Abstract
Circadian rhythmicity is a fundamental process that synchronizes behavioral cues with metabolic homeostasis. Disruption of daily cycles due to jet lag or shift work results in severe physiological consequences including advanced aging, metabolic syndrome, and even cancer. Our understanding of the molecular clock, which is regulated by intricate positive feedforward and negative feedback loops, has expanded to include an important metabolic transcriptional coregulator, Steroid Receptor Coactivator-2 (SRC-2), that regulates both the central clock of the suprachiasmatic nucleus (SCN) and peripheral clocks including the liver. We hypothesized that an environmental uncoupling of the light-dark phases, termed chronic circadian disruption (CCD), would lead to pathology similar to the genetic circadian disruption observed with loss of SRC-2. We found that CCD and ablation of SRC-2 in mice led to a common comorbidity of metabolic syndrome also found in humans with circadian disruption, non-alcoholic fatty liver disease (NAFLD). The combination of SRC-2−/− and CCD results in a more robust phenotype that correlates with human non-alcoholic steatohepatitis (NASH) and hepatocellular carcinoma (HCC) gene signatures. Either CCD or SRC-2 ablation produces an advanced aging phenotype leading to increased mortality consistent with other circadian mutant mouse models. Collectively, our studies demonstrate that SRC-2 provides an essential link between the behavioral activities influenced by light cues and the metabolic homeostasis maintained by the liver.
Keywords: SRC-2, non-alcoholic fatty liver disease, NAFLD, chronic circadian disruption, liver, metabolism
Circadian rhythmicity is the manifestation of a central process that synchronizes the self-sustaining endogenous molecular clock of an organism to exogenous cues in the environment (i.e., light, temperature, and food). The suprachiasmatic nucleus (SCN), located in the hypothalamus, houses the primary mammalian endogenous clock responsible for processing light stimuli and coordinating secondary tissue clocks for physiological processes including metabolism, locomotor activity, endocrine secretion, and cardiovascular performance. Synchronization of these various systems allows an organism to properly coordinate the transition from day-to-night with sleep-wake and feeding-fasting behaviors to ensure survival.
The importance of these coordinated cycles is highlighted in humans subjected to shift work or jet lag conditions who exhibit severe physiological consequences including increased insomnia during the night and drowsiness during the day, increased psychological disorders, decreased work productivity, and decreased cognitive function (Cho et al., 2000; Tapp and Natelson, 1989). Consistent with the effects of circadian disruption in humans, chronic jet lag in mice increases morbidity and mortality (Davidson et al., 2006). Furthermore, chronic circadian disruption (CCD) occurs in a large subset of full-time wage earners in the United States, with nearly 15% currently employed as shift workers who collectively have an increased risk of developing breast, prostate, or colon cancers (US Department of Labor, 2005; Wang et al., 2011; Megdal et al., 2005). Shift work is also associated with increased metabolic disorders including obesity, hypertriglyceridemia, hypertension, and aberrant hormone homeostasis (Karlsson et al., 2001; Scheer et al., 2009; Kiessling et al., 2010; Parsons et al., 2015). Comorbidities of the metabolic syndrome are often observed in the CCD population, which include type 2 diabetes mellitus, cardiovascular disease, and the most prevalent chronic liver disease in developed countries, non-alcoholic fatty liver disease (NAFLD) (Wang et al., 2011; Lin et al., 2014; Lin and Chen, 2015; Sookoian and Pirola, 2009). The increased incidence of NAFLD that accompanies CCD also creates a predisposition for cirrhosis and hepatocellular carcinoma (HCC) and has even been reported to accelerate aging and increase mortality (Froy, 2013; Kondratova and Kondratov, 2012).
In the liver, the molecular circadian clock is regulated by a tightly controlled transcriptional-translational feedback system that involves the basic helix-loop-helix (bHLH)-PAS transcription factors, brain and muscle-Arnt-like 1 (BMAL1), circadian locomotor output cycles kaput (CLOCK), and the CLOCK homolog, neuronal PAS domain protein 2 (NPAS2). BMAL1 forms a heterodimer with either CLOCK or NPAS2 on E-box DNA response elements to facilitate transcription of cryptochrome1/2 (Cry1/2) and period1/2 (Per1/2) genes (Partch et al., 2013). Accumulation of the Per1/2 and Cry1/2 proteins feed back and inhibit the transcriptional activity of the BMAL1 heterodimer. Ancillary regulatory loops are formed by BMAL1 heterodimers to promote transcription of opposing nuclear receptor (NR) programs such as retinoic acid receptor–related orphan receptor (RORα) and reverse erythroblastosis virus α/β (Rev-erb α/β) gene expression (Akashi and Takumi, 2005; Cho et al., 2012; Bugge et al., 2012). In addition to directly regulating NR expression, the BMAL1 heterodimer participates in a self-regulatory feedback loop that controls the expression of powerful transcriptional coactivators including Steroid Receptor Coactivator-2 (SRC-2) that regulates the transcriptional amplitude of the BMAL1 heterodimers (Stashi et al., 2014).
A variety of circadian defects are observed upon perturbation of the primary molecular components of the circadian machinery in mice. In fact, an assortment of severe metabolic defects have been reported in Bmal1−/−, Rev-erbα/β−/−, CLOCK−/−, and SRC-2−/− mice (Kondratov, 2006; Cho et al., 2012; Eckel-Mahan et al., 2012; Hatanaka et al., 2010; Chopra et al., 2008; Chopra et al., 2011; Fleet et al., 2015). Work from our laboratory has shown that SRC-2 is a potent coactivator for the BMAL1:CLOCK heterodimer that coordinates gene expression events to balance circadian and metabolic homeostasis (Stashi et al., 2014). We have shown that SRC-2 ablation alters normal circadian locomotor behavior and creates a predisposition for metabolic dysfunction and hepatic cancer (O’Donnell et al., 2012; Stashi et al., 2014). More recently, our laboratory has reported that recruitment of SRC-2 by BMAL1 and/or ROR to circadian and metabolic gene promoters is required for chromatin decondensation, which facilitates the repressive actions of Rev-erb α/β to maintain proper gene amplitude and circadian homeostasis (Zhu et al., 2015). Given the epidemiological findings that circadian disruption tightly correlates with an increased incidence of metabolic disease, we investigated the metabolic and behavioral consequences of SRC-2 ablation in a mouse model of CCD.
Herein, we report that mice devoid of SRC-2 fail to adapt to the stress of CCD. Phenotypically, SRC-2−/− mice develop histopathological hallmarks of NAFLD (Kleiner et al., 2005), which are consistent with epidemiological observations made in humans subjected to jet lag or shift work (Wang et al., 2011; Lin et al., 2014; Lin and Chen, 2015; Sookoian and Pirola, 2009). Transcriptomic analyses of livers from SRC-2−/− mice subjected to CCD versus normal light (N/L) conditions revealed NAFLD as a predominant pathway influenced by loss of SRC-2 function. Integration of our transcriptomic data with the gene expression profiles from human HCC patients demonstrates that expression of gene signatures found in SRC-2−/− mice is predictive of increased patient mortality. Further validating these findings, we report that loss of SRC-2 and CCD both lead to advanced signs of aging and decreased survival in mice. Taken together, our findings highlight the importance of SRC-2 for coordinating circadian rhythmicity with environmental cues to preserve a balance between daily cycles and metabolic homeostasis.
MATERIALS AND METHODS
Animals
The generation of the SRC-2−/− mice has been described previously (Chopra et al., 2011; Gehin et al., 2002; Bunger et al., 2000). Congenic WT and SRC-2−/− male littermate mice (8–12 weeks old) from a heterozygous breeding strategy were used for all studies. The Animal Care and Research Committee at Baylor College of Medicine approved all animal studies. WT and SRC-2−/− mice were maintained on a normal 12-h light and 12-h dark schedule (N/L). WT and SRC-2−/− male littermate mice were exposed to CCD for ~52 weeks. CCD is defined by 2 alternating light schedules, one with lights on from 0600 h to 1800 h and a second with lights on from 2200 h to 1000 h as previously described (Kettner et al., 2015), except mice were maintained on the phase advance for 6 days and on the phase delay for 8 days. WT and SRC-2−/− mice subjected to CCD were sacrificed at ZT4 and ZT18 on the first day of the phase delay cycle, which mirrors the control cohorts that were maintained on N/L for the duration of the study.
Gene Expression Profiling
For gene expression profiling of mouse liver cells, total RNA was extracted using the Purelink Mini Kit (Life Technologies, Waltham, MA) following the manufacturer’s instructions. RNA was reverse transcribed and microarray hybridization was performed using the Illumina Gene Expression MouseWG-6 v2.0 Expression BeadChip Kit at the Laboratory for Translational Genomics at Baylor College of Medicine. Microarray scanned images were imported into the Illumina GenomeStudio for data quality control, and the transcriptomic profile data were quantile-normalized by the Bioconductor lumi package (Cho et al., 2000; Du et al., 2008; Tapp and Natelson, 1989; Davidson et al., 2006) (Illumina, San Diego, CA). Differences in gene expression between different genotypes or CCD conditions were inferred using the Bioconductor limma package (Smyth, 2004) (p < 0.05) and imposing a fold change exceeding 1.5× using the R statistical system. Hierarchical clustering and heat maps were generated using the R statistical system.
SRC-2-dependent Gene Signatures in Mouse Liver Cells
A previously published signature of SRC-2−/− transcriptional response in mouse liver tissue (Jeong et al., 2006) was partitioned into up-regulated and down-regulated genes and used for direct comparison against transcriptomic signatures from SRC-2−/− mice maintained on N/L or CCD conditions.
Gene Set Enrichment Analysis
Gene set enrichment analysis (GSEA) was carried out using the GSEA software package (Subramanian et al., 2005) to assess the degree of similarity among the studied gene signatures (Greenawalt et al., 2011; Roessler et al., 2010; Ahrens et al., 2013; Roessler et al., 2012; Hatoum et al., 2013). Patient information from Roessler et al. (2010) is located in Supplementary Table S1. For each SRC-2 transcriptomic response produced under various CCD conditions, genes were ranked by the fold change between SRC-2−/− mice and the WT samples. For the previously published SRC-2−/− mouse liver gene signature, we used separately the up-regulated and down-regulated genes. Normalized enrichment scores (NES) and adjusted q values were computed using the GSEA method based on 1000 random permutations of the ranked genes.
Histology and Immunohistochemistry
For histological staining, liver tissue was fixed overnight with 10% formalin at 4°C. Fixed samples were embedded in paraffin, sectioned, and stained with hematoxylin and eosin (H&E), TUNEL (Millipore, Billerica, MA, ApopTag Peroxidase In Situ Apoptosis Kit S7100), or trichrome for light microscopy. Immunohistochemistry was performed for Ki-67 (Neomarkers/Thermo Scientific, Waltham, MA, RM-9106) and F4/80 (AbD Serotec, Atlanta, GA, C1:A3-1). Slides were deparaffinized and blocked in endogenous peroxidase (50 mL of methanol containing 1 mL of 30% hydrogen peroxide) and washed in distilled water. Antigen retrieval was performed for 20 min in antigen retrieval buffer and blocked with serum (Rodent Block M). Ki-67 (1:200 dilution) and F4/80 (1:50 dilution) primary antibodies were incubated for 30 min. DAB Plus was used for detection, and slides were briefly counterstained in CAT hematoxylin (diluted 1:3 in distilled water).
Quantitative Polymerase Chain Reaction (qPCR)
Total mRNA was isolated from liver with Purelink RNA Mini Kit (Life Technologies, Waltham, MA). Reverse transcription was carried out using Superscript VILO (Life Technologies, Waltham, MA) per the manufacturer’s instructions. For gene expression analyses, qPCR was performed using the Taqman system with sequence-specific primers and the Universal Probe Library (Roche, Indianapolis, IN). All data were analyzed with 18S rRNA as the endogenous control. All qPCR primer sequences are available upon request.
Isolation of Mouse Bone Marrow from Long Bones
Mouse femurs and tibias were dissected and placed on ice. A 26-gauge needle with cold Hank’s Balanced Salt Solution (HBSS+) buffer was used to flush the bone marrow (BM) into a clean petri dish on ice. The flushed BM was homogenized into a single-cell suspension using an 18-gauge needle and filtered through a 40-μm cell strainer. Cells were pelleted by centrifugation at 400 × g, 4°C for 10 min. Cells were resuspended and incubated in 10 mL 1× red blood cell lysis buffer for 10 min at room temperature (RT) and then repelleted and resuspended in HBSS+.
Isolation of Mouse Spleen
The spleen was dissected and rinsed in PBS, minced, and dissociated in HBSS+ using an 18-gauge needle (10×) and then a 26-gauge needle (10×) to generate a single cell suspension. The suspension was filtered using a 40-μm filter, followed by red blood cell lysis as described above, and then resuspended.
Isolation on Mouse Peripheral Blood
Peripheral blood (PB) samples were placed in 150 μL of 2% dextran solution and 150 μL of heparin solution (10 mg/mL). PB settled until the supernatant appeared white. The supernatant was placed in a 5-mL FACS tube, and 1 mL of 1× red blood cell lysis buffer was added at RT for 10 min.
Hematopoietic Lineage Analysis of BM, Spleen, and PB
Single-color antibody controls were prepared using BM collected from the femur/tibia (eFluor450 only, PE-Cy7 only). To each blood sample, 2 mL of HBSS+ was added, and samples were centrifuged at 200 × g for 10 min. The resulting supernatant was discarded, and the pellet was resuspended in 100 μL of lineage antibody cocktail: CD45-APC (1:100 dilution), B220-eFluor450 (1:100 dilution), B220-PE-Cy7 (1:100 dilution), CD4-eFluor450 (1:100 dilution), CD8a-eFluor 450 (1:100 dilution), Gr-1-PE-Cy7 (1:200 dilution), Mac-1-PE-Cy7 (1:200 dilution) (eBioscience, San Diego, CA). Single-color BM controls were stained by resuspending the cells in 100 μL of HBSS+ and adding 1 μL of either B220-eFluor450 or B220-PE-Cy7 antibody. Cells were incubated for 10 min and centrifuged as described previously (200 × g, 10 min). Cells were resuspended in HBSS+, and propidium iodide (PI) was added to a final concentration of 2 μg/mL.
Samples were analyzed on the LSRII flow cytometer (BD Biosciences, San Jose, CA). Cells were first visualized and gated under forward (FSC) versus side-scatter (SSC). The target WBC population was gated for PI-negative (live) cells, and the CD45+ population was gated to display the live CD45+ target population as B220/Mac-1-PE-Cy7 on the vertical y-axis and CD4/B220-eFluor450 on the horizontal x-axis. B-cells were called as B220-eFluor450+ and B220-PE-Cy7+, T-cells were called as CD4/CD8-eFluor450+, and myeloid cells were gated as Mac-1/Gr-1-PE-Cy7+.
Statistics
Results are displayed as the mean ± SEM. Graphpad Prism 6 was used to make statistical comparisons between 2 different genotype groups (WT and SRC-2−/−) using individual 2-tailed unpaired Student t test with differences of p < 0.05 considered as significant.
RESULTS
SRC-2 Modulates Behavioral Changes in Response to CCD
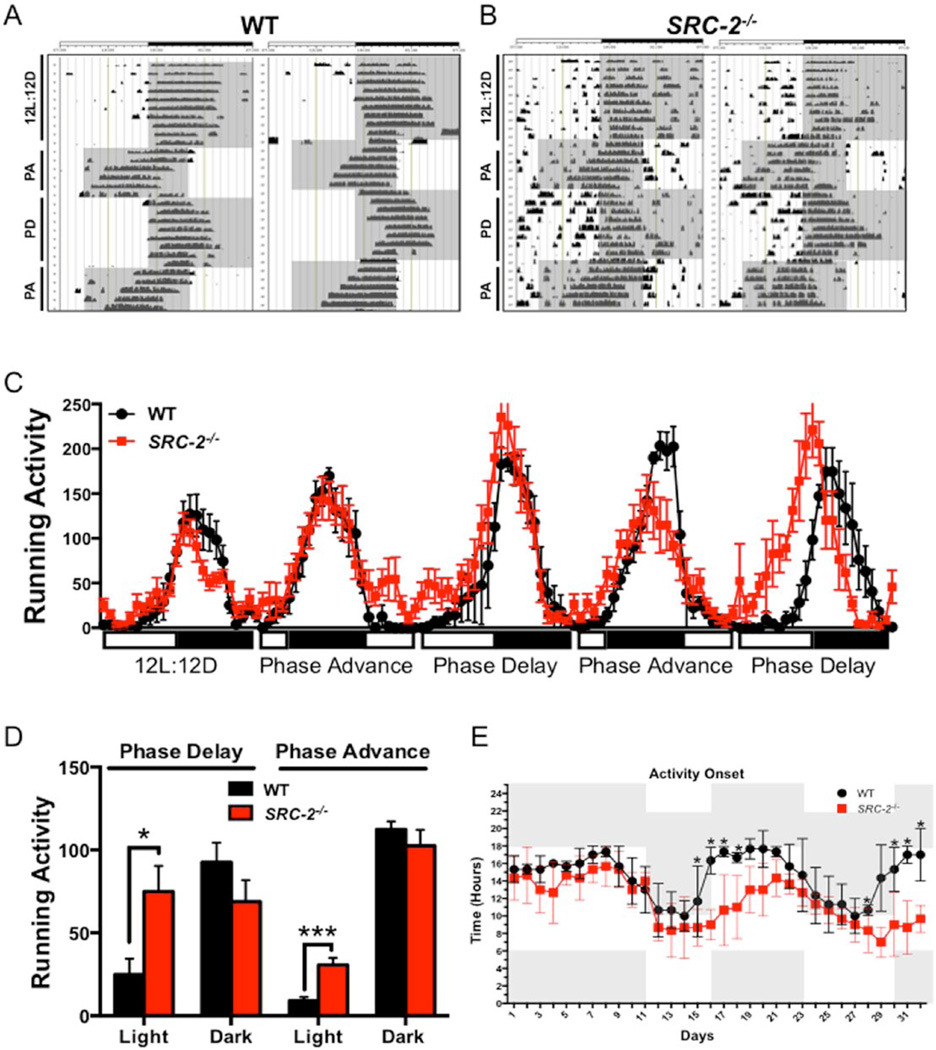
A growing body of evidence demonstrates that the functions of core circadian clock members BMAL1, CLOCK, PER1/2, and CRY1/2 are disrupted in mouse models of CCD (Barclay et al., 2012; Reddy et al., 2002; Kettner et al., 2015). We therefore examined how SRC-2, a coactivator of BMAL1:CLOCK transcriptional activity, contributes to this known disrupted circadian metabolic phenotype. Running wheel analyses demonstrate that WT mice readily adapt their locomotor activity in response to 8-h advances or 8-h delays in light phase, known as a circadian disruption (Fig. 1A). Conversely, SRC-2−/− mice show a blunted response to changes in light phase advances and delays, suggesting that circadian arrhythmia persists in SRC-2−/− mice irrespective of light cues (Fig. 1B) (Stashi et al., 2014). Quantification of running wheel activity further supports that WT mice more readily adapt to light changes and increased running wheel activity after the phase shift, whereas SRC-2−/− mice are unable to fully adapt to alterations in the light cycle (Fig. 1C). Aggregate analysis of the phase delay and phase advance cycles demonstrates that SRC-2−/− mice spend significantly more active time in the light phase compared with their WT littermates (Fig. 1D). The average activity onset in response to circadian disruption reveals that SRC-2−/− mice fail to readily adjust to changes in the light-dark phases, whereas WT mice have an activity onset that follows the beginning of the dark phase (Fig. 1E). Collectively, these data indicate that SRC-2−/− mice display disrupted activity in the light phase as opposed to WT mice that have a distinct activity onset that is dictated by light-dark cycling. These observations confirm our previous findings that SRC-2−/− mice display disrupted circadian rhythmicity and demonstrate their inability to adapt to the stress of CCD.
Figure 1.
SRC-2−/− mice have impaired response to CCD. (A) Representative running wheel activity actograms of 2 WT mice during 12-h light and 12-h dark (12L:12D), 8-h phase advance (PA), and 8-h phase delay (PD). (B) Representative running wheel actograms of 2 SRC-2−/− mice during 12L:12D, PA, and PD. (C) Average running wheel activity, defined as wheel turns, for each ZT of 24 h for WT and SRC-2−/− mice under 12L:12D conditions and alternating PA and PD cycles. (D) Average running wheel activity, defined as total wheel turns, in the light and dark phases of WT and SRC-2−/− mice in response to the PD and PA conditions. (E) Average circadian hour that WT and SRC-2−/− mice demonstrate initial activity onset during the 12L:12D and alternating PA and PD conditions throughout the running wheel experiment. All data were collected at approximately 20 weeks of either normal light (N/L) or chronic circadian disruption (CCD) as indicated. Data are graphed as the mean ± SEM. *p < 0.05. **p < 0.01. ***p < 0.001. (N = 5).
CCD Negatively Affects Body Composition and Liver Histopathology
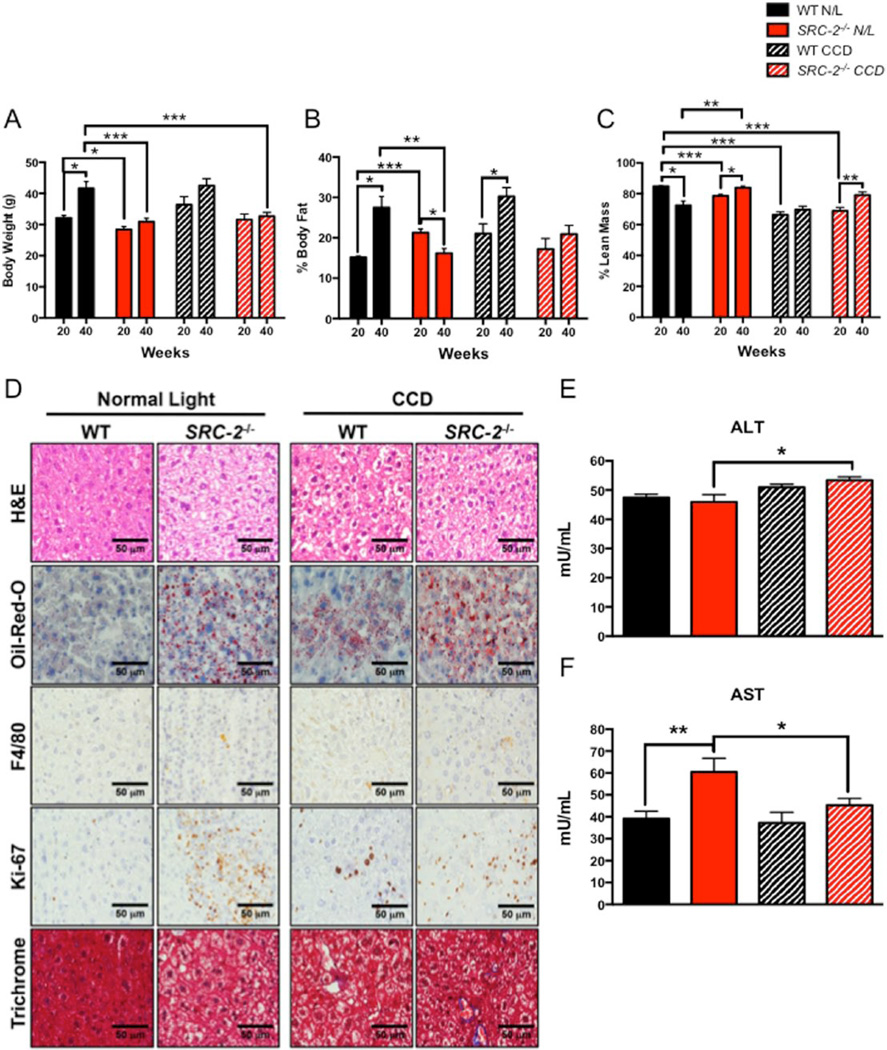
Given that epidemiologic data have established a strong correlation between perturbed circadian rhythmicity (i.e., shift work or jet lag) and poor metabolic outcomes such as metabolic syndrome and NAFLD (Lin et al., 2014; Lin and Chen, 2015), we analyzed body composition during the progression of CCD at 20 weeks and 40 weeks in WT and SRC-2−/− mice. Consistent with previous reports, SRC-2−/− mice show reduced total body weight compared with WT mice (Fig. 2A) (Gehin et al., 2002; Chopra et al., 2008). At 40 weeks of N/L, we found a significant increase in total body weight and percent body fat composition along with a reduction in percent lean mass in WT mice compared with the 20-week time point (Fig. 2A–C). At 40 weeks of CCD, WT mice also show increased body weight and percent body fat but no change in percent lean mass (Fig. 2A–C). In contrast, SRC-2−/− mice show no increase in total body weight, a reduction in percent body fat, and an increase in lean mass when maintained on N/L (Fig. 2A–C). In response to CCD, SRC-2−/− mice fail to increase body weight or percent fat mass and, surprisingly, show an increase in percent lean mass (Fig. 2A–C).
Figure 2.
Phenotypic characterization of SRC-2−/− mice during N/L and CCD conditions. (A) Body weight (grams), (B) percent body fat, and (C) percent lean mass of WT and SRC-2−/− mice subjected to N/L or CCD for 20 and 40 weeks, respectively. (D) Comparative 40× liver histology and immunohistochemistry for H&E, Oil-Red-O, F4/80, Ki-67, and trichrome staining from WT and SRC-2−/− mice subjected to N/L and CCD. (E) Plasma ALT of WT and SRC-2−/− mice subjected to N/L and CCD. (F) Plasma AST of WT and SRC-2−/− mice subjected to N/L and CCD. Data are graphed as the mean ± SEM. *p < 0.05. **p < 0.01. ***p < 0.001. (N = 3–4).
H&E and Oil Red O staining of livers isolated from WT and SRC-2−/− mice shows that loss of SRC-2 leads to accumulation of neutral lipid in N/L conditions, while CCD results in lipid sequestration in both WT and SRC-2−/− livers (Fig. 2D, Suppl. Fig. S1A,B). Immunohistochemical staining for F4/80, a marker for macrophage infiltration, remains unchanged in SRC-2-null livers compared with WT mice on N/L conditions (Fig. 2D and Suppl. Fig. S1A,C). In response to CCD, only WT livers, but not SRC-2−/− livers, display elevated macrophage infiltration that is consistent with observations made in genetic models of chronic circadian disruption that have increased inflammatory markers (Mazzoccoli et al., 2014) (Fig. 2D and Suppl. Fig. S1A,C). Proliferative (Ki-67) and fibrotic (Masson’s trichrome and collagen) markers are increased in the livers of SRC-2−/− mice on N/L, which are both key indices of steatohepatitis (Fig. 2D and Suppl. Fig. S1A,D,E,F). Consistent with previous reports, CCD also potentiates NAFLD progression in WT livers as indicated by increased cell proliferation and inflammation (Fig. 2D and Suppl. Fig. S1A,C,D).
Levels of alanine and aspartate aminotransferases (ALT and AST) are often used as indicators of hepatic stress, although it is important to note that static measurements of ALT and AST levels show poor correlation in patients clinically diagnosed with NAFLD, suggesting that these biomarkers are insufficient to capture the severity of hepatic steatosis (Browning et al., 2004; Obika and Noguchi, 2012). Consistent with these patient data, WT mice subjected to CCD show no significant increase in either ALT (Fig. 2E) or AST (Fig. 2F). Conversely, mice null for SRC-2 exposed to CCD show a nominal increase in ALT (Fig. 2E) and a reduction in AST (Fig. 2F) levels compared with N/L. Based on the histological and immunohistochemical data, these findings suggest that CCD induces a NAFLD-like phenotype in WT mice, which mirrors the increased fat accumulation, inflammation, and fibrosis observed upon SRC-2 ablation during both N/L and CCD conditions.
Gene Expression Profiling of N/L versus CCD Conditions Identifies a NAFLD-like Signature in SRC-2−/− Mice
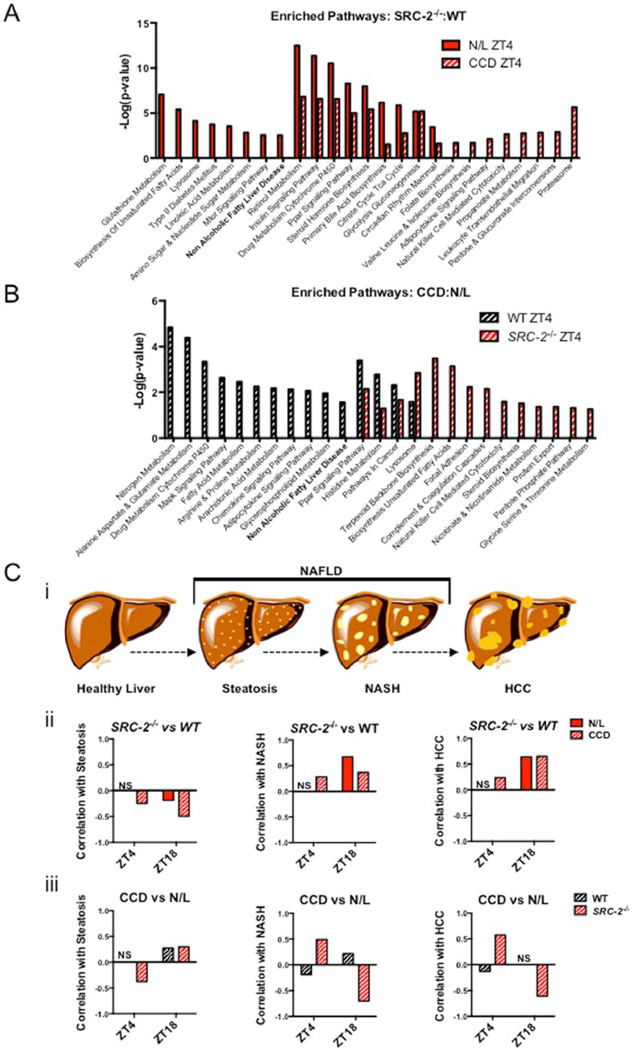
To investigate the effects of CCD in the absence of SRC-2, we generated transcriptomic profiles from livers of WT and SRC-2−/− mice exposed to N/L or CCD conditions for ~52 weeks at 2 zeitgeber times (ZT4 and ZT18). During N/L conditions, WT and SRC-2−/− mice show a large number of significant gene expression changes at both ZT4 and ZT18 (Suppl. Fig. S2A). In response to CCD, the number of genes that are significantly changed decreased by 80% and therefore demonstrate that disruption of circadian rhythmicity due to ablation of SRC-2 partially recapitulates the environmentally induced mRNA expression changes of CCD (Suppl. Fig. S2A). To further characterize the programmatic gene expression changes that occur in SRC-2−/− mice subjected to CCD, we performed gene ontology analysis of genes significantly changed in the SRC-2−/− mice compared with WT as well as mice in CCD relative to N/L at both ZT4 and ZT18. We identified NAFLD as a top common ontological process that was enriched consistently at ZT4 and ZT18 when we compared either SRC-2−/− mice to WT or CCD relative to N/L (Fig. 3A,B and Suppl. Fig. S2B,C). To determine whether CCD and SRC-2 ablation resemble human NAFLD gene expression profiles, we used human microarray datasets that span the progression of NAFLD from steatosis, NASH, and ultimately HCC (Figure 3C(i)). First, we calculated the Pearson correlations between the human steatosis, NASH, and HCC datasets and found no correlation between the human NASH and steatosis gene expression signatures, an inverse correlation between the human HCC and steatosis, and a positive correlation between the human NASH and HCC microarrays (Suppl. Fig. S2D). These findings suggest that the gene expression profile of NASH is more similar to that of HCC than steatosis, as previously reported (Starmann et al., 2012). Comparison of gene signatures from human hepatic steatosis, NASH, and HCC with those generated from WT and SRC-2−/− mice exposed to N/L or CCD demonstrates that loss of SRC-2 in both N/L and CCD at ZT4 and ZT18 inversely correlates with steatosis (Fig. 3C(ii)). Conversely, the gene expression profile of SRC-2−/− liver positively correlates with NASH and HCC, although not in N/L conditions at ZT4 (Fig. 3C(ii)). Analyses of CCD gene signatures in WT and SRC-2−/− mice show minimal correlation with steatosis at either ZT4 or ZT18 (Fig. 3(iii)). Consistent with our gene ontology analyses, CCD transcriptomic profiles correlate well with the NASH and HCC signatures upon ablation of SRC-2 (Fig. 3C(iii)). The negative correlation of CCD with NASH and HCC gene expression in the SRC-2−/− at ZT18 may be attributed to the relatively low expression of SRC-2 at ZT18 or the general impact of SRC-2 on gene expression amplitude as reflected by the 2 zeitgeber times tested (Stashi et al., 2014; Zhu et al., 2015). Based on the phenotype and gene expression correlations, CCD alone appears to induce a steatotic-like gene program, whereas ablation of SRC-2 produces more NASH- and HCC-like genotypes. Our findings suggest that the combination of CCD and SRC-2 ablation results in a stronger programmatic shift toward the NASH and HCC expression pattern.
Figure 3.
NAFLD gene expression in the SRC-2−/− CCD model. (A) Gene ontology pathways enriched in liver microarray data from SRC-2−/− mice relative to WT mice at either ZT4 during N/L or ZT4 during CCD. (B) Gene ontology pathways enriched in CCD relative to N/L in ZT4 WT mice or ZT4 SRC-2−/− mice. (C) (i) A schematic representation of NAFLD progression. Pearson r correlation of (ii) SRC-2−/− mice enriched over WT mice during N/L (red) or CCD (red-white) at ZT4 and ZT18 or (iii) CCD enriched relative to N/L in WT (black-white) and SRC-2−/− (red-white) mice at ZT4 and ZT18 compared with human steatosis, NASH, and HCC hepatic microarray datasets (left to right).
Gene Set Enrichment Analysis of NASH and SRC-2−/− Transcriptomes
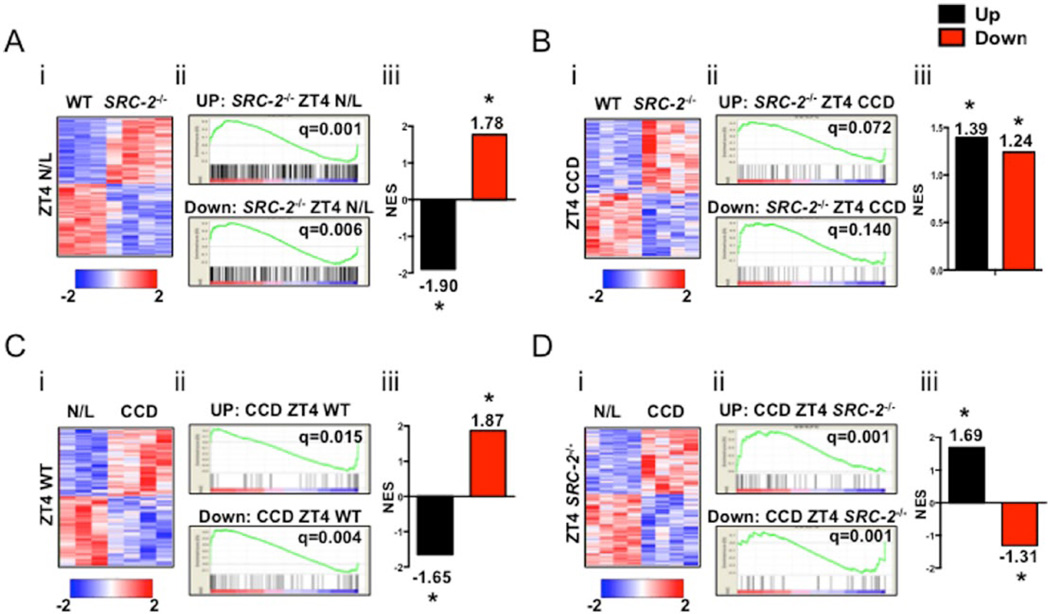
Given our observations that loss of SRC-2 results in histological and correlative gene signatures in liver that are consistent with NASH, we further defined the directionality of gene expression changes due to either CCD or loss of SRC-2 by performing GSEA to compare human NASH microarray datasets with transcriptomic signatures of WT and SRC-2−/− mice in N/L or CCD at ZT4 and ZT18. Heat map representations of gene expression differences between WT and SRC-2−/− mice (Fig. 4A,B(i)) and N/L and CCD (Fig. 4C,D(i)) at both ZT4 and ZT18 revealed opposing expression patterns (Suppl. Fig. S3A-D(i)). GSEA of the SRC-2−/− hepatic microarray relative to WT mice in N/L demonstrated an inverse correlation with human NASH mRNA expression data in that up-regulated genes in NASH versus healthy livers are down-regulated in SRC-2−/− murine livers relative to WT (Fig. 4A(ii,iii)). This observation is consistent with the non-significant Pearson coefficient in the aggregate gene expression correlation between human NASH and SRC-2−/− microarrays from Figure 3C(ii). The up-regulated genes in the hepatic microarray from SRC-2−/− mice exposed to CCD conditions positively correlate with the NASH human microarrays, although we observed an inverse correlation in the down-regulated genes between the 2 datasets (Fig. 4B(ii,iii)). These simultaneous positive and negative GSEA scores perhaps clarify the relatively small positive correlation between NASH and CCD SRC-2−/− gene signatures observed in Figure 3C(ii).
Figure 4.
Gene set enrichment analysis (GSEA) of murine CCD hepatic transcriptomic data compared with human HCC microarray data. (A) (i) Heat map representation of liver microarray in WT and SRC-2−/− mice at ZT4 N/L. (ii) GSEA plot representation of upregulated and down-regulated genes in common between the HCC human hepatic microarray dataset and enriched relative to WT in the SRC-2−/− ZT4 N/L hepatic microarray. (iii) Normalized enrichment score of up-regulated and down-regulated genes from A(ii). (B) (i) Heat map representation of liver microarray in WT and SRC-2−/− mice at ZT4 CCD. (ii) GSEA plot representation of up-regulated and down-regulated genes in common between the HCC human hepatic microarray dataset and enriched relative to WT in the SRC-2−/− ZT4 CCD hepatic microarray. (iii) Normalized enrichment score of up-regulated and down-regulated genes from B(ii). (C) (i) Heat map representation of liver microarray in N/L and CCD WT mice at ZT4. (ii) GSEA plot representation of up-regulated and down-regulated genes in common between the HCC human hepatic microarray dataset and enriched relative to N/L in the CCD ZT4 WT mice hepatic microarray. (iii) Normalized enrichment score of up-regulated and down-regulated genes from C(ii). (D) (i) Heat map representation of liver microarray in N/L and CCD SRC-2−/− mice at ZT4. (ii) GSEA plot representation of up-regulated and down-regulated genes in common between the HCC human hepatic microarray dataset and enriched relative to N/L in the CCD ZT4 SRC-2−/− mice hepatic microarray. (iii) Normalized enrichment score of up-regulated and down-regulated genes from D(ii).
GSEA of CCD versus N/L liver transcriptomic profiles in WT mice at ZT4 also demonstrated an inverse correlation between up-regulated and downregulated genes in the NASH human hepatic microarray (Fig. 4C(ii,iii)), which accounts for the overall negative gene expression correlation found between the human NASH and CCD WT mice in Figure 3C(iii). This inverse correlation suggests that WT mice in CCD express a more steatotic gene program that negatively correlates with NASH and HCC human microarray expression profiles (see Suppl. Fig. S2D). Most notably, GSEA of CCD versus N/L with transcriptomic profiles of SRC-2−/− mice at ZT4 revealed a positive correlation with up-regulated and down-regulated genes in the NASH human datasets (Fig 4D(ii,iii)). These findings substantiate the strong positive Pearson correlation observed in Figure 3C(iii). Moreover, these data highlight similarities with the NASH gene signature as well as directionality of those genes affected between CCD and SRC-2 ablation, suggesting that both insults share a common mechanism. Alternatively, at ZT18 we observed only a positive correlation with up-regulated genes in SRC-2−/− mice in N/L relative to WT and down-regulated genes in CCD compared with N/L (Suppl. Fig. S3A(iii),C(iii)). All other ZT18 comparisons between up-regulated and down-regulated genes revealed inverse correlations (Suppl. Fig. S3A-D(iii)).
Validation of Candidate Gene Expression between CCD Conditions and SRC-2 Ablation
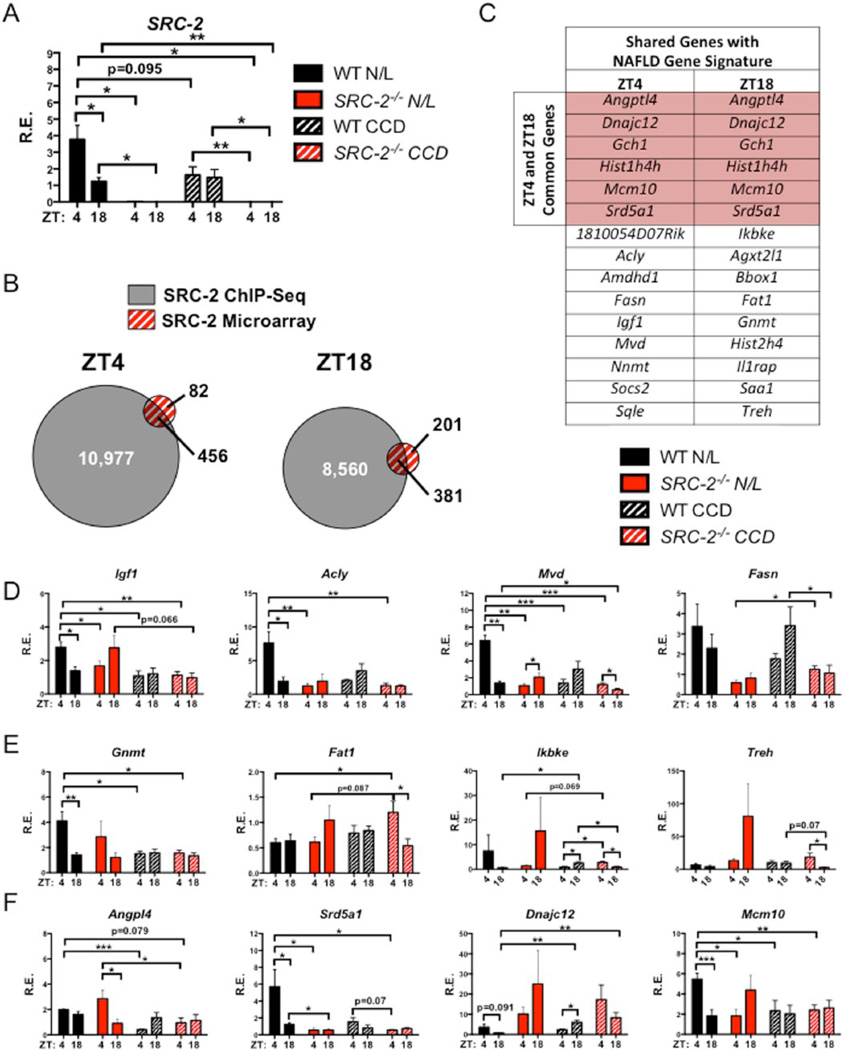
Given the strong directional correlation of the NASH gene signature with those genes affected by either CCD or SRC-2 ablation, we next sought to determine whether the expression of individual genes perturbed in human NASH and HCC are recapitulated by loss of SRC-2 and/or exposure to CCD in mice. An initial analysis of SRC-2 expression revealed an amplitude difference between ZT4 and ZT18 in WT mice on N/L conditions, but this effect is blunted in WT mice exposed to CCD (Fig. 5A). We previously reported that aberrations in SRC-2 expression perturb metabolic and circadian gene expression that results in a variety of metabolic consequences observed in NAFLD, including disturbances in lipid and glucose homeostasis (Stashi et al., 2014; Fleet et al., 2015). Here, we sought to characterize the effect of SRC-2 ablation and/or CCD on the expression of hepatic genes involved in the progression to NASH. We integrated published hepatic SRC-2 ChIP-Seq datasets (ZT4 and ZT18) with transcriptomic data obtained from the livers of the SRC-2−/− mice subjected to CCD at ZT4 and Z18 to identify candidate genes that are directly regulated by SRC-2 (Fig. 5B) (Stashi et al., 2014). Our integrated analyses revealed 456 genes at ZT4 and 381 genes at ZT18 as putative direct SRC-2 target genes (Fig. 5B). Next, we overlapped these putative SRC-2 target genes with available human microarray datasets from patients with steatosis, NASH, and HCC to identify candidate genes implicated in the progression of fatty liver disease (Fig. 5C). Analysis of these datasets identified a subset of 65 genes we termed the “Steatosis/NASH/HCC Overlap” that were found in all 3 datasets, and 231 genes referred to as the “NASH/HCC Overlap” that were shared between the NASH and HCC datasets only (Suppl. Fig. S4A). This integrated analysis identified 15 putative SRC-2 target genes in common with the NASH/HCC Overlap at both ZT4 and ZT18 (Fig. 5C). Similarly, integration of the human Steatosis/NASH/HCC Overlap with putative SRC-2 target genes identified 12 genes shared at ZT4 and ZT18 (Suppl. Fig. S4B).
Figure 5.
Hepatic transcriptomic analysis of WT and SRC-2−/− mice subjected to N/L and CCD. (A) Liver mRNA expression of SRC-2 in WT and SRC-2−/− mice subjected to N/L and CCD at ZT4 and ZT18. (B) Overlap of hepatic ZT4 (left) and ZT18 (right) SRC-2 ChIP-Seq and microarray data from SRC-2−/− mice maintained on N/L at ZT4 and ZT18. (C) Table of common genes shared between the SRC-2 ChIP-Seq and microarray from ZT4 and ZT18 from (B) that are found in common with human liver microarrays from patients with NASH and HCC. (D) mRNA expression of genes from (C) that represent ZT4 genes that are found in common with human patients with NASH and HCC in WT and SRC-2−/− mice subjected to N/L and CCD at ZT4 and ZT18. (E) mRNA expression of genes from (C) that represent ZT18 genes that are found in common with human patients with NASH and HCC in WT and SRC-2−/− mice maintained on N/L and CCD conditions at ZT4 and ZT18. (F) mRNA expression of genes from (C) that represent ZT4 and ZT18 genes that are found in common with human patients with NASH and HCC in WT and SRC-2−/− mice subjected to N/L and CCD at ZT4 and ZT18. Data are graphed as the mean ± SEM. *p < 0.05. **p < 0.01. ***p < 0.001. (N = 3–4).
Selective validation of putative SRC-2 target genes from our integrated analyses revealed specific amplitude changes upon either the loss of SRC-2 or exposure to CCD. More specifically, from the genes at ZT4 that overlap with NASH/HCC, SRC-2 ablation and CCD caused amplitude effects in Igf1, Acly, Mvd, and Fasn (Fig. 5D). Additionally, Igf1 and Fasn expression was similar to HCC patients with poor survival (Table 1). Genes at ZT18 with altered amplitudes due to CCD or loss of SRC-2 that overlap with NASH/HCC and show poor prognosis in HCC patients include Gnmt and Fat1 (Fig. 5E). Ikbke and Treh also showed differential expression during CCD conditions or upon loss of SRC-2, but these genes fail to correlate with HCC patient outcome (Fig. 5E and Table 1). Of the 15 ZT4 and ZT18 genes listed in Figure 5C, we found 6 that were shared between ZT4 and ZT18, which include Dnajc12, Hist1h4h, Srd5a1, Gch1, Angptl4, and Mcm10 (Fig. 5C, highlighted in red). Of these 6 genes, we identified changes in Angp14, Srd5a1, Dnajc12, and Mcm10 in SRC-2−/− and CCD cohorts whose expression patterns correlate with poor survival in HCC patients (Fig. 5F and Table 1). The ZT4 and ZT18 genes that overlap with the Steatosis/NASH/HCC Overlap and whose expression patterns in HCC patients correlate with poor prognosis include Csad, Cyp7a1, Gpd1, and Slc38a2 (Suppl. Fig. S5E). From our qPCR validation, we have demonstrated that loss of SRC-2 and CCD perturbs the cyclic expression of genes at ZT4 and ZT18 and also largely recapitulates the gene expression found in HCC patients with poor disease-free survival. From these analyses, we have determined that loss of SRC-2 cycling, whether accomplished by genetic manipulation or CCD, results in gene expression profiles that are similar to those found in patients with NAFLD and HCC.
Table 1.
Correlative SRC-2 Target Genes
| NAFLD/HCC Overlap | ||
|---|---|---|
| Better Survival in HCC Patients with: | ||
| Higher Expression | Lower Expression | No Data |
| Igf1 | Dnajc12 | Ikbke |
| Socs2 | Acly | P4ha2 |
| Aldh6a1 | Fasn | Hist1h4h |
| Srd5a1 | Sqle | C9 |
| Gch1 | Robo1 | Il1rap |
| Angptl4 | Mcm10 | Bbox1 |
| Gnmt | Fabp5 | Nnmt |
| Agxt2l1 | Col4a2 | Amdhd1 |
| Sds | Mvd | Hist1h4i |
| Acadsb | Hist2h4 | |
| Fat1 | 1810054D07Rik | |
| Treh | Saa1 | |
| Slco2a1 | ||
| NAFLD Gene Signature | ||
| Better Survival in HCC patients with: | ||
| Higher Expression | Lower Expression | No Data |
| Gpd1 | Agtr1a | Slc45a3 |
| Csad | Slc38a2 | LOC100039795 |
| Cyp7a1 | Fdps | Hal |
| Thrsp | ||
| Hpgd | ||
| Hsd17b2 | ||
| Axud1 | ||
Ablation of SRC-2 and CCD Show Advanced Aging and Increased Mortality
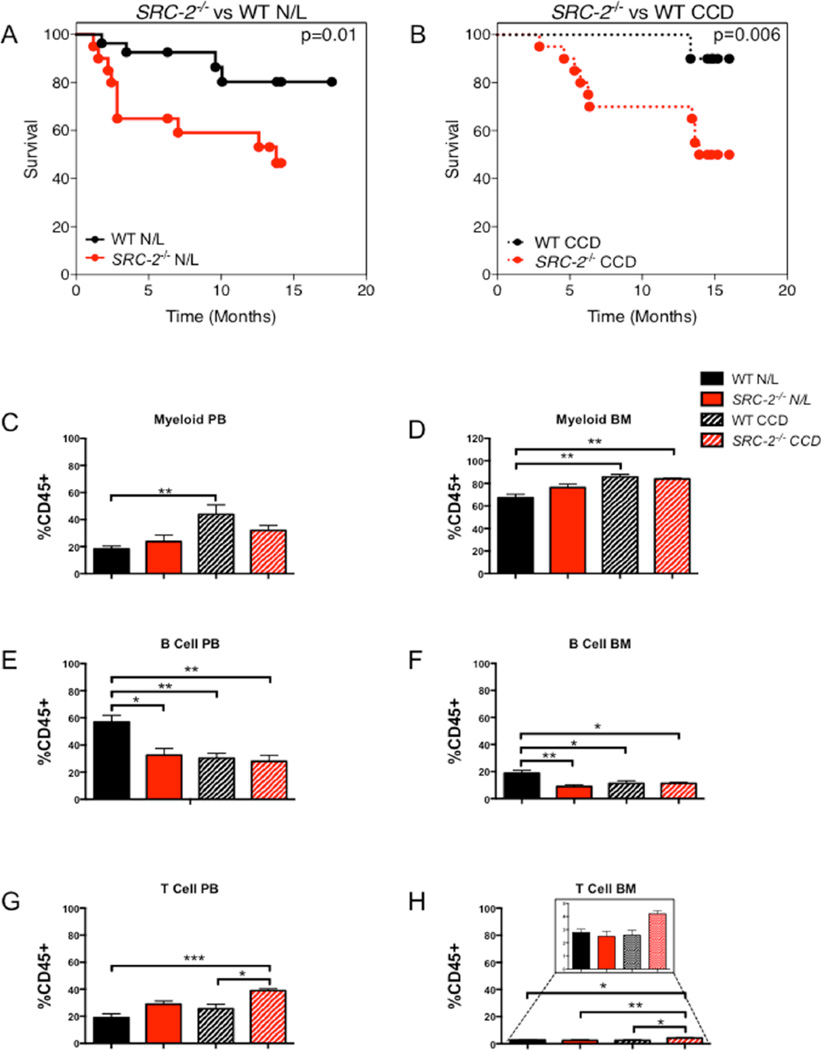
Disruption of circadian rhythmicity has been shown to negatively affect lifespan (Davidson et al., 2006; Froy, 2013; Yu and Weaver, 2011; Kondratova and Kondratov, 2012), and increased morbidity and mortality have been observed in several circadian mutant models (Dubrovsky et al., 2010; Kondratov, 2006). In fact, Bmal1−/− mice show enhanced signs of aging in PB with increased myelopoiesis and decreased lymphopoiesis (Kondratov, 2006). In line with other circadian knockout models, ablation of SRC-2 in N/L and CCD relative to WT mice results in significantly increased mouse mortality (Fig. 6A,B). Similar to the signs of advanced aging in Bmal1−/− mice, we observed an increase in the cells of the myeloid linage in PB and BM (Fig. 6C,D) and decreased B-cell counts (Fig. 6E,F) in SRC-2−/− mice compared with WT. CCD recapitulates the aging hematopoietic phenotype with respect to the myeloid and B-cell linages, as we observed increased myeloid and decreased B-cells in both the PB and BM with respect to WT mice on N/L (Fig. 6C,D). Aging is known to decrease lymphocytes (Kondratov, 2006), and we found a reduction in B-cells (PB and BM) upon loss of SRC-2 (Fig. 6E,F). Conversely, we found that loss of SRC-2 collectively increases the number of T-cells in PB, while SRC-2−/− mice in CCD conditions have an increase in T-cell number in BM (Fig. 6G,H). It is known that Kupffer cells and natural killer cells, both products of the myeloid linage, along with natural killer T-cells, increase in NAFLD (Ganz and Szabo, 2013). Consistent with these findings, SRC-2 ablation in the CCD model shows increased myeloid and T-cells in PB and BM, which likely reflect the compound effect of advanced aging and NAFLD inflammatory responses (Fig. 6G,H). Analysis of the spleen for myeloid, B-cells, and T-cells reveals no defect in clearance but rather implicates proliferation as the major contributing difference observed upon SRC-2 ablation (Suppl. Fig. S5A–C). Our hematopoietic analyses revealed that both individually and combined, SRC-2−/− and CCD reflect advanced signs of aging as well as NAFLD-like phenotypes.
Figure 6.
Loss of SRC-2 results in advanced aging and increased mortality. (A) Kaplan-Meier plot of WT and SRC-2−/− mice mortality rates maintained in N/L conditions. (B) Kaplan-Meier plot of WT and SRC-2−/− mice mortality rates subjected to CCD conditions. (C) Percent of myeloid PB cells in WT and SRC-2−/− mice in N/L or CCD. (D) Percent of myeloid BM cells in WT and SRC-2−/− mice subjected to N/L or CCD. (E) Percent of B-cells in PB in WT and SRC-2−/− mice subjected to N/L or CCD. (F) Percent of B-cells in BM in WT and SRC-2−/− mice subjected to N/L or CCD. (G) Percent of T-cells in PB in WT and SRC-2−/− mice subjected to N/L or CCD. (H) Percent of T-cells in BM in WT and SRC-2−/− mice maintained in N/L or CCD conditions. Data are graphed as the mean ± SEM. *p < 0.05. **p < 0.01. ***p < 0.001. (N = 3–4).
Ablation of SRC-2 and CCD Predict Poor Disease-free Survival in HCC
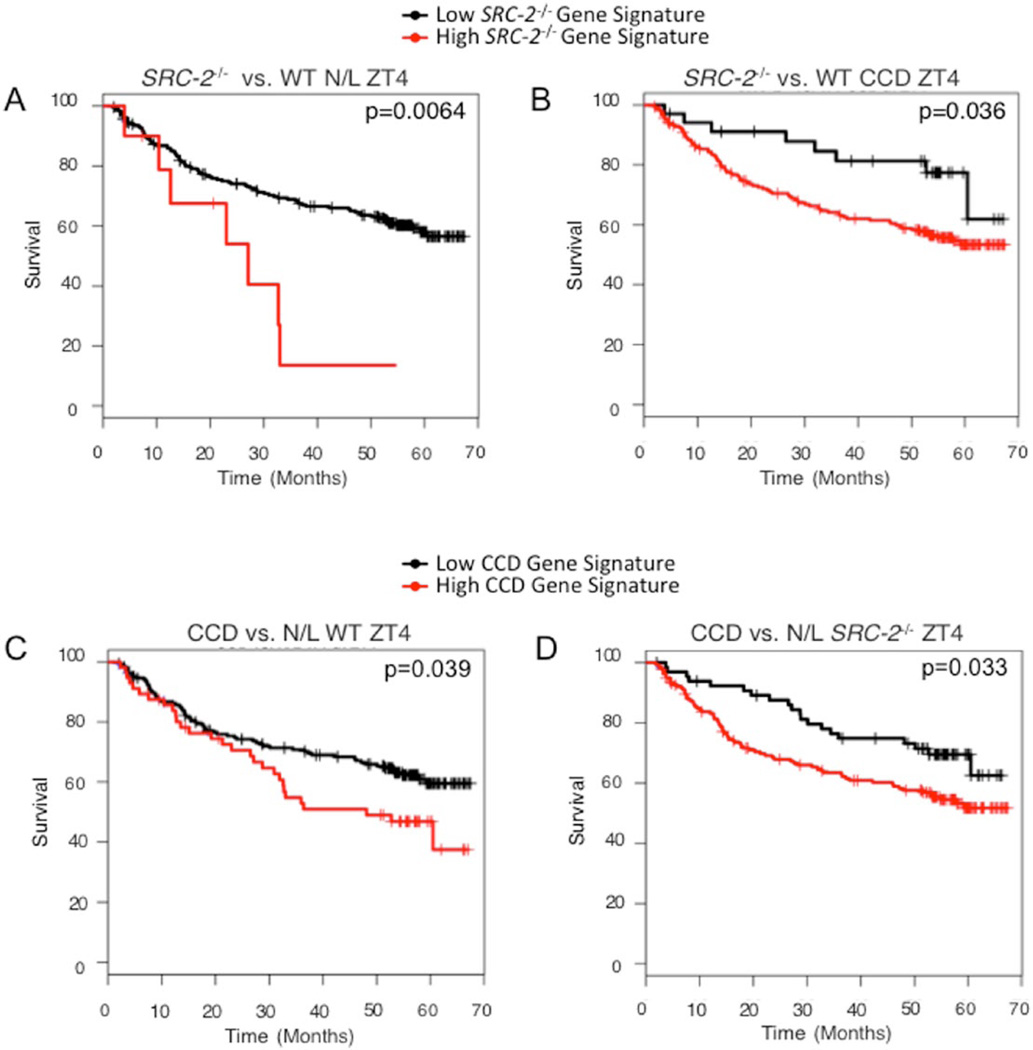
Published findings have defined SRC-2 as a suppressor of hepatic tumorigenesis, and low SRC-2 expression is a predictor of advanced mortality in HCC patients (O’Donnell et al., 2012). We also observed that loss of SRC-2 results in increased mortality in both N/L and CCD (see Fig. 6A,B). Moreover, the hepatic gene signatures of SRC-2−/− mice exposed to CCD are sufficient to stratify human HCC survival data in the same manner. HCC patients with a gene signature resembling that of SRC-2−/− mice in either N/L or CCD have significantly reduced survival rates (Fig. 7A,B). Additionally, patients with a gene signature mirroring those of mice subjected to CCD show reduced survival rates (Fig. 7C,D). Taken together, these findings demonstrate that the gene signatures produced from either SRC-2 ablation or CCD conditions are sufficient to stratify HCC patients into distinct prognostic groups. These data suggest that disruptions in circadian rhythmicity, whether genetic or environmental, result in increased morbidity and mortality in mice and show strong prognostic power in data obtained from human subjects.
Figure 7.
SRC-2−/− and CCD gene signatures stratify human HCC patient survival. (A) Kaplan-Meier survival plots of human HCC patients stratified based on gene expression of SRC-2−/− mice relative to WT at ZT4 subjected to N/L conditions (SRC-2−/− Gene Signature). (B) Kaplan-Meier survival plots of human HCC patients stratified based on gene expression of SRC-2−/− mice relative to WT at ZT4 in response to CCD. (C) Kaplan-Meier survival plots of human HCC patients stratified based on gene expression of CCD relative to N/L in SRC-2−/− mice at ZT4. (D) Kaplan-Meier survival plots of human HCC patients stratified based on gene expression of CCD relative to N/L in WT mice at ZT4.
DISCUSSION
Models of Circadian Disruption
Maintenance of circadian rhythmicity is a fundamental process that becomes disrupted in individuals subjected to shift work or jet lag conditions. Chronic circadian disruption (CCD) accelerates aging and increases the risk for cancer, metabolic syndrome, type 2 diabetes, obesity, and NAFLD (Wang et al., 2011; Megdal et al., 2005; Karlsson et al., 2001; Scheer et al., 2009; Kiessling et al., 2010; Parsons et al., 2015; Lin et al., 2014; Lin and Chen, 2015; Sookoian and Pirola, 2009). For this study, we chose a complex CCD model wherein the phase advance and phase delay periods were imbalanced (see Methods), thereby creating a dynamic system for stressing the circadian rhythmicity of mice. One key element of our CCD model was that after 52 weeks of circadian disruption, mice were sacrificed on the first day of their phase delay cycle, which importantly made them comparable to cohorts maintained on N/L but also induced an acute stress condition. Using this model, we show that while WT mice behaviorally adapt to changes in light exposure, mice devoid of SRC-2 fail to respond to circadian perturbation with impaired activity during the light phase, and both CCD and loss of SRC-2 result in increased hepatic steatosis, inflammation, and fibrosis.
SRC-2 Integrates Metabolic and Circadian Gene Expression
Phenotypic and molecular characterization of the SRC-2−/− mice have identified hallmarks of premature aging, including mitochondrial dysfunctions, disruption in circadian rhythmicity, and metabolic alterations (Duteil et al., 2010; Stashi et al., 2014; Chopra et al., 2008). Recent publications from our laboratory have defined SRC-2 as an essential transcriptional coactivator that coordinates circadian rhythmicity with metabolic homeostasis (Stashi et al., 2014; Zhu et al., 2015). SRC-2 is required for maintaining central clock (SCN) synchrony with the peripheral liver clock (Stashi et al., 2014). Mice devoid of SRC-2 have hepatic metabolic phenotypes that resemble those observed in Bmal1−/− mice, which include aberrant glycolysis, gluconeogenesis, and lipid metabolism (Hatanaka et al., 2010; Kettner et al., 2015; Chopra et al., 2008; Chopra et al., 2011; Fleet et al., 2015; Rudic et al., 2004; Zhang et al., 2014; Shimba et al., 2011). Alterations in hepatic glucose and lipid homeostasis are hallmarks of metabolic syndrome, and like the Bmal1−/− mice, SRC-2−/− mice display the obesity-associated comorbidity, NAFLD. SRC-2 functions in concert with a variety of transcription factor machinery to regulate the expression of hepatic gene programs that govern metabolic homeostasis. Ablation of SRC-2 attenuates the rhythmic 24-h oscillation of the genes encoding the core circadian clock genes Bmal1, Clock, Per1, Per2, Cry1, and Rev-erb/β (Stashi et al., 2014). Mechanistically, SRC-2 serves as a transcriptional amplifier of circadian transcription factors (i.e., BMAL1, CLOCK, ROR) leading to relaxation of chromatin structure that preserves the proper oscillatory expression of metabolic and circadian genes programs. During normal daily cycles, BMAL1 or ROR preferentially bind to closed chromatin and recruit interacting partners SRC-2 and PBAF, which remodels chromatin to an open configuration (Zhu et al., 2015). The open chromatin structure coincides with preferential dismissal of BMAL1/ROR and recruitment of REV-ERB α/β whereby SRC-2 facilitates the repressive phase of circadian gene expression (Zhu et al., 2015). Herein, we provide evidence for how loss of the circadian coactivator SRC-2 results in the hepatic milieu of aberrant glucose and lipid metabolism that contributes to the development and progression of NAFLD.
Phenotypic vs. Transcriptomic Signatures
The complicated pathogenesis of NAFLD is illustrated in our SRC-2−/− and CCD models. Individually, loss of SRC-2 or CCD results in similar alterations in hepatic gene expression, but the combination of the 2 circadian insults produces a robust transcriptomic profile that resembles the stages of fatty liver disease progression. Consistent with our histological observations, hepatic transcriptomic data from CCD and SRC-2−/− mice show enrichment for gene signatures associated with human NAFLD outcomes, particularly NASH and HCC. Although individually, SRC-2 ablation and CCD partially resemble the directionality of human NASH and HCC gene signatures, exposure of SRC-2−/− mice to CCD results in a hepatic gene expression profile that strongly correlates with NASH/HCC patient data. These findings are somewhat paradoxical when one considers that the combined stress of CCD and SRC-2 ablation fails to produce additive phenotypic effects (i.e., activity, body composition, serum biochemistry, histology). While each of these phenotypic readouts is valuable for understanding the fractional contribution of genetic or environmentally induced circadian stress, these data are limited because of the inability to directly integrate them with patient data. Importantly, the transcriptomic approach used in this study provided a robust and unbiased dataset for seamless integration with hepatic gene signatures from humans harboring progressive forms of hepatic pathology (i.e., NAFLD < NASH < HCC).
Insights for Human Liver Disease
Left unabated, NAFLD is a well-established predictor of hepatocellular carcinoma, which increases morbidity and mortality and may contribute to the decreased survival observed in SRC-2−/− mice. Consistent with these findings, we and others have shown that SRC-2 functions as a hepatic tumor suppressor (O’Donnell et al., 2012), and here we demonstrate that gene signatures associated with loss of SRC-2 and CCD also confer poor disease-free survival in human HCC patient data. Collectively, these data suggest that prolonged CCD and ablation of SRC-2 create a predisposition for HCC. In fact, case studies of shift workers reveal that these individuals are at a greater risk for liver damage and liver cancer than those exposed to normal light schedules (Lin et al., 2014; Lin and Chen, 2015; Echave Llanos and Nash, 1970). Collectively, our data demonstrate that SRC-2 is at the nexus of metabolic and circadian regulation of liver gene programs. Ablation of SRC-2 phenocopies the outcomes of CCD showing a transcriptomic switch toward NAFLD/HCC progression, increased aging and mortality as well as the hepatic steatotic phenotype observed in many circadian gene knockout models (Mazzoccoli et al., 2014). Our findings perhaps identify a potential screening mechanism for people who experience forms of circadian disruption since those with metabolic signatures of the clock mutants, such as perturbations in SRC-2 expression, are at greater risk for developing NAFLD and other metabolic comorbidities such as HCC that influence patient survival. Our studies establish SRC-2 as a molecular link between the genetic and environmental factors that contribute to the metabolic and tumorigenic comorbidities associated with chronic circadian disruption.
Acknowledgments
We thank Rainer Lanz for lessons on relational database management. We thank Sean McGuire for critical reading of the manuscript. We thank Dr. Corey Reynolds from the Mouse Phenotyping Core at BCM for technical support. This research was supported by grants from the NIH (F31DK107254 to T.F., F31CA171350 to E.S., P01 DK59820 and 3U19DK062434-10W1 to B.W.O.) and Diabetes Research Center (DRC) grant NIDDK-P30 DK079638. Partial support was provided by NIH grant GM033976. Additionally, research support was provided by the Welch Foundation (Q1521) and from the Center for the Advancement of Science in Space (CASIS) Integrated OMICs Award to B.Y. and B.W.O. Partial support for this work was provided by a grant to Baylor College of Medicine from the Howard Hughes Medical Institutes through the Med into Grad Initiative, and to the Medical Scientist Training Program NIGMS T32 GM7330.
Biographies

Tiffany Fleet
Baylor College of Medicine

Kathrina Marcelo
Baylor College of Medicine
Cristian Coarfa
Baylor College of Medicine

Brian York
Baylor College of Medicine
Footnotes
CONFLICT OF INTEREST
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Supplementary online material is available on the journal’s website at http://jbr.sagepub.com/supplemental.
REFERENCES
- Ahrens M, Ammerpohl O, von Schönfels W, Kolarova J, Bens S, Itzel T, Teufel A, Hermann A, Brosch M, Hinrichsen H, et al. DNA methylation analysis in nonalcoholic fatty liver disease suggests distinct disease-specific and remodeling signatures after bariatric surgery. Cell Metab. 2013;18(2):296–302. doi: 10.1016/j.cmet.2013.07.004. [DOI] [PubMed] [Google Scholar]
- Akashi M, Takumi T. The orphan nuclear receptor rorα regulates circadian transcription of the mammalian core-clock Bmal1. Nat Struct Mol Biol. 2005;12(5):441–448. doi: 10.1038/nsmb925. [DOI] [PubMed] [Google Scholar]
- Barclay JL, Husse J, Bode B, Naujokat N, Meyer-Kovac J, Schmid SM, Lehnert H, Oster H. Circadian desynchrony promotes metabolic disruption in a mouse model of shiftwork. PLoS One. 2012;7(5):e37150. doi: 10.1371/journal.pone.0037150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning JD, Szczepaniak LS, Dobbins R, Nuremberg P, Horton JD, Cohen JC, Grundy SM, Hobbs HH. Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology. 2004;40(6):1387–1395. doi: 10.1002/hep.20466. [DOI] [PubMed] [Google Scholar]
- Bugge A, Feng D, Everett LJ, Briggs ER, Mullican SE, Wang F, Jager J, Lazar MA. Rev-Erb and Rev-Erb coordinately protect the circadian clock and normal metabolic function. Genes Dev. 2012;26(7):657–667. doi: 10.1101/gad.186858.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103(7):1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Han, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, Chong LW, DiTacchio L, Atkins AR, Glass CK, et al. Regulation of circadian behaviour and metabolism by REV-ERB-A and REV-ERB-B. Nature. 2012;485(7396):123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho K, Ennaceur A, Cole JC, Suh CK. Chronic jet lag produces cognitive deficits. J Neurosci. 2000;20(6):RC66. doi: 10.1523/JNEUROSCI.20-06-j0005.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra AR, Louet J, Saha P, An J, DeMayo F, Xu J, York B, Karpen S, Finegold M, Moore D, et al. Absence of the SRC-2 coactivator results in a glycogenopathy resembling Von Gierke’s disease. Science. 2008;322(5906):1395–1399. doi: 10.1126/science.1164847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra AR, Kommagani R, Saha P, Louet JF, Salazar C, Song J, Jeong J, Finegold M, Viollet B, DeMayo F, et al. Cellular energy depletion resets whole-body energy by promoting coactivator-mediated dietary fuel absorption. Cell Metab. 2011;13(1):35–43. doi: 10.1016/j.cmet.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson AJ, Sellix MT, Daniel J, Yamazaki S, Menaker M, Block GD. Chronic jet-lag increases mortality in aged mice. Curr Biol. 2006;16(21):R914–R916. doi: 10.1016/j.cub.2006.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P, Kibbe WA, Lin SM. Lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24(13):1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- Dubrovsky YV, Samsa WE, Kondratov RV. Deficiency of circadian protein CLOCK reduces lifespan and increases age-related cataract development in mice. Aging. 2010;2(12):936–944. doi: 10.18632/aging.100241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duteil D, Chambon C, Ali F, Malivindi R, Zoll J, Kato S, Geny B, Chambon P, Metzger D. The transcriptional coregulators TIF2 and SRC-1 regulate energy homeostasis by modulating mitochondrial respiration in skeletal muscles. Cell Metab. 2010;12(5):496–508. doi: 10.1016/j.cmet.2010.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echave Llanos JM, Nash RE. Mitotic circadian rhythm in a fast-growing and a slow-growing hepatoma: mitotic rhythm in hepatomas. J Natl Cancer Inst. 1970;44(3):581–585. [PubMed] [Google Scholar]
- Eckel-Mahan KL, Patel VR, Mohney RP, Vignola KS, Baldi P, Sassone-Corsi P. Coordination of the transcriptome and metabolome by the circadian clock. Proc Natl Acad Sci U S A. 2012;109(14):5541–5546. doi: 10.1073/pnas.1118726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleet T, Zhang B, Lin F, Zhu B, Dasgupta S, Stashi E, Tackett B, Thevanather S, Rajapakshe KI, Gonzales N, et al. SRC-2 orchestrates polygenic inputs for fine-tuning glucose homeostasis. Proc Natl Acad Sci U S A. 2015;112(44):E6068–E6077. doi: 10.1073/pnas.1519073112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froy O. Circadian aspects of energy metabolism and aging. Ageing Res Rev. 2013;12(4):931–940. doi: 10.1016/j.arr.2013.09.002. [DOI] [PubMed] [Google Scholar]
- Ganz M, Szabo G. Immune and inflammatory pathways in NASH. Hepatol Int. 2013;7(Suppl 2):771–781. doi: 10.1007/s12072-013-9468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehin M, Mark M, Dennefeld C, Dierich A, Gronemeyer H, Chambon P. The function of TIF2/GRIP1 in mouse reproduction is distinct from those of SRC-1 and P/CIP. Mol Cell Biol. 2002;22(16):5923–5937. doi: 10.1128/MCB.22.16.5923-5937.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenawalt DM, Dobrin R, Chudin E, Hatoum IJ, Suver C, Beaulaurier J, Zhang B, Castro V, Zhu J, Sieberts KS, et al. A survey of the genetics of stomach, liver, and adipose gene expression from a morbidly obese cohort. Genome Res. 2011;21(7):1008–1016. doi: 10.1101/gr.112821.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatanaka F, Matsubara C, Myung J, Yoritaka T, Kamimura N, Tsutsumi S, Kanai A, Suzuki Y, Sassone-Corsi P, Aburatani H, et al. Genome-wide profiling of the core clock protein BMAL1 targets reveals a strict relationship with metabolism. Mol Cell Biol. 2010;30(24):5636–5648. doi: 10.1128/MCB.00781-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatoum IJ, Greenawalt DM, Cotsapas C, Daly MJ, Reitman ML, Kaplan LM. Weight loss after gastric bypass is associated with a variant at 15q26.1. Am J Hum Genet. 2013;92(5):827–834. doi: 10.1016/j.ajhg.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong JW, Kwak I, Lee KY, White LD, Wang XP, Brunicardi PC, O’Malley BW, DeMayo FJ. The genomic analysis of the impact of steroid receptor coactivators ablation on hepatic metabolism. Mol Endocrinol. 2006;20(5):1138–1152. doi: 10.1210/me.2005-0407. [DOI] [PubMed] [Google Scholar]
- Karlsson B, Knutsson A, Lindahl B. Is there an association between shift work and having a metabolic syndrome? Results from a population based study of 27,485 people. Occup Environ Med. 2001;58(11):747–752. doi: 10.1136/oem.58.11.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettner NM, Mayo SA, Hua J, Lee C, Moore DD, Fu L. Circadian dysfunction induces leptin resistance in mice. Cell Metab. 2015;22(3):448–459. doi: 10.1016/j.cmet.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiessling S, Eichele G, Oster H. Adrenal glucocorticoids have a key role in circadian resynchronization in a mouse model of jet lag. J Clin Invest. 2010;120(7):2600–2609. doi: 10.1172/JCI41192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiner DE, Brunt EM, Van Natta M, Behling C, Contos MJ, Cummings OW, Ferrell LD, Liu YC, Torbenson MS, Unalp-Arida A, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41(6):1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- Kondratov RV. Early aging and age-related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev. 2006;20(14):1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratova AA, Kondratov RV. The circadian clock and pathology of the ageing brain. Nat Rev Neurosci. 2012;13(5):325–335. doi: 10.1038/nrn3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Chen PC. Persistent rotating shift work exposure is a tough second hit contributing to abnormal liver function among on-site workers having sonographic fatty liver. Asia Pac J Public Health. 2015;27(2):NP1765–NP1774. doi: 10.1177/1010539512469248. [DOI] [PubMed] [Google Scholar]
- Lin YC, Hsieh IC, Chen PC. Long-term day-and-night rotating shift work poses a barrier to the normalization of alanine transaminase. Chronobiol Int. 2014;31(4):487–495. doi: 10.3109/07420528.2013.872120. [DOI] [PubMed] [Google Scholar]
- Mazzoccoli G, Vinciguerra M, Oben J, Tarquini R, De Cosmo S. Non-alcoholic fatty liver disease: the role of nuclear receptors and circadian rhythmicity. Liver Int. 2014;34(8):1133–1152. doi: 10.1111/liv.12534. [DOI] [PubMed] [Google Scholar]
- Megdal SP, Kroenke CH, Laden F, Pukkala E, Schernhammer ES. Night work and breast cancer risk: a systematic review and meta-analysis. Eur J Cancer. 2005;41(13):2023–2032. doi: 10.1016/j.ejca.2005.05.010. [DOI] [PubMed] [Google Scholar]
- O’Donnell KA, Keng VW, York B, Reineke EL, Seo C, Fan C, Silverstein KA, Schrum CT, Xie WR, Mularoni L, et al. A sleeping beauty mutagenesis screen reveals a tumor suppressor role for Ncoa2/Src-2 in liver cancer. Proc Natl Acad Sci U S A. 2012;109(21):E1377–E1386. doi: 10.1073/pnas.1115433109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obika M, Noguchi H. Diagnosis and evaluation of nonalcoholic fatty liver disease. Exp Diabetes Res. 2012;2012:145754. doi: 10.1155/2012/145754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons MJ, Moffitt TE, Gregory AM, Goldman-Mellor S, Nolan PM, Poulton R, Caspi A. Social jet-lag, obesity and metabolic disorder: investigation in a cohort study. Int J Obes (Lond) 2015;39(5):842–848. doi: 10.1038/ijo.2014.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partch CL, Green CB, Takahashi JS. Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 2013;24(2):90–99. doi: 10.1016/j.tcb.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AB, Field MD, Maywood ES, Hastings MH. Differential resynchronisation of circadian clock gene expression within the suprachiasmatic nuclei of mice subjected to experimental jet lag. J Neurosci. 2002;22(17):7326–7330. doi: 10.1523/JNEUROSCI.22-17-07326.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler S, Long EL, Budhu A, Chen Y, Zhao X, Ji J, Walker R, Jia HL, Ye QH, Qin LX, et al. Integrative genomic identification of genes on 8p associated with hepatocellular carcinoma progression and patient survival. Gastroenterology. 2012;142(4):957.e12–966.e12. doi: 10.1053/j.gastro.2011.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler S, Jia HL, Budhu A, Forgues M, Ye QH, Lee JS, Thorgeirsson SS, Sun Z, Tang ZY, Qin LX, et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010;70(24):10202–10212. doi: 10.1158/0008-5472.CAN-10-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB, Fitzgerald GA. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2(11):e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer FAJL, Hilton MF, Mantzoros CS, Shea SA. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci U S A. 2009;106(11):4453–4458. doi: 10.1073/pnas.0808180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimba S, Ogawa T, Hitosugi S, Ichihashi Y, Nakadaira Y, Kobayashi M, Tezuka M, Kosuge Y, Ishige K, Ito Y, et al. Deficient of a clock gene, brain and muscle Arnt-like protein-1 (BMAL1), induces dyslipidemia and ectopic fat formation. PLoS One. 2011;6(9):e25231. doi: 10.1371/journal.pone.0025231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- Sookoian S, Pirola CJ. Shift work and subclinical atherosclerosis: recommendations for fatty liver disease detection. Atherosclerosis. 2009;207(2):346–347. doi: 10.1016/j.atherosclerosis.2009.06.008. [DOI] [PubMed] [Google Scholar]
- Starmann J, Falth M, Spindelbock W, Lanz KL, Lackner C, Zatloukal K, Trauner M, Sultmann H. Gene expression profiling unravels cancer-related hepatic molecular signatures in steatohepatitis but not in steatosis. PLoS One. 2012;7(10):e46584. doi: 10.1371/journal.pone.0046584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stashi E, Lanz RB, Mao J, Michailidis G, Zhu B, Kettner NM, Putluri N, Reineke EL, Dasgupta S, Dean A, et al. SRC-2 Is an essential coactivator for orchestrating metabolism and circadian rhythm. Cell Rep. 2014;6(4):633–645. doi: 10.1016/j.celrep.2014.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapp WN, Natelson BH. Circadian rhythms and patterns of performance before and after simulated jet lag. Am J Physiol. 1989;257(4 Pt 2):R796–R803. doi: 10.1152/ajpregu.1989.257.4.R796. [DOI] [PubMed] [Google Scholar]
- United States Department of Labor. Workers on Flexible and Shift Schedules in May 2004. Washington, DC: United States Department of Labor; 2005. [Google Scholar]
- Wang XS, Armstrong MEG, Cairns BJ, Key TJ, Travis RC. Shift work and chronic disease: the epidemiological evidence. Occup Med. 2011;61(2):78–89. doi: 10.1093/occmed/kqr001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu EA, Weaver CR. Disrupting the circadian clock: gene-specific effects on aging, cancer, and other phenotypes. Aging. 2011;3(5):479–493. doi: 10.18632/aging.100323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Tong X, Arthurs B, Guha A, Rui L, Kamath A, Inoki K, Yin L. Liver clock protein BMAL1 promotes de novo lipogenesis through insulin-mTORC2-AKT signaling. J Biol Chem. 2014;289(37):25925–25935. doi: 10.1074/jbc.M114.567628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Gates LA, Stashi E, Dasgupta S, Gonzales N, Dean A, Dacso CC, York B, O’Malley BW. Coactivator-dependent oscillation of chromatin accessibility dictates circadian gene amplitude via REV-ERB loading. Mol Cell. 2015;60(5):769–783. doi: 10.1016/j.molcel.2015.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]