis. Purification and plating assays for bacteriophages φMH2K and φX174, used as S value markers, have been previously described (3, 9).
ELISA protocols, MAbs, and polyclonal antiserum.
Enzyme-linked immunosorbent assay (ELISA) protocols and the monoclonal antibodies (MAbs) used in the assays have previously been described (8). The VP3 gene of Chp2 was cloned into the expression vector pRSETA as previously described (8). Recombinant VP3 expressed in Escherichia coli was purified from host proteins by metal affinity chromatography using the six-histidine tag introduced by the N-terminal fusion peptide as described by the manufacturer (Invitrogen Life Technologies). Hyperimmune antisera to purified recombinant VP3 were produced in Wistar rats. A primary immunization with 100 μg of VP3 was administered subcutaneously in complete Freund's adjuvant followed by five boosts (100 μg each) in incomplete Freund's adjuvant at 10-day intervals.
The ELISA to detect Chp2 using MAb 40 was used to detect VP3 in sucrose gradient fractions: briefly 3 μl of each gradient fraction was added to 100 μl of 0.05 M carbonate-bicarbonate buffer and used to coat immunoassay trays overnight. VP3 was detected by the same protocol with polyclonal serum to VP3. Bound anti-VP3 antibodies were detected with an anti-rat-horseradish peroxidase conjugate (ISL, Paignton, United Kingdom) and tetramethylbenzidine substrate.
Estimation of Chp2 DNA in sucrose gradient fractions by quantitative PCR.
The absolute number of Chp2 phage genomes in each sample was determined by real-time, quantitative PCR on an ABI PRISM 7700 sequence detection system (Applied Biosystems), using TaqMan chemistry. Primer and probe sequences were designed within the Chp2 VP2 gene: forward primer 1931F (1931CTGCGAAGCAAATTGCTAGAGAG1953), reverse primer 2044R (2044AAGCTAACATAGGGTTAAGGCCAG2021) and fluorescent, dual-labeled probe 1957T (car-boxyfluorescein-ATGGCTTTTCAGGAGCGCATGTCTAACA-carboxytetramethylrhodamine). Sucrose gradient fractions were diluted 100-fold in deionized water, and 5 μl was used in each PCR, which also contained the primers, probe, and TaqMan Universal PCR mix (Applied Biosystems). Forty-cycle PCR was performed according to the manufacturer's instructions. A recombinant plasmid, comprising the entire Chp2 genome cloned by its unique BamHI site into pUC18, was accurately quantified by A260 measurement, and serial dilutions were used as assay standards. All standard dilutions contained pBR322 (100 μg/ml) to block adsorption of DNA to the polypropylene tubes.
EM.
Gradient fractions were adsorbed onto Formvar-coated carbon grids for 2 min and then negatively stained with 0.75% phosphotungstic acid (pH 6.0) for 10 s and air dried. Electron microscopy (EM) grids were examined with a Hitachi H7000 transmission EM.
RESULTS AND DISCUSSION
Biochemical characterization of particles with assembled capsids.
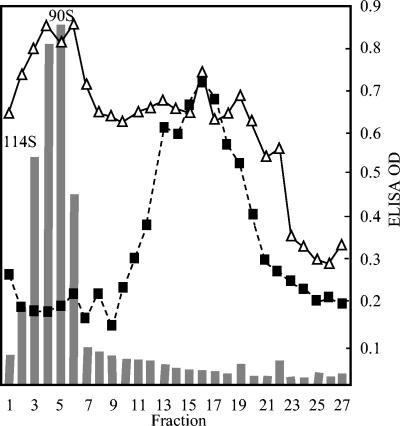
Lysates of Chp2-infected C. abortus were generated and processed by two protocols described in Material and Methods and layered onto 5 to 30% sucrose gradients. Duplicate gradients also containing 106 particles of φMH2K (90S) and φX174 (114S), used as S value markers, were also analyzed to ensure that the presence of the other bacteriophages did not lead to false positives in subsequent ELISA and quantitative PCR assays. Differences in the two purification protocols and the presence of other phages in the duplicate gradients did not affect the results of the subsequent assays and analyses.
After centrifugation, each gradient was divided into 50 0.1-ml fractions, which were assayed for the presence of the coat protein VP1 and the hypothesized scaffolding protein VP3. The MAb to the coat protein (MAb 40) only recognizes assembled coat protein (8). Thus, only virions and particles with assembled capsids would be detected. Assembled Chp2 coat protein was detected in fractions 3 to 7 and 12 to 17 (Fig. 1). Fraction 1 represents the bottom of the gradient. However, VP3 was only associated with the material in the slower-sedimenting peak (Fig. 1). S values were estimated by determining the location of bacteriophages φX174 (114S) and φMH2K (90S) from the gradient in direct plating assays and were found in greatest concentration in fractions 2 and 5, respectively (positions indicated in the figure). Thus, the faster-moving VP1-containing particles sediment at 90S, like mature φMH2K particles, and are likely to be Chp2 virions (see below). The VP3-containing particles were estimated to sediment at 55S.
FIG. 1.
Sedimentation profile of gradient-purified Chp2-related particles. Each fraction was assayed by ELISA for the presence of capsid (VP1) with MAb 40 (triangles, solid line) and procapsids with polyclonal antiserum to recombinant VP3 (squares, dashed line). Quantitative PCR (Taqman) was used to identify virions by the presence of genomic DNA (histogram). OD, optical density.
By the classical definition (16), a scaffolding protein is found in the viral procapsid, the empty assembly intermediate into which the genome is packaged, but not in the mature virion. To investigate further the particles detected by ELISAs, quantitative PCR assays were performed (Fig. 1, histogram). Whereas material from the 90S peak yielded a clear signal, the 55S material was not associated with DNA above background levels, suggesting the 55S particle represented the Chp2 procapsid.
Verification of particle identity by EM.
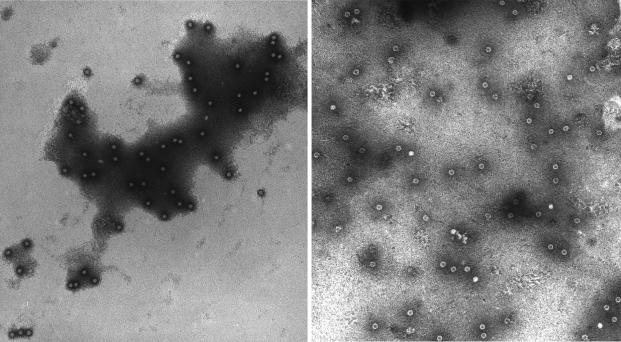
The 90S and 55S particles were examined by EM (Fig. 2). While the 55S particles were stain penetrated, the 90S particles were not. Stain penetration usually indicates a hollow structure, one in which DNA does not inhibit the internal saturation of the particle, and/or a structure with pores, as is often the case with viral procapsids, but not virions (2). Forty particles from each fraction were measured on electron micrographs, and the respective average diameters of the virion and procapsid were found to be 20.0 ± 0.1 and 23.4 ± 0.3 nm, respectively. The φX174 procapsid-to-virion transition involves a radial collapse of the maturing particle (6, 7, 19), rather than the expansion seen in many double-stranded DNA (dsDNA) viruses (15, 17, 21). The additional morphogenetic requirement of an external scaffolding protein during φX174 assembly has generally been believed to be responsible for this morphogenetic eccentricity. The external scaffolding protein outwardly restrains coat protein pentamers, thus preventing their contact in the procapsid (2). While the N terminus of the φX174 internal scaffolding protein may self-associate across two fold axes of symmetry, N-terminal sequences are not required for procapsid morphogenesis (20). The results of the analyses presented here demonstrate that capsid collapse is a general property of Microviridae assembly and is not solely the consequence of an external scaffolding protein, which the Chp2-like phages appear to lack.
FIG. 2.
Negative-stain EM picture of particles. (Left panel) Fraction 5 (virion). (Right panel) Fraction 16 (putative procapsid).
Conclusions.
Although the results of the analyses presented here do not directly determine an internal or external location for Vp3 within the procapsid, several observations strongly argue for an internal location. (i) The amino acid sequences of the Chp-2 like Vp3s are more closely related to the φX174-like internal scaffolding proteins than the external scaffolding proteins. (ii) Key amino acids known to make contacts between the φX174 internal scaffolding and coat proteins are conserved in the Chp2-like Vp3 and coat proteins (18). (iii) An internal location better explains two other observations about the particles described here and differences from the well-characterized φX174-like phages (10, 13). First, the difference in values of sedimentation rates between the Chp2 virion and procapsid is considerably larger than that observed for φX174, 114S and 108S, respectively. The φX174 procapsid contains 240 copies of the external scaffolding protein, which constitutes approximately 30% of the particles' mass. Secondly, no assembly intermediate corresponding to the φX174 provirion was detected. The φX174 provirion intermediate is found between the procapsid and virion in the assembly pathway as packaged particles that have lost the internal scaffolding protein, due to DNA packaging, but retain the external scaffolding protein. Studies on the morphogenesis of the Chp2-like phages will be useful in understanding Microviridae morphogenesis in the absence of an external scaffolding protein. The identity of both the early assembly intermediates and the molecular details of procapsid morphogenesis remain to be determined. Be that as it may, the ability to isolate procapsids is a critical step toward our longer-term aim of developing an in vitro packaging extract based on the chlamydiaphages.
Acknowledgments
This research was supported by research grants from the Wellcome Trust 063882 (I.N.C.), NSF MCB0234976 (B.A.F.), and USDA-Hatch funds to the University of Arizona.
REFERENCES
- 1.Aoyama, A., R. K. Hamatake., and M. Hayashi. 1981. Morphogenesis of φX174: in vitro synthesis of infectious phage from purified viral components. Proc. Natl. Acad. Sci. USA 78:7285-7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernal, R. A., S. Hafenstein, N. H. Olson, V. D. Bowman, P. R. Chipman, T. S. Baker, B. A. Fane, and M. G. Rossmann. 2003. Structural studies of bacteriophage alpha3 assembly. J. Mol. Biol. 325:11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brentlinger, K. L., S. Hafenstein, C. R. Novak, B. A. Fane, R. Borgon, R. McKenna, and M. Agbandje-McKenna. 2002. Microviridae, a family divided: isolation, characterization, and genome sequence of φMH2K, a bacteriophage of the obligate intracellular parasitic bacterium Bdellovibrio bacteriovorus. J. Bacteriol. 184:1089-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burch, A. D., and B. A. Fane. 2003. Analyses of putative conformation switching and cross-species inhibitory domains in Microviridae external scaffolding proteins. Virology 310:64-71. [DOI] [PubMed] [Google Scholar]
- 5.Chipman, P. R., M. Agbandje-McKenna, J. Renaudin, T. S. Baker, and R. McKenna. 1998. Structural analysis of the Spiroplasmavirus, SpV4: implications for evolutionary variation to obtain host diversity among the Microviridae. Structure 6:135-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dokland, T., R. McKenna, L. L. Ilag, B. R. Bowen, N. L. Incardona, B. A. Fane, and M. G. Rossmann. 1997. Structure of a viral assembly intermediate with molecular scaffolding. Nature 389:308-313. [DOI] [PubMed] [Google Scholar]
- 7.Dokland, T., R. A. Bernal, A. D. Burch, S. Pletnev, B. A. Fane, and M. G. Rossman. 1999. The role of scaffolding proteins in the assembly of the small, single-stranded DNA virus φX174. J. Mol. Biol. 288:595-608. [DOI] [PubMed] [Google Scholar]
- 8.Everson, J. S., S. A. Garner, B. Fane, B.-L. Liu, P. R. Lambden, and I. N. Clarke. 2002. Biological properties and cell tropism of Chp2, a bacteriophage of the obligate intracellular bacterium Chlamydophila abortus. J. Bacteriol. 184:2748-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fane, B. A., and M. Hayashi. 1991. Second-site suppressors of a cold-sensitive prohead accessory protein of bacteriophage φX174. Genetics 128:663-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fane, B. A., and P. E. Prevelige, Jr. 2003. Mechanism of scaffolding-assisted viral assembly. Adv. Protein Chem. 64:259-299. [DOI] [PubMed] [Google Scholar]
- 11.Garner, S. A., J. S. Everson, P. R. Lambden, B. A. Fane, and I. N. Clarke. 2004. Isolation, molecular characterisation and genome sequence of a bacteriophage (Chp3) from Chlamydophila pecorum. Virus Genes 28:207-214. [DOI] [PubMed] [Google Scholar]
- 12.Hafenstein, S., and B. A. Fane. 2002. φX174 genome-capsid interactions influence the biophysical properties of the virion: evidence for a scaffolding-like function for the genome during the final stages of morphogenesis. J. Virol. 76:5350-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayashi, M., A. Aoyama, D. L. Richardson, and M. N. Hayashi. 1998. Biology of bacteriophage φX174, p 1-71. In R. Calendar (ed.), The bacteriophages, vol. 2. Plenum Publishing Corporation, New York, N.Y.
- 14.Hsia, R. C., L. M. Ting, and P. M. Bavoil. 2000. Microvirus of Chlamydia psittaci strain Guinea pig inclusion conjunctivitis: isolation and molecular characterization. Microbiology 146:1651-1660. [DOI] [PubMed] [Google Scholar]
- 15.Jardine, P. J., and D. H. Combs. 1998. Capsid expansion follows the initiation of DNA packaging in bacteriophage T4. J. Mol. Biol. 284:661-672. [DOI] [PubMed] [Google Scholar]
- 16.King, J., and S. Casjens. 1974. Catalytic head assembling protein in virus morphogenesis. Nature 251:112-119. [DOI] [PubMed] [Google Scholar]
- 17.Lata, R., J. F. Conway, N. Cheng, R. L. Duda, R. W. Hendrix, W. R. Wikoff, J. E. Johnson, H. Tsuruta, and A. C. Steven. 2000. Maturation dynamics of a viral capsid: visualization of transitional intermediate states. Cell 100:253-263. [DOI] [PubMed] [Google Scholar]
- 18.Liu, B. L., J. S. Everson, B., Fane, P. Giannikopoulou, E. Vretou, P. R. Lambden, and I. N. Clarke. 2000. Molecular characterization of a bacteriophage (Chp2) from Chlamydia psittaci. J. Virol. 74:3464-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKenna, R., D. Xia, P. Willingmann, L. L. Ilag, S. Krishnaswamy, M. G. Rossmann, N. H. Olson, T. S. Baker, and N. L. Incardona. 1992. Atomic structure of single-stranded DNA bacteriophage φX174 and its functional implications. Nature 355:137-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Novak, C. R., and B. A. Fane. 2004. Functions of the N terminus of the phiX174 internal scaffolding protein, a protein encoded in an overlapping reading frame in a two scaffolding protein system. J. Mol. Biol. 335:383-390. [DOI] [PubMed] [Google Scholar]
- 21.Prasad, B. V., P. E. Prevelige, E. Marietta, R. O. Chen, D. Thomas, J. King, and W. Chiu. 1993. Three-dimensional transformation of capsids associated with genome packaging in a bacterial virus. J. Mol. Biol. 231:65-74. [DOI] [PubMed] [Google Scholar]
- 22.Read, T. D., C. M. Fraser, R. C. Hsia, and P. M. Bavoil. 2000. Comparative analysis of Chlamydia bacteriophages reveals variation localized to a putative receptor binding domain. Microb. Comp. Genomics 5:223-231. [DOI] [PubMed] [Google Scholar]
- 23.Saikku, P., M. Leinonen, K. Mattila, M. R. Ekman, M. S. Nieminen, P. H. Makela, J. K. Huttunen, and V. Valtonen. 1988. Serological evidence of an association of a novel chlamydia, TWAR, with chronic coronary heart disease and acute myocardial infarction. Lancet ii:983-986. [DOI] [PubMed] [Google Scholar]
- 24.Schacter, J., and C. R. Dawson. 1990. The epidemiology of trachoma predicts more blindness in the future. Scand. J. Infect. Dis. Suppl. 69:55-62. [PubMed] [Google Scholar]
- 25.Storey, C. C., M. Lusher, and S. J. Richmond. 1989. Analysis of the complete nucleotide sequence of Chp1, a phage which infects avian Chlamydia psittaci. J. Gen. Virol. 70:3381-3390. [DOI] [PubMed] [Google Scholar]
- 26.Thylefors, B., A. D. Negrel, R. Pararajasegaram, and K. Y. Dadzie. 1995. Global data on blindness. Bull. W. H. O. 73:115-121. [PMC free article] [PubMed] [Google Scholar]