Abstract
FtsA, a member of the ATPase superfamily that includes actin and bacterial actin homologs, is essential for cell division of Escherichia coli and is recruited to the Z ring. In turn, recruitment of later essential division proteins to the Z ring is dependent on FtsA. In a polar recruitment assay, we found that FtsA can recruit at least two late proteins, FtsI and FtsN, to the cell poles independently of Z rings. Moreover, a unique structural domain of FtsA, subdomain 1c, which is divergent in the other ATPase superfamily members, is sufficient for this recruitment but not required for the ability of FtsA to localize to Z rings. Surprisingly, targeting the 1c subdomain to the Z ring by fusing it to FtsZ could partially suppress a thermosensitive ftsA mutation. These results suggest that subdomain 1c of FtsA is a completely independent functional domain with an important role in interacting with a septation protein subassembly.
Cytokinesis in Escherichia coli is a complex process that relies on the intricate timing and placement of the Z ring. FtsZ assembly into the Z ring at mid-cell is proposed to provide a scaffold upon which at least 12 essential cell division proteins, including FtsA, ZipA, FtsEX, FtsK, FtsQ, FtsL, YgbQ, FtsW, FtsI, and FtsN, are recruited in a mostly linear order of dependency (6, 27). The resulting putative protein complex or divisome is required for the synthesis of the division septum and subsequent formation of new cell poles. FtsA, the second protein recruited to the Z ring, is a membrane-associated cytosolic protein (25) that is a member of a large family of ATPases that include actin, Hsp70, sugar kinases, and MreB (4). FtsA localization to mid-cell depends on prior assembly of FtsZ into the Z ring (3, 29) and is mediated via the C terminus of FtsZ (20, 32). Although an essential component of the divisome, with homologs identified in over 20 bacterial genera (21), the actual function of FtsA remains elusive.
FtsA is essential for cell division and for the recruitment of all other known essential proteins to the Z ring except for FtsZ and ZipA (6). ZipA is also required for recruitment of later proteins (17), although the need for ZipA in cell division, and therefore in late recruitment, can be bypassed by a gain-of-function mutation in FtsA (13). This result suggests that despite the established linear dependency of protein recruitment to the division septum, and some idea of binary protein interactions (10), most of the direct protein-protein relationships in the assembly process have yet to be shown conclusively. For example, the protein being recruited might directly interact with the immediately preceding protein in the pathway or a protein that localized earlier, such as FtsZ.
Recruitment of FtsA to the septum may help to stabilize the Z ring (24) as well as link FtsZ to integral membrane septal proteins such as FtsI and FtsN, which are recruited late in the division process (1, 31). FtsI, also known as PBP3, is a bitopic membrane protein with periplasmic transpeptidase activity involved exclusively in synthesis of septal peptidoglycan (22, 23). The formation of filamentous cells after inactivation of FtsI indicates that Z-ring constriction is inhibited until all septum synthesis components are recruited to the division complex (26). Domain-swapping experiments have shown that proper recruitment of FtsI may provide a checkpoint signal, as both the cytoplasmic and transmembrane domains are important for FtsI function, whereas the equivalent portions of other essential bitopic proteins FtsL, FtsQ, and FtsN are not (1, 9, 15).
Is there a potential domain of FtsA that might be implicated in the interaction with later proteins such as FtsI? One such candidate domain is subdomain 1c, which, as deduced from the recent crystal structure of Thermotoga maritima FtsA (30), is conserved among other FtsAs and occupies a different spatial position within the molecule compared to other members of the ATPase superfamily, including MreB (29). Recently, subdomain 1c has been shown to be essential for FtsA function (7). In a novel adaptation of a bacterial two-hybrid assay based on polar recruitment of proteins to the cell poles by DivIVA (11), we show here that FtsA, as well as subdomain 1c, can recruit downstream cell division proteins FtsI and FtsN to the cell poles independently of the Z ring. The independent activity of this domain was further suggested by the ability of an FtsZ-1c chimera to partially complement a thermosensitive ftsA mutant. These results suggest that one key role of FtsA, and subdomain 1c in particular, is to interact either directly or indirectly with an FtsI-FtsN subassembly.
MATERIALS AND METHODS
Growth media and conditions.
E. coli cells were grown in Luria broth (LB) medium supplemented with chloramphenicol (Cm) at 20 μg/ml, kanamycin at 50 μg/ml, ampicillin (Ap) at 100 μg/ml, or tetracycline at 10 μg/ml as needed. All cells were grown at 30°C unless otherwise stated. For FtsA-GFP and FtsAΔ1c-GFP localization studies, an overnight culture was diluted 1:200 in LB plus Cm, grown to early logarithmic phase, and induced with 40 μM IPTG (isopropyl-β-d-thiogalactoside) for 1.5 h. For localization of green fluorescent protein (GFP) fusions to FtsZ, FtsK, FtsI, FtsQ, FtsA, or FtsW in the bacterial two-hybrid polar recruitment assay, cells were grown to early log phase and induced simultaneously with 0.05% arabinose and either 10, 20, 40, 100, 80, or 80 μM IPTG, respectively, for 1.5 to 2 h. Arabinose induced expression of the DivIVA-FtsA or DivIVA-1c fusions, while IPTG induced expression of the GFP fusions.
Plasmid and strain construction.
Oligonucleotide primers used in plasmid constructions are shown in Table 1. To disrupt subdomain 1c of FtsA, two PCR products corresponding to the segments of FtsA N-terminal and C-terminal to the 1c subdomain were amplified, using primers A16 and AC1 for the C-terminal portion and A7 and A15 for the N-terminal portion. The two PCR products were cut with KpnI-BamHI (N terminus) and BamHI-AscI (C terminus) and cloned into pET28-FtsA (13) cut with KpnI-AscI to create an ftsA gene with an in-frame partial deletion of the 1c subdomain marked with a BamHI site (pWM1832). The resulting FtsAΔ1c protein contained a Gly-Ser substitution in place of residues R126-I161. The pET28-FtsA derivatives expressed FtsA at complementing levels from weak T7-independent vector transcription (13). The pBAD-FtsAΔ1c plasmid (pWM1941) and pLac-FtsAΔ1c-GFP fusion (pWM1909) were constructed by cloning the BglII-AscI fragment from pWM1832 into BglII-AscI-cut pBAD-FtsA (13), which is a pBAD33 derivative (14), and pWM633 (pAG-lacIq) (19), respectively. CH2/pDB280 (16), kindly supplied by Piet de Boer, was used as the FtsA depletion strain to test the functionality of pWM1832.
TABLE 1.
Oligonucleotide primers used in plasmid constructions
| Primer | Sequence (5′→3′) |
|---|---|
| AC1 | GGTTCTAGAATACGGCTCCTGAGCA |
| A7 | AACATATGATCAAGGCGACGGAC |
| A15 | AAGGATCCATGCTCATCGCGCACACGC |
| A16 | AAGGATCCACATGTCACAACGATATGG |
| A13 | AATCTAGAATGATCAAGGCGACGGACAG |
| A14 | TTCTGCAGCTCTCCGATTTGTGCCTGTC |
| Azip3 | TTTCTAGACTTTCTGGTAAGCACATCAG |
| Azip4 | CTTAAAGCTTGTTCAACCGCTTTGACGATG |
| FtsK1 | TTGAGCTCCCTGGAGAGCCTTTCTTGA |
| FtsK3 | AATCTAGAAAAGGTATCTACCGGC |
| FtsW1 | CAGAATTCAACAACAACAACATGCGTTTATCTCTCCCTC |
| FtsW2 | CCCTAAGCTTTCATCGTGAACCTCGTACAAACGC |
| QA2 | AAGGTACCGTCGGCCTCATAAAA |
| Q4 | TTCTGCAGTATGTCGCAGGCTGCTCT |
| FtsN1 | CGAGAATTCAACAACAACGCACAACGAGATTATG |
| FtsN2 | ACGAAGCTTTCAACCCCCGGCGGCGAG |
The pBAD-DivIVA-FtsA fusion plasmid (pWM1806) was constructed by replacing gfp in pWM1461 (also known as pZD1396, which is pBAD33-DivIVA-GFP [11]) with full-length ftsA amplified from chromosomal DNA with primers A13 and A14 and inserted as an XbaI-Pst1 fragment. The pBAD-DivIVA-1c fusion plasmid (pWM1814) was constructed by amplifying the entire subdomain 1c of ftsA (residues L83-E176 with several flanking residues) with pET28-FtsA (14) as template and primers Azip3 and Azip4 and replacing the XbaI-HindIII fragment containing gfp in pWM1461 with the resulting 0.3-kb XbaI-HindIII PCR product. In pWM1814, the 1c subdomain attached to DivIVA included 94 residues of FtsA (L83 to E176), along with an additional 17-amino-acid C-terminal tail encoded by the vector portion of pBAD33 (QAWLFWRMREDFQPDTD) and a 2-amino-acid N-terminal linker (SR). The pBAD-DivIVA plasmid (pWM2045) was made by filling in the XbaI site of pWM1461, creating a full-length DivIVA gene under pBAD control.
To make GFP-FtsK (pWM1801), a 2.7-kb SacI-XbaI fragment encoding residues 1 to 859 of the 1,329-amino-acid protein encoded by ftsK was amplified from E. coli genomic DNA with primers FtsK1 and FtsK3 and inserted into pDSW207 (31), which is compatible with the pBAD33 derivatives and has an IPTG-inducible pTrc promoter. To make GFP-FtsQ (pWM1800), full-length ftsQ was cloned from pWM708 and inserted into pDSW207 as an NcoI-HindIII fragment. Plasmid pWM708 was made by amplifying genomic ftsQ with QA2 and Q4 primers, cleaving the PCR product internally with PstI and PvuII, and cloning into a PstI-EcoRV-cut derivative of pBC(SK+). To make GFP-FtsW (pWM1818), full-length ftsW was amplified with primers FtsW1 and FtsW2 and cloned as an EcoRI-HindIII fragment into pDSW209 (31), a derivative of pDSW207 with a weakened pTrc promoter. To make GFP-FtsN (pWM1152), full-length ftsN was amplified with FtsN1 and FtsN2 primers and cloned as an EcoRI-HindIII fragment into pDSW207. A GFP-FtsA fusion compatible with pBAD33 (pWM1333) was made by cloning full-length ftsA downstream of GFP in pDSW209 with SacI and BamHI. To make the FtsZ-1c fusion in pBAD33, the SacI-XbaI fragment from pBAD-DivIVA-1c (pWM1814) containing divIVA was replaced with the SacI-XbaI fragment from pZG (19) containing full-length ftsZ lacking its stop codon.
To construct the ftsA12(ts) GFP-FtsI strain WM1793, WM1488 (EC436), a kind gift from David Weiss (31) containing gfp-ftsI (Apr) in single copy on the chromosome, was transduced to Tetr with a P1 phage grown on WM1115, a strain containing ftsA12(ts) and a linked leu:Tn10 (13). About 70% of Apr Tetr transductants could grow at 30°C but not 42°C, and one was chosen to be WM1793. The ftsZ84(ts) mutation was introduced into Cmr Apr strains carrying pWM1814 and either GFP-FtsN or GFP-FtsI by transducing the recipient strain with a P1 lysate grown on WM1109 (13) (ftsZ84 leu:Tn10), selecting for Tetr Cmr Apr transductants, and screening for those that grew at 30°C but not at 42°C. E. coli strains used included TX3772, a lacY-deficient derivative of the wild-type (WT) E. coli strain MG1655 (28), and Top10 (F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG).
The strain for DivIVA overproduction, WM1831, was BL21(DE3) (Novagen) carrying pWM1831, which contains the entire Bacillus subtilis divIVA gene from pWM1461 (11) inserted as an NheI-XhoI fragment into pET28a. This resulted in an N-terminal hexahistidine tag on DivIVA, with some intervening vector sequences, which was then used to purify the protein on an affinity resin as described previously (13).
Immunoblotting and protein quantitation.
To quantitate the levels of GFP-FtsI, GFP-FtsN, and DivIVA fusion proteins under the polar recruitment assay conditions, cells were harvested after growth with the appropriate inducers and checked for typical localization patterns, and then total protein from a known amount of cells was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis along with a concentration range of protein standards. These standards were either purified GFP or DivIVA, depending on whether GFP fusions or DivIVA fusions were being quantitated. The gels were then blotted onto nitrocellulose membranes and probed with anti-GFP (13) for the GFP-FtsN or GFP-FtsI analysis or with anti-DivIVA for DivIVA fusion protein analysis. Both antibodies were rabbit polyclonal preparations. Incubation with secondary anti-rabbit antibody conjugated to horseradish peroxidase was followed by chemiluminescence detection and quantitation of band intensities (13). Protein per cell values were estimated from the number of molecules of protein in the bands relative to those in the purified standards and divided by the cell equivalents loaded on the original gel.
Microscopy and image acquisition.
Microscopic examination of immobilized live cells and immunofluorescence of fixed cells with anti-FtsZ was performed as described previously (8). Fluorescence intensity plots were made with ImageJ freeware from the National Institutes of Health, starting from ∼0.5 μm outside each cell pole.
RESULTS
A deletion of subdomain 1c of FtsA blocks its full function but not its localization to Z rings.
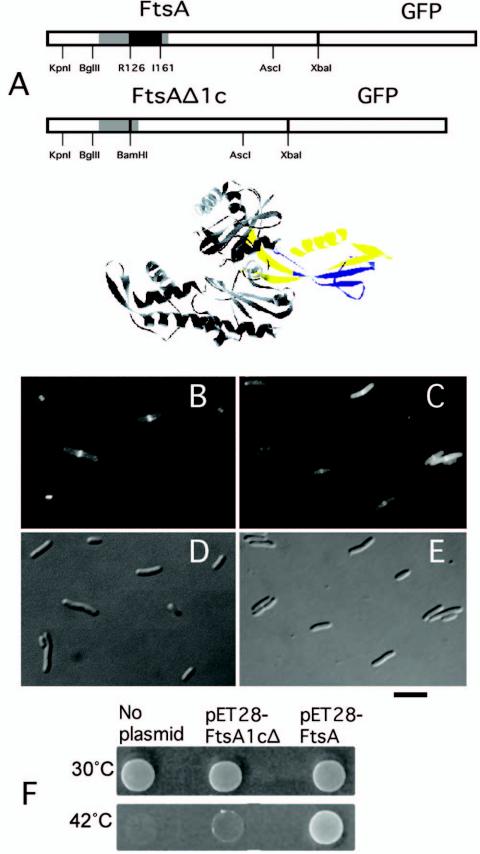
To investigate whether subdomain 1c was required for localizing FtsA to the Z ring or for interacting with downstream proteins, we constructed an ftsA gene with an in-frame deletion of part of the subdomain, removing sequences encoding 36 amino acids. This ftsAΔ1c construct was then fused to GFP, replacing WT FtsA in pLac-FtsA-GFP carried on pAG-lacIq (Fig. 1A). When induced with IPTG, the FtsAΔ1c-GFP fusion was able to localize to mid-cell in the majority of cells examined (Fig. 1C and E), although the level of fluorescence elsewhere in the cell was significantly higher than for FtsA-GFP (Fig. 1B and D). This higher background fluorescence was likely caused by instability of the GFP-tagged deleted protein, as indicated in immunoblots probed with anti-GFP (data not shown).
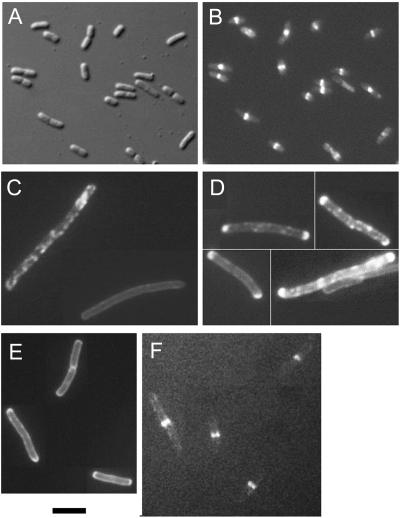
FIG. 1.
Effects of a deletion of subdomain 1c on FtsA function. (A) At the top are two linear schematics of FtsA-GFP and FtsAΔ1c-GFP fusions, showing restriction sites used in their construction. Black = deleted part of the 1c subdomain; gray = all of subdomain 1c; white = rest of FtsA and GFP. Restriction sites are shown below, in addition to the amino acid residues at the boundary of the deleted region of subdomain 1c in FtsAΔ1c. Below the schematics, the Thermotoga crystal structure of FtsA is shown, with subdomain 1c in yellow and the portion of this domain deleted to make FtsAΔ1c shown in blue. (B to E) Cells expressing FtsA-GFP from pWM633 (B and D) or FtsAΔ1c-GFP from pWM1909 (C and E) were grown in 40 μM IPTG for 4 h, and live cells were examined by fluorescence (B and C) or Nomarski (D and E) microscopy. (F) Complementation of an ftsA null strain (CH2/pDB280) by FtsA but not by FtsAΔ1c. CH2/pDB280 cells containing no extra plasmid, pET28-FtsA, or pET28-FtsAΔ1c were grown at 30°C, diluted 1:10, spotted onto LB plates, and incubated at the temperatures shown. At 42°C, the Cmr pDB280 plasmid containing ftsA was lost, depleting cellular FtsA. Bar, 5 μm.
To test whether the FtsAΔ1c protein was otherwise functional, we decided not to use the GFP fusion, because FtsA-GFP poorly complements ftsA mutants (19). Instead, ftsAΔ1c was used to replace the WT ftsA present on plasmids pET28-FtsA and pBAD-FtsA, which synthesize an FtsA protein with an N-terminal histidine tag (13) capable of complementing ftsA12(ts) or FtsA depletion strains under restrictive conditions in the absence of arabinose (Fig. 1F and data not shown.). However, pET28-FtsAΔ1c was unable to complement the FtsA depletion strain at 42°C (Fig. 1F). Moreover, pBAD-FtsAΔ1c failed to complement an ftsA12(ts) mutant at 42°C at several arabinose concentrations ranging from 0 to 0.2% (data not shown), suggesting that higher concentrations of FtsAΔ1c were not compensatory. Taken together, these results indicated that subdomain 1c is not required for localization of FtsA to the Z ring. The lack of complementation by FtsAΔ1c, even when expressed at higher levels in the presence of arabinose, also suggested that disruption of subdomain 1c prevents proper function of the protein.
Mislocalization of FtsA to the cell poles targets FtsI and FtsN to the poles.
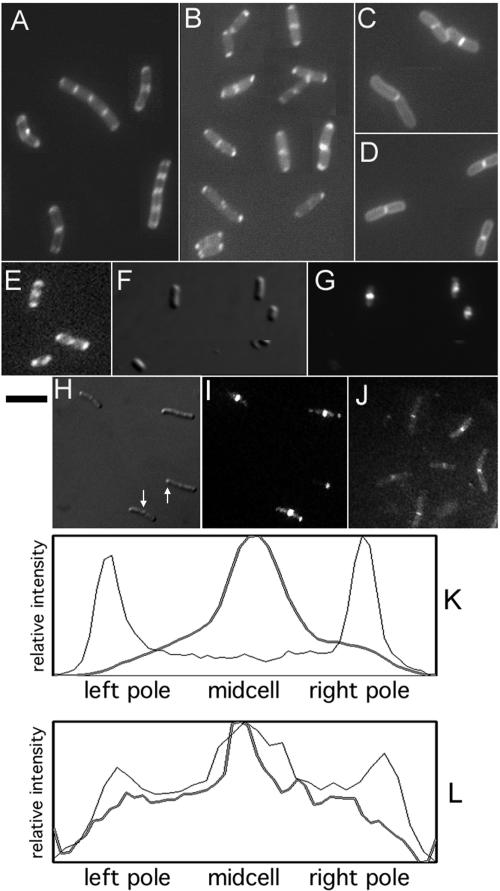
Because of the potential problems in interpreting the effects of the deletion of subdomain 1c, we took a different strategy to define how FtsA, and subdomain 1c in particular, might interact with downstream cell division proteins. To do this, we adapted a bacterial two-hybrid assay (11) that uses the DivIVA protein to recruit a protein complex to the cell poles. We reasoned that recruitment of later cell division proteins to the cell poles by FtsA, especially if independent of FtsZ and the Z ring, would make a much more compelling case for a direct role of FtsA in the interactions with late septation proteins. We started with a pBAD-DivIVA-GFP plasmid that, upon induction with arabinose, resulted in a fluorescence localization pattern at the poles of >95% of E. coli cells (Fig. 2E). As has been shown previously (12), fluorescence was also observed at mid-cell in longer cells that were closer to the next division (Fig. 2E, two top cells). The fluorescence intensity pattern along the length of a typical short cell expressing DivIVA-GFP is shown in Fig. 2K (light line); it is clear that the intensity at the poles far exceeded the fluorescence at other positions.
FIG. 2.
Localization of FtsA to the cell poles targets other cell division proteins. Cells were induced with arabinose (E), IPTG (C and D), or arabinose plus IPTG (A, B, and F to J) and examined by fluorescence microscopy. (A and C) DivIVA-FtsA(pWM1806) plus GFP-FtsI (in WM1488). (B and D) DivIVA-FtsA(pWM1806) plus GFP-FtsN (pWM1152) in TX3772. (E) DivIVA-GFP(pWM1461) in Top10. (F and G) IFM staining for FtsZ on cells used for panel B (Nomarski [F] or fluorescence [G]). (H and I) DivIVA-FtsA(pWM1806) plus FtsZ-GFP in TX3772 (Nomarski [HN] or fluorescence [I]). (J) DivIVA-FtsA(pWM1806) plus GFP-FtsQ(pWM1800) in TX3772. (K and L) Fluorescence intensity profiles along the length of a representative cell from panel E (K, light line), panel G (K, heavy line), panel B (L, light line), and panel J (L, heavy line). Arrows in panel H highlight visible inclusion bodies. Bar, 5 μm.
The GFP was then replaced with FtsA, and the pBAD-DivIVA-FtsA plasmid was introduced into strains containing GFP fusions to various cell division proteins under the control of IPTG. We found that GFP fusions to FtsI and FtsN could be recruited to the poles of 30 to 50% of cells upon addition of both arabinose and IPTG (Fig. 2A and B, respectively). A fluorescence profile of a typical cell coexpressing DivIVA-FtsA and GFP-FtsN is shown in Fig. 2L, showing a peak of fluorescence at mid-cell and secondary peaks at both poles. In contrast, induction with IPTG alone resulted in fluorescent bands at mid-cell, whereas the rest of the cell including the poles had only weak, uniform fluorescence (Fig. 2C and D). We almost never saw the unipolar and sometimes bipolar localization of GFP-FtsI that was observed previously upon protein overproduction (5, 31), perhaps because we used a shorter induction time and examined live cells instead of fixed cells. With arabinose only, fluorescence was either not detectable or observed as a generalized peripheral pattern throughout the cells, not at mid-cell or the poles (data not shown). Cells coexpressing DivIVA-FtsA and GFP alone from pDSW207 exhibited uniform fluorescence, with no significant peaks or valleys (data not shown).
FtsZ-GFP, normally only localized at mid-cell, also partially relocalized to the poles when DivIVA-FtsA was coexpressed (Fig. 2H and I). However, immunofluorescence microscopy (IFM) staining for native FtsZ in cells expressing DivIVA-FtsA showed only weak polar fluorescence and cells that were not elongated (Fig. 2F and G), suggesting that native FtsZ was not strongly relocalized to the poles by DivIVA-FtsA. A fluorescence profile of one such cell is shown in Fig. 2K. These results suggest that DivIVA-FtsA cannot displace significant quantities of FtsZ from the Z ring, but that excess FtsZ in the form of FtsZ-GFP can be attracted to the poles via DivIVA-FtsA. Inclusion bodies were sometimes observed with FtsZ-GFP (Fig. 2H), but not with GFP fusions to FtsI or FtsN. These results are consistent with known weak interactions between FtsZ and FtsA and indicate that the polar recruitment assay is sufficiently sensitive to detect the known interactions among these proteins under certain conditions.
The recruitment of GFP-FtsI and GFP-FtsN to the cell poles, on the other hand, suggested that a subassembly of the division machinery was relocated to a new location in the cell because of the relocation of FtsA. As recruitment of GFP-FtsN to the division site depends on FtsI (B. D. Corbin and W. Margolin, unpublished data), this result also suggested that the FtsI recruited to the poles by DivIVA-FtsA might be functional for recruitment.
The ability of DivIVA-FtsA to attract FtsN and FtsI to the cell poles prompted us to test whether FtsA might also be able to recruit other late division proteins to the poles. Plasmids containing GFP fusions to FtsW, FtsQ, or a large N-terminal fragment of FtsK, all of which were derivatives of the same plasmid used to express GFP-FtsN, were used to coexpress these fusions with DivIVA-FtsA. Induction with IPTG alone resulted in fluorescent bands at the middle of most cells for all three fusions (data not shown), indicating that the fusion proteins were expressed and able to localize. However, addition of arabinose to induce coexpression of DivIVA-FtsA did not cause relocalization of the mid-cell fluorescence. A typical field of cells (Fig. 2J) and a fluorescence profile of a typical cell (Fig. 2L) coexpressing DivIVA-FtsA and GFP-FtsQ are shown. In contrast to cells with GFP-FtsN, it is clear that there is no more fluorescence at the poles than at other positions. These results suggest that GFP fusions to FtsW, FtsK, or FtsQ, while able to localize to mid-cell in many cells, were not detectably recruited to the poles by DivIVA-FtsA. This also suggests that the presence of small amounts of FtsZ at the poles is not sufficient to attract detectable levels of FtsW, FtsK or FtsQ under these conditions.
FtsA subdomain 1c is sufficient to recruit FtsI and FtsN to the poles when it is fused to DivIVA.
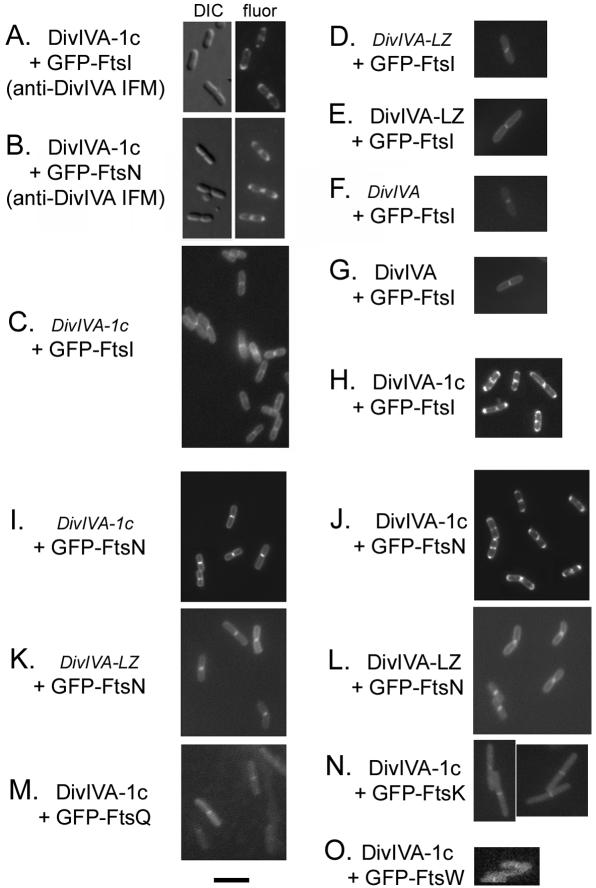
Because FtsAΔ1c localized to mid-cell but failed to complement, it suggested that subdomain 1c might be important for interacting with later proteins. We therefore investigated whether it might be sufficient to recruit GFP-FtsI and GFP-FtsN to the poles. A 94-amino-acid segment including all of subdomain 1c was used to replace the full-length FtsA in the DivIVA-FtsA construct to make DivIVA-1c. This plasmid was then tested in the same polar recruitment assay as described above. We first confirmed that the DivIVA-1c chimeric proteins exhibited the same localization pattern as DivIVA under our assay conditions. Cells expressing DivIVA-1c and either GFP-FtsI or GFP-FtsN were fixed and examined by IFM with anti-DivIVA antibodies. Cell poles and midpoints displayed strong staining relative to the rest of the cell, indicating that DivIVA-1c was targeted to the same sites as DivIVA-GFP (Fig. 3A and B). IFM with anti-FtsA polyclonal antibodies gave similar patterns (data not shown). Control cells not expressing DivIVA-1c showed no clear localization pattern in IFM experiments (data not shown), confirming that the IFM patterns seen with DivIVA-1c-expressing cells were specific.
FIG. 3.
Polar recruitment of FtsN and FtsI by subdomain 1c of FtsA. Individual or representative fields of cells examined for GFP fluorescence are shown. (A and B) IFM, using anti-DivIVA, of cells induced with IPTG plus arabinose, expressing DivIVA-1c (pWM1814) plus GFP-FtsI (in WM1488) (A) or DivIVA-1c (pWM1814) plus GFP-FtsN (pWM1152) (B). (C to O) GFP fluorescence from live cells. Regular typeface indicates that expression of the fusion protein was induced by arabinose (all DivIVA derivatives) or IPTG (all GFP fusions), while italics indicate that the plasmid encoding the fusion protein was present but the expression of the fusion was not induced. (C and H) DivIVA-1c (pWM1814) plus GFP-FtsI (in WM1488); (D and E) DivIVA-LZ (pZD1396) plus GFP-FtsI (in WM1488); (F and G) DivIVA (pWM2045) plus GFP-FtsI (in WM1488); (I and J) DivIVA-1c (pWM1814) plus GFP-FtsN (pWM1152) in TX3772; (K and L) DivIVA-LZ (pZD1396) plus GFP-FtsN (pWM1152) in TX3772; (M) DivIVA-1c (pWM1814) plus GFP-FtsQ (pWM1800) in TX3772; (N) DivIVA-1c (pWM1814) plus GFP-FtsK (pWM1801) in TX3772; (O) DivIVA-1c (pWM1814) plus GFP-FtsW (pWM1818) in TX3772. Bar, 5 μm.
Remarkably, just like DivIVA-FtsA, DivIVA-1c was able to recruit GFP-FtsI and GFP-FtsN to the poles of 30 to 50% of cells with both arabinose and IPTG (Fig. 3H and J). IPTG alone, on the other hand, resulted in mid-cell fluorescence only, with no polar fluorescence (Fig. 3C and I). These results indicated that the polar fluorescence was dependent on arabinose-induced expression of DivIVA-1c. To be sure that the 1c subdomain and not DivIVA was responsible for the polar recruitment, we replaced DivIVA-1c with DivIVA(pWM2045) or DivIVA-LZ (leucine zipper) in pZD1396 (11). When DivIVA-LZ or DivIVA was coexpressed with GFP-FtsN or GFP-FtsI, no polar fluorescence was observed (Fig. 3D to G, K, and L), indicating that the polar fluorescence was a result of interaction with the FtsA or subdomain 1c moiety and not with DivIVA itself.
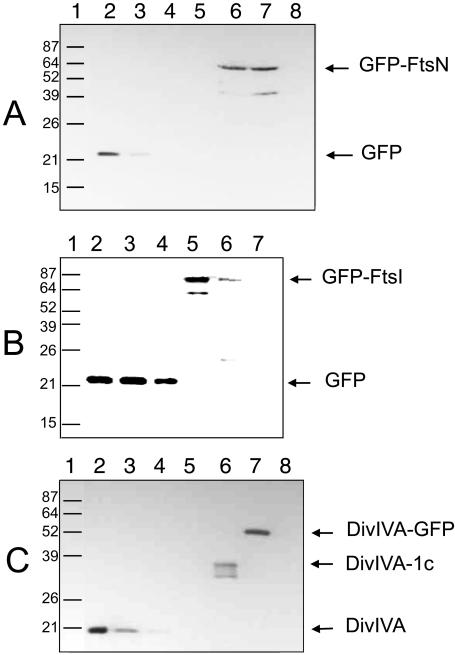
Immunoblot analysis of cells used for the microscopy experiments indicated that GFP-FtsN, GFP-FtsI, and DivIVA-1c were present at ∼8,000, 200, and 106 molecules per cell, respectively, when polar recruitment was observed with IPTG plus arabinose (Fig. 4). GFP-FtsN levels were unchanged in the absence of arabinose, although some protein degradation was detectable in the presence of arabinose (Fig. 4A). GFP-FtsI displayed significant protein degradation, particularly in the presence of arabinose, where the full-length fusion was less abundant compared to cells in the absence of arabinose (Fig. 4B). This rules out the possibility that increased levels of GFP fusion proteins resulted in their increased localization at cell poles in the presence of DivIVA-1c.
FIG. 4.
Immunoblotting of GFP and DivIVA fusion proteins in cells under assay conditions. Immunoblots of sodium dodecyl sulfate-polyacrylamide gel electrophoresis separations of cellular extracts or purified protein standards are shown, probed with rabbit polyclonal anti-GFP (A and B) or anti-DivIVA (C). (A) Immunoblotting of GFP-FtsN. Lane 1, protein molecular mass markers in kilodaltons; lanes 2 to 5, purified GFP protein (74, 45, 30, and 15 ng, respectively); lane 6, pWM1152 (GFP-FtsN) plus pWM1814 (DivIVA-1c) in TX3772 cells induced with IPTG; lane 7, same as lane 6 but induced with IPTG and arabinose; lane 8, cell lysate from TX3772. (B) Immunoblotting of GFP-FtsI. Lane 1, protein molecular mass markers in kilodaltons; lanes 2 to 4, purified GFP protein (120, 60, and 30 ng, respectively); lane 5, pWM1814 (DivIVA-1c) in WM1488 (GFP-FtsI) induced with IPTG; lane 6, same as lane 5 but induced with IPTG and arabinose; lane 7, cell lysate from TX3772 cells. (C) Immunoblotting of DivIVA-1c. Lane 1, protein molecular mass markers in kilodaltons; lanes 2 to 5, purified DivIVA protein (3,000, 2,000, 1,000, and 500 ng, respectively); lane 6, pWM1814 (DivIVA-1c) in WM1488 (GFP-FtsI) induced with arabinose; lane 7, DivIVA-GFP expressed from pWM1461 in Top10; lane 8, cell lysate from TX3772 cells.
Like DivIVA-FtsA, DivIVA-1c was not able to recruit several other late cell division proteins to the cell poles, at least not detectably. When GFP fusions to FtsQ, FtsK, or FtsW were expressed by IPTG induction, the majority of cells showed a fluorescent band or focus at mid-cell. When DivIVA-1c was coexpressed with the GFP fusions by induction with arabinose, this pattern did not change significantly (Fig. 3M to O).
Polar recruitment by subdomain 1c is independent of FtsZ and FtsA.
One of the important potential conclusions from the above results is that FtsA can interact with FtsI and FtsN independently of FtsZ or the Z ring. Because FtsA and FtsZ interact and the potential interaction between subdomain 1c and FtsZ is unknown, we wanted to rule out that FtsZ was involved in this polar recruitment.
To make a case for FtsZ independence, cells coexpressing DivIVA-FtsA or DivIVA-1c and GFP-FtsN or GFP-FtsI were stained for FtsZ by IFM. While 20 to 50% of cells prior to fixation exhibited peaks of polar fluorescence resulting from the recruitment of GFP-FtsI or GFP-FtsN to the poles by DivIVA-FtsA or DivIVA-1c, >80% of the cells stained with FtsZ showed only mid-cell fluorescence. This frequency of mid-cell staining is fairly typical for FtsZ IFM studies. Nevertheless, <5% of the cells exhibited detectable fluorescence at the poles, which was usually very weak (Fig. 5A and B). Therefore, if FtsZ were being attracted to the poles by DivIVA-FtsA or DivIVA-1c, it was at a frequency and intensity insufficient to explain the high frequency and intensity of polar fluorescence observed in the same cells with GFP-FtsI or GFP-FtsN.
FIG. 5.
Polar recruitment of FtsN and FtsI by subdomain 1c is independent of FtsZ and FtsA. (A and B) TX3772 cells coexpressing DivIVA-1c (pWM1814) and GFP-FtsN (pWM1152) with arabinose plus IPTG were fixed and stained for FtsZ by IFM. Panels A and B show the same field of cells visualized by Nomarski and fluorescence. (C to E) Cells were grown at 30°C, induced with IPTG (C) or arabinose plus IPTG (D and E), then shifted to 42°C for 20 min to inactivate FtsZ, and examined for GFP fluorescence. (D) GFP-FtsN (pWM1152) plus DivIVA-1c (pWM1814) plus ftsZ84(ts) in TX3772; (E) GFP-FtsI in WM1488 plus DivIVA-1c (pWM1814) plus ftsZ84(ts) in TX3772. (F) Cells induced with arabinose plus IPTG, coexpressing DivIVA-1c (pWM1814) and GFP-FtsA (pWM1333). Bar, 5 μm.
To provide further evidence that FtsA-mediated interaction with FtsI and FtsN was independent of the Z ring, we introduced the thermosensitive ftsZ84 allele by P1 transduction into the strains containing DivIVA-1c and GFP-FtsN or GFP-FtsI to test if the polar fluorescence would still localize upon inactivation of FtsZ assembly. When the growth temperature is shifted from 30 to 42°C, Z rings in ftsZ84(ts) cells rapidly disappear and cells immediately stop dividing and form nonseptate filaments (2). We coexpressed DivIVA-1c and GFP-FtsN or GFP-FtsI by inducing with IPTG and arabinose at 30°C and then shifted to 42°C for 20 min. Despite the inactivation of FtsZ, strong polar fluorescence of GFP-FtsN or GFP-FtsI was still observed in 30 to 50% of the cells (Fig. 5D to E). This pattern is similar to that observed with DivIVA-GFP in an ftsZ(ts) mutant (12). IFM of these cells coexpressing DivIVA-1c and GFP-FtsI confirmed that Z rings were absent in nearly all the cells (data not shown). Expression of GFP-FtsN but not DivIVA-1c in ftsZ84(ts) cells at 42°C resulted in peripheral fluorescence with no bias towards the cell poles (Fig. 5C), confirming that DivIVA-1c was necessary for the preferential polar localization of the GFP fusion protein in the absence of functional FtsZ.
It was recently proposed that subdomain 1c, in addition to the C terminus (33), is involved in dimerization of FtsA (7). If this were true, then one possible mechanism to explain the independent recruitment activity of DivIVA-1c would be that WT FtsA might actually bind to the 1c moiety on the DivIVA-1c protein, resulting in the recruitment of FtsI and FtsN by FtsA itself and not by subdomain 1c. To address whether such a bridging interaction might be involved, we coexpressed GFP-FtsA and DivIVA-1c to determine whether any fluorescence was detectable at the cell poles, which would be indicative of an FtsA-DivIVA-1c interaction. Whereas GFP-FtsA fluorescence localized to mid-cell in 50 to 70% of cells examined, GFP-FtsA fluorescence was not detectable at the cell poles above background levels (Fig. 5F). Although we cannot rule out the possibility that the GFP tag inhibits the interaction between FtsA and subdomain 1c, the simplest interpretation of the data is that FtsA is not present in sufficient quantities at the poles to cause the intense polar fluorescence observed with GFP-FtsN or GFP-FtsI recruited there by DivIVA-1c. These results suggest that subdomain 1c interacts with GFP-FtsI and GFP-FtsN by itself and not with the help of full-length FtsA.
Tethering FtsA subdomain 1c to FtsZ partially complements an ftsA(ts) cell division defect.
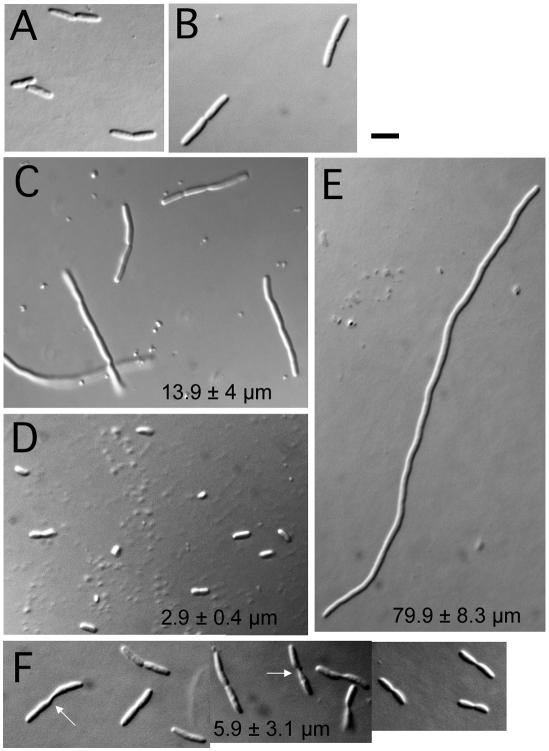
If the 1c subdomain of FtsA were truly an independent protein interaction module as suggested by the previous results, we predicted that it might be able to recruit proteins to the division site in the absence of functional FtsA if it were tethered to another cell division protein. To test this idea, we fused subdomain 1c to the C terminus of FtsZ, which normally interacts with FtsA. The FtsZ-1c construct was cloned into pBAD33, similar to the DivIVA-1c construction. This plasmid, pBAD-FtsZ-1c, was introduced into the GFP-FtsI strain that also contained the ftsA12(ts) allele in the chromosome (WM1793). WM1793 cells, like others containing ftsA12(ts), divided fairly normally at 30°C (Fig. 6A and B) but became highly filamentous at 37 and 42°C (Fig. 6E and data not shown; see also reference 3).
FIG. 6.
Tethering of subdomain 1c to FtsZ can partially suppress filamentation in an ftsA(ts) mutant, as shown in Nomarski images of WM1793 (ftsA12) cells containing or lacking the pBAD-FtsZ-1c plasmid and grown under the stated conditions. pBAD-FtsZ-1c (A) or no plasmid (B) at 30°C; no plasmid (C) or pBAD-FtsZ-1c (D) shifted from 30°C to 37°C for 90 min; no plasmid (E) or pBAD-FtsZ-1c (F) shifted from 30°C to 37°C for 3 h. Panel F is a composite of several fields of cells, emphasizing cells having difficulty dividing; arrows highlight elongated septa. Average cell size statistics from multiple fields of cells under each condition for panels C to F are listed in the appropriate panel. Bar, 5 μm.
Remarkably, however, this plasmid partially rescued the filamentous cell phenotype of WM1793 at 37°C in the absence of arabinose. When cells were shifted from 30 to 37°C for only 90 min, WM1793 cells carrying pBAD-FtsZ-1c (Fig. 6D) were the same average length as WT cells, while cells without the plasmid or with pBAD33 vector were on average four to six times longer (Fig. 6C). After a longer incubation at 37°C for 2 h, cells carrying pBAD-FtsZ-1c were somewhat longer than WT cells, but cells without the plasmid were on average over 10 times longer (data not shown). Finally, after 3 h, cells without pBAD-FtsZ-1c were on average 13 times longer than cells with the plasmid (Fig. 6E and F). WM1793 cells with just pBAD33 vector or with pBAD-FtsZ lacking the 1c extension formed long filaments in the absence of arabinose, similar to those without the plasmid (data not shown). Induction of GFP-FtsI with IPTG showed that most septa in the cells containing pBAD-FtsZ-1c at 37°C were fluorescent (data not shown). Comparison with known quantities of purified FtsZ on immunoblots probed with anti-FtsZ indicated that the amount of FtsZ-1c protein sufficient to suppress ftsA12(ts) at 37°C was ∼200 to 300 molecules per cell (data not shown), similar to estimated levels of native FtsA and approximately 100-fold less than that for WT FtsZ (18).
These results indicated that cell division was largely but not completely restored by FtsZ-1c at 37°C. Interestingly, the longer cells expressing FtsZ-1c after extended incubation at 37°C often exhibited blunt or twisted septa (Fig. 6F), providing evidence that septation in cells with FtsZ-1c was not completely normal. In addition, we were not able to observe significant suppression of cell filamentation or colony formation by FtsZ-1c at 42°C with a range of arabinose concentrations, suggesting either that the FtsA12 protein has an additional defect at 42°C or that the FtsZ-1c chimera is itself thermosensitive. Nevertheless, the partial suppression of filamentation by FtsZ-1c when a mutant FtsA is thermoinactivated supports the idea that the 1c subdomain of FtsA has independent activity.
DISCUSSION
Our complementation and localization results strongly suggest that subdomain 1c of FtsA is dispensable for localization of FtsA to the Z ring but is essential for some other function. To test whether this function was in protein-protein interactions, we used a DivIVA-mediated polar recruitment assay. We found that FtsA as well as subdomain 1c interacted with a divisome subassembly that includes FtsI and FtsN. Control experiments in which DivIVA alone was coexpressed with GFP-FtsI or GFP-FtsN, or in which GFP-FtsI or GFP-FtsN were expressed without a DivIVA fusion, did not result in polar fluorescence, suggesting that the interaction is specific. The curved appearance of the polar fluorescence coupled with mid-cell fluorescence typical of DivIVA localization also indicated that the interaction was a specific DivIVA-mediated polar interaction and not a nonspecific polar accumulation of inclusion bodies.
What distinguishes this assay from the established mid-cell localization assays is that interactions among cell division proteins can be explored in the absence of the Z ring. In this study, several independent experiments, including a polar recruitment assay performed in the absence of functional FtsZ, suggested that the interaction between subdomain 1c of FtsA and late septation proteins FtsI and FtsN is independent of the Z ring. This suggests that subdomain 1c can interact with later cell division proteins in FtsZ-independent subassemblies.
Because localization of GFP-FtsN, like that of FtsN, depends upon FtsI, polar relocalization of GFP-FtsN by DivIVA-FtsA or DivIVA-1c may occur via recruitment by FtsI. It is unlikely that FtsA interacts with FtsN directly, given the dispensability of the cytoplasmic tail of FtsN. While it is possible that subdomain 1c is important for the formation of FtsA dimers, as was proposed recently, we favor the idea that the primary function of subdomain 1c is to specifically contact another protein, either FtsI or another protein that interacts with FtsI. A recent bacterial two-hybrid study showed an interaction between FtsA and FtsI, which is consistent with our findings (10).
Recruitment of FtsI and FtsN to the Z ring requires not only FtsA but also all the other essential septation proteins in the intervening assembly pathway. Yet, in the mislocalized recruitment system used here, several of the latter proteins, such as FtsK, FtsQ, and FtsW, were not detectably targeted to the cell poles by DivIVA-FtsA or DivIVA-1c. Moreover, cells expressing DivIVA-FtsA were not filamentous, indicating that the polar recruitment of cell division proteins does not titrate out a limiting septation factor from the Z ring. One possible explanation for this latter observation is based on the fact that DivIVA normally localizes to both the division septum and cell poles. As a result, DivIVA-FtsA or DivIVA-1c may be able to recruit these other proteins to mid-cell, where they could potentially assist in septation, negating the predicted titration effect.
A potential explanation for the lack of polar recruitment of FtsK, FtsQ, and FtsW stems from the dispensability of the essential cell division protein ZipA in the presence of an R286W mutation in FtsA (13). Although ZipA is required to recruit a number of later septation proteins (17), the dispensability of ZipA in an ftsA mutant indicates that ZipA does not directly recruit the later proteins but instead enhances the ability of another septal component to recruit them and that this function could be reproduced by the altered FtsA. One possible model, by analogy, is that FtsA is not directly involved in recruitment of downstream proteins other than FtsI and FtsN but instead enhances the ability of another septal component to recruit them. The Z-ring-independent polar recruitment of FtsI and FtsN by FtsA, on the other hand, indicates that FtsA may have a direct role in recruitment of these two late proteins to the Z ring. Alternatively, FtsA may not normally recruit FtsI and FtsN to the Z ring until it is activated by another later septation protein, such as FtsW. In our system, it might be that polar targeting by DivIVA bypasses this activation requirement.
Another intriguing possible explanation of our data is that the polar recruitment of FtsI and FtsN by FtsA reflects a potentially regulatory interaction between FtsI-FtsN and FtsA that normally occurs after the entire septal protein complex is in place. This model is attractive because it might give a clue as to how the Z ring receives a signal from the completed septal machinery that it is ready to contract. Such a trigger must exist, but it is unknown. Work is in progress to address these various models, as well as to define further the protein-protein interactions implicated in this work.
Acknowledgments
We thank T. Xue for help with making plasmid constructs, D. Weiss for EC436, P. de Boer for CH2/pDB280, K. Rose for DivIVA purification, and Z. Ding and P. Christie for pZD1396.
This work was supported by a grant from the National Institutes of Health (R01-GM61074).
REFERENCES
- 1.Addinall, S. G., C. Cao, and J. Lutkenhaus. 1997. FtsN, a late recruit to the septum in Escherichia coli. Mol. Microbiol. 25:303-309. [DOI] [PubMed] [Google Scholar]
- 2.Addinall, S. G., C. Cao, and J. Lutkenhaus. 1997. Temperature shift experiments with an ftsZ84(Ts) strain reveal rapid dynamics of FtsZ localization and indicate that the Z ring is required throughout septation and cannot reoccupy division sites once constriction has initiated. J. Bacteriol. 179:4277-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Addinall, S. G., and J. Lutkenhaus. 1996. FtsA is localized to the septum in an FtsZ-dependent manner. J. Bacteriol. 178:7167-7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bork, P., C. Sander, and A. Valencia. 1992. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc. Natl. Acad. Sci. USA 89:7290-7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd, D., D. S. Weiss, J. C. Chen, and J. Beckwith. 2000. Towards single-copy gene expression systems making gene cloning physiologically relevant: lambda InCh, a simple Escherichia coli plasmid-chromosome shuttle system. J. Bacteriol. 182:842-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buddelmeijer, N., and J. Beckwith. 2002. Assembly of cell division proteins at the E. coli cell center. Curr. Opin. Microbiol. 5:553-557. [DOI] [PubMed] [Google Scholar]
- 7.Carettoni, D., P. Gomez-Puertas, L. Yim, J. Mingorance, O. Massidda, M. Vicente, A. Valencia, E. Domenici, and D. Anderluzzi. 2003. Phage-display and correlated mutations identify an essential region of subdomain 1C involved in homodimerization of Escherichia coli FtsA. Proteins 50:192-206. [DOI] [PubMed] [Google Scholar]
- 8.Corbin, B. D., X.-C. Yu, and W. Margolin. 2002. Exploring intracellular space: function of the Min system in round-shaped Escherichia coli. EMBO J. 21:1988-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dai, K., Y. Xu, and J. Lutkenhaus. 1996. Topological characterization of the essential Escherichia coli cell division protein FtsN. J. Bacteriol. 178:1328-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Lallo, G., M. Fagioli, D. Barionovi, P. Ghelardini, and L. Paolozzi. 2003. Use of a two-hybrid assay to study the assembly of a complex multicomponent protein machinery: bacterial septosome differentiation. Microbiology 149:3353-3359. [DOI] [PubMed] [Google Scholar]
- 11.Ding, Z., Z. Zhao, S. J. Jakubowski, A. Krishnamohan, W. Margolin, and P. J. Christie. 2002. A novel cytology-based, two-hybrid screen for bacteria applied to protein-protein interaction studies of a type IV secretion system. J. Bacteriol. 184:5572-5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edwards, D. H., H. B. Thomaides, and J. Errington. 2000. Promiscuous targeting of Bacillus subtilis cell division protein DivIVA to division sites in Escherichia coli and fission yeast. EMBO J. 19:2719-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geissler, B., D. Elraheb, and W. Margolin. 2003. A gain of function mutation in ftsA bypasses the requirement for the essential cell division gene zipA in Escherichia coli. Proc. Natl. Acad. Sci. USA 100:4197-4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guzman, L. M., D. S. Weiss, and J. Beckwith. 1997. Domain-swapping analysis of FtsI, FtsL, and FtsQ, bitopic membrane proteins essential for cell division in Escherichia coli. J. Bacteriol. 179:5094-5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hale, C. A., and P. A. de Boer. 1999. Recruitment of ZipA to the septal ring of Escherichia coli is dependent on FtsZ and independent of FtsA. J. Bacteriol. 181:167-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hale, C. A., and P. A. de Boer. 2002. ZipA is required for recruitment of FtsK, FtsQ, FtsL, and FtsN to the septal ring in Escherichia coli. J. Bacteriol. 184:2552-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu, C., J. Stricker, and H. P. Erickson. 1998. FtsZ from Escherichia coli, Azotobacter vinelandii, and Thermotoga maritima—quantitation, GTP hydrolysis, and assembly. Cell Motil. Cytoskeleton 40:71-86. [DOI] [PubMed] [Google Scholar]
- 19.Ma, X., D. W. Ehrhardt, and W. Margolin. 1996. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc. Natl. Acad. Sci. USA 93:12998-13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma, X., and W. Margolin. 1999. Genetic and functional analysis of the conserved C-terminal core domain of Escherichia coli FtsZ. J. Bacteriol. 181:7531-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Margolin, W. 2000. Themes and variations in prokaryotic cell division. FEMS Microbiol. Rev. 24:531-548. [DOI] [PubMed] [Google Scholar]
- 22.Mercer, K. L., and D. S. Weiss. 2002. The Escherichia coli cell division protein FtsW is required to recruit its cognate transpeptidase, FtsI (PBP3), to the division site. J. Bacteriol. 184:904-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen-Disteche, M., C. Fraipont, N. Buddelmeijer, and N. Nanninga. 1998. The structure and function of Escherichia coli penicillin-binding protein 3. Cell. Mol. Life Sci. 54:309-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pichoff, S., and J. Lutkenhaus. 2002. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 21:685-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pla, J., A. Dopazo, and M. Vicente. 1990. The native form of FtsA, a septal protein of Escherichia coli, is located in the cytoplasmic membrane. J. Bacteriol. 172:5097-5102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pogliano, J., K. Pogliano, D. S. Weiss, R. Losick, and J. Beckwith. 1997. Inactivation of FtsI inhibits constriction of the FtsZ cytokinetic ring and delays the assembly of FtsZ rings at potential division sites. Proc. Natl. Acad. Sci. USA 94:559-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmidt, K. L., N. D. Peterson, R. J. Kustusch, M. C. Wissel, B. Graham, G. J. Phillips, and D. S. Weiss. 2004. A predicted ABC transporter, FtsEX, is needed for cell division in Escherichia coli. J. Bacteriol. 186:785-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsui, H. C., G. Feng, and M. E. Winkler. 1997. Negative regulation of mutS and mutH repair gene expression by the Hfq and RpoS global regulators of Escherichia coli K-12. J. Bacteriol. 179:7476-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van den Ent, F., L. A. Amos, and J. Löwe. 2001. Prokaryotic origin of the actin cytoskeleton. Nature 413:39-44. [DOI] [PubMed] [Google Scholar]
- 30.van Den Ent, F., and J. Löwe. 2000. Crystal structure of the cell division protein FtsA from Thermotoga maritima. EMBO J. 19:5300-5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiss, D. S., J. C. Chen, J. M. Ghigo, D. Boyd, and J. Beckwith. 1999. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL J. Bacteriol. 181:508-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan, K., K. H. Pearce, and D. J. Payne. 2000. A conserved residue at the extreme C terminus of FtsZ is critical for the FtsA-ftsZ interaction in Staphylococcus aureus. Biochem. Biophys. Res. Commun. 270:387-392. [DOI] [PubMed] [Google Scholar]
- 33.Yim, L., G. Vandenbussche, J. Mingorance, S. Rueda, M. Casanova, J. M. Ruysschaert, and M. Vicente. 2000. Role of the carboxy terminus of Escherichia coli FtsA in self-interaction and cell division. J. Bacteriol. 182:6366-6373. [DOI] [PMC free article] [PubMed] [Google Scholar]