Abstract
The genes encoding Shiga toxin (Stx), the major virulence factor of Shiga toxin-producing Escherichia coli, are carried in the genomes of bacteriophages that belong to the lambdoid family of phages. Previous studies demonstrated that induction of prophages encoding stx significantly enhances the production and/or release of Stx from the bacterium. Therefore, factors that regulate the switch between lysogeny and lytic growth, e.g., repressor, operator sites, and associated phage promoters, play important roles in regulating the production and/or release of Stx. We report the results of genetic and biochemical studies characterizing these elements of the Stx-encoding bacteriophage 933W. Like λ, 933W has three operator repeats in the right operator region (OR), but unlike λ and all other studied lambdoid phages, which have three operator repeats in the left operator region (OL), 933W only has two operator repeats in OL. As was observed with λ, the 933W OR and OL regions regulate transcription from the early PR and PL promoters, respectively. A lysogen carrying a 933W derivative encoding a noncleavable repressor fails to produce Stx, unlike a lysogen carrying a 933W derivative encoding a cleavable repressor. This finding provides direct evidence that measurable expression of the stx genes encoded by a 933W prophage requires induction of that prophage with the concomitant initiation of phage gene expression.
Shiga toxin-producing Escherichia coli (STEC) strains are a group of food-borne pathogens that can infect humans, causing a range of illnesses including hemorrhagic colitis and life-threatening sequelae such as hemolytic uremic syndrome (26, 50). Shiga toxins (Stx), a group of cytopathic toxins first identified in Shigella dysenteriae (reviewed in reference 39), are essential virulence factors of STEC, eliciting many of the pathological features associated with STEC infections (42). Stx holotoxins, classified as AB5 toxins, are composed of one enzymatic A subunit and five receptor-binding B subunits (reviewed in reference 38). The A subunit is an rRNA N-glycosidase, catalyzing the removal of a specific adenine residue from the 28S rRNA of the 60S ribosomal subunit, resulting in inhibition of protein synthesis in the target cell (48). STEC produce two types of Stx proteins: Stx1, which is essentially identical to Stx of S. dysenteriae, and Stx2, which differs in sequence but not in function from Stx1 (38).
In most identified STEC strains, the toxin genes, stxAB, are located in the genomes of prophages that resemble the coliphage λ (7, 22, 24, 27, 40, 58). Lambdoid phages share similar genome organization and schemes of transcription control (3, 20). The λ prophage remains in a quiescent state due to binding of the cI-encoded repressor protein to the right and left operator sites, in this way inhibiting transcription from the phage early promoters PR and PL, respectively (47). Prophage induction, which leads to lytic growth, results from removal of repression. This event occurs primarily through RecA-mediated repressor autocleavage following activation of the bacterial SOS response by a DNA-damaging agent (32). During lytic growth, transcription from the early PR promoter modified through the action of the phage-encoded N protein transcends a number of terminators, resulting in expression of the Q protein (reviewed in reference 14). Q, in turn, modifies transcription initiating at the late PR′ promoter, which, as shown for λ, results in synthesis of a ∼26-kb mRNA that encodes all late functions (52). The location of the stx genes, downstream of PR′ and upstream of the lysis cassette, is conserved among most identified Stx-encoding prophages (27, 37, 43, 51, 58) (see Fig. 1A). Thus, prophage induction ultimately results in Q-modified transcription initiating at PR′ that transcends the tR′ terminator, leading to expression of downstream genes that include stx and those encoding lysis functions. No obvious mode of release of Stx by the intact bacterium has been identified (38). However, it has been suggested, with some corroborating evidence, that phage-mediated lysis provides the route for Stx release (36, 60). Studies with an stx2-encoding phage provide compelling evidence that Stx production and release can occur primarily from a small fraction of a STEC population that undergoes prophage induction (60). At least in that case, the repressor-operator interactions must play a major role in the process of Stx production and release.
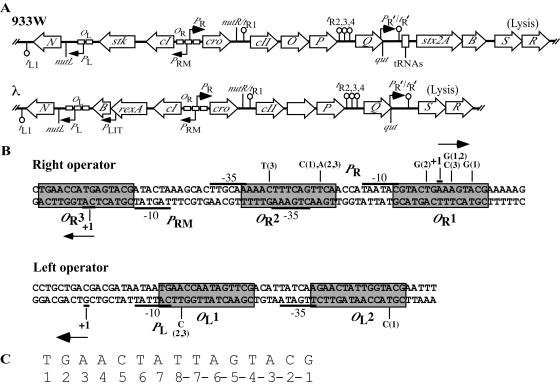
FIG. 1.
(A) Map of the regulatory region of the bacteriophage 933W genome showing relevant genes (not drawn to scale) based on sequence analysis (43). The open reading frames are drawn as arrows pointed in the direction of transcription inferred from studies of transcription of analogous genes of λ. Transcription terminators are represented as lollipop structures. For comparison, a map of the regulatory region of bacteriophage λ is included below the phage 933W map. (B) Sequence of phage 933W OR and OL and associated promoters. Phage promoters PR, PL, and PRM were identified by primer extension studies reported in this work (Fig. 4). Identified transcription start sites of the promoters are designated by +1, and arrows illustrate the direction of transcription initiating at the described promoters. Operator sites are boxed in gray. Putative operator repeats in both OL and OR were assigned by sequence analysis (11, 43; also see the text). Nucleotide changes found in λimm933Wvir mutants are listed above the wild-type sequence in OR and below the wild-type sequence in OL. The numbers in parentheses listed with the nucleotide changes refer to the vir mutant in which these changes are present: 1, λimm933Wvir-5; 2, λimm933Wvir-6; 3, λimm933WvirT-3A. (C) Consensus sequence derived from an alignment of the five 933W operator-binding repeats. Listed below the sequence is the numbering of the nucleotide positions in the sequence as used in this work.
The operator regions in lambdoid phages flank the cI gene and are usually composed of three related sequences with partial dyad symmetry (45). The details of repressor-operator interactions have been elucidated for λ primarily by the work of Ptashne and his many collaborators (45). The λ cI-encoded repressor binds as a dimer to the operator repeats but not with equal affinities (reviewed in reference 29). The CI dimers cooperatively bind to OR1 and OR2 to form a tetramer, repressing PR, and OL1 and OL2 to form a tetramer, repressing PL (see Fig. 1). Moreover, recent studies have shown that interaction of the tetramers bound at OL with those bound at OR further stabilizes repressor binding (8, 49). However, there are variations in the arrangement of operator repeats in other lambdoid phages (4). For example, phage H-19B, carrying stx1, possesses four operator sites in OR (55). In addition, analyses of repressor interactions with OR suggest that H-19B repressor binding may differ from that observed in λ (55). Studies conducted with a genetic tool designed to identify lysogens in which prophages spontaneously induce indicate that H-19B and 933W induce more readily than lambdoid phages that do not encode Stx (33). The apparent central role of the repressor-operator interaction in Stx expression by phages carrying stx2 led us to investigate the repressor-operator region of phage 933W, which carries stx2, and directly assess its role in regulating Stx expression.
MATERIALS AND METHODS
Media and strain construction.
Table 1 lists the strains and phages used in these studies. Bacteria were cultured in Luria broth (LB) at 37°C, unless otherwise stated. Mitomycin C, chloramphenicol, spectinomycin, ampicillin, tetracycline, and kanamycin were used at concentrations of 2, 25, 80, 100, 15, and 50 μg/ml, respectively, unless otherwise stated.
TABLE 1.
Bacterial strains and phages used in this work
| Strain or phage | Relevant descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| K37 | N99 | NIH collection |
| K9680 | K37(λimm933W) | This laboratory |
| K9675 | K37(933W) | 57 |
| K10570 | K37 mini-λ; Tcr | 6 |
| K10575 | K10570(933W); Tcr | This laboratory |
| K10599 | K10575 cured of mini-λ | This laboratory |
| K10595 | K37(933WcI-K178N); cI(Ind−) mutant | This laboratory |
| K9537 | W3110 ΔlacU169 gal490 λcI857Δ(cro-bioA) sun::cat; Cmr | D.L. Court |
| K9837 | K9537(λimm933W); Cmr | This laboratory |
| K9838 | K9837 pGB2-plac-NH-19B; Cmr Spr | This laboratory |
| K9835 | K9680 pGB2-plac-NH-19B; Spr | This laboratory |
| K10693 | K37(λimm933WcI-K178N); cI(Ind−) mutant; Knr | This laboratory |
| K10710 | K37(λimm933WcI-K178N); cI(Ind−) mutant; λ resistant; mutS::Ampr, Knr | This laboratory |
| K10717 | K37(λimm933WcI-K178N, T169I); cI(Ind−) ts mutant; Knr | This laboratory |
| K10786 | BL21(DE3) pET-33b(+) (Novagen)+cI; encodes rCI; Knr | This laboratory |
| Phages | ||
| 933WcIind | 933WcI-K178N ind mutant | This laboratory |
| λcI857Kanr | A viable λcI857 carrying a Kanr cassette | Ryland Young |
| λimm933W | Hybrid phage of λ with immunity region of 933W | This laboratory |
| λimm933Wvir | λimm933W virulent mutant | This laboratory |
| λimm933WcIind Kanr | λimm933WcI-K178N Ind− mutant | This laboratory |
| λimm933WcIind ts Kanr | λimm933WcI-K178N, T168I; induces at 42°C | This laboratory |
Cmr, chloramphenicol resistant; Spr, spectinomycin resistant; Tcr, tetracycline resistant; Knr, kanamycin resistant.
(i) Construction of lysogens
933W and λimm933W lysogens were constructed essentially by procedures developed for λ (17).
(ii) Isolation of λimm933Wvir.
A ∼250-bp DNA fragment containing the putative OR region of 933W was mutagenized using the Mn-dITP PCR method (63). The primers used in PCR amplification were 5′-GGCTATAGCCATTCCCCTAC-3′ and 5′-CGCTAACTCCACAAGCCTTC-3′. Isolated PCR products were electroporated into competent cells (64) of K9838, a contiguous dilysogen with the hybrid prophage λimm933W and a defective λcI857 prophage. In addition, K9838 carries a plasmid that constitutively expresses the N protein of H-19B, which is functionally the same as N of 933W (33). The defective prophage encodes the thermally sensitive λ CI857 repressor, the λ Red recombination functions, and the λ Xis and Int proteins (64). The Xis and Int proteins act to excise the λimm933W prophage from the bacterial chromosome (18). Following electroporation, the culture was incubated at 32°C in 1 ml of LB for approximately 1.5 h. The culture was diluted to 10 ml and incubated overnight at 39°C. Incubation at the high temperature allows for constitutive expression from the defective prophage of both the Red recombination and Int and Xis excision functions. This resulted in recombination of the electroporated DNA with the λimm933W prophage and excision of the prophage from the bacterial chromosome.
Partially virulent phages, with mutations in OR that reduce repressor binding, were isolated from the lysate as plaque formers on a lawn formed by K9835, a λimm933W lysogen that also carries a plasmid that constitutively expresses the N protein. Lysates of λimm933W partial vir mutants, confirmed by their ability to grow on lawns formed by K9835 and K37, but not K9680 (a λimm933W lysogen), were then made from single plaques. Spontaneous fully virulent mutants were selected from lysates of partially virulent mutants as plaque formers on lawns of K9680.
(iii) Construction of λimm933Wvir with targeted mutations in OR.
Partially virulent λimm933W mutants with a designed mutation in OR2 were constructed using the λ Red system to cross the mutation from a synthesized single-stranded oligonucleotide (10) into the λimm933W prophage in K9838. This procedure was conducted essentially as described above for the isolation of λimm933W partial vir mutants. The sequence of the oligonucleotide was 5′-CATGAGTACGATACTAAAGCACTTGCAAAAACTTT-CAGTACAACCATAA-3′. Spontaneous fully virulent mutants were selected from partially virulent mutant lysates as plaque formers on lawns of K9680.
(iv) Construction of 933WcI(Ind−).
A 933W prophage with an ind mutation was constructed using the mini-λ red recombination system (6, 30). Based on previous studies with other SOS-inducible repressors (56), Lys codon 178 was changed from AAG to an Asn codon, AAC, in strain K10575, a 933W lysogen. The single-stranded DNA oligonucleotide 5′-ATGATCCAGATGCCTTTGGTCTTCGTGTGAAAGGAGACGCAATGTGGCCCAGAATAAAATCAGGAGAATATGTACTC-3′, containing the nucleotide substitution, was used to generate this codon change. Lysogens containing the Lys-to-Asn codon change in the 933W cI gene were identified by the mismatch amplification mutation assay PCR technique (5) using primers that yield a product only if the DNA contains the specified codon change. The strain with the wild-type cI and the strain with the Lys-to-Asn codon change in cI were cured of their mini-λ by a heat pulse at 42°C (6), generating strains K10599 and K10595, respectively.
(v) Construction of λimm933WcI(Ind−).
A λimm933WcIind hybrid phage was constructed by crossing the cI(Ind−) allele from the prophage in K10595 to an infecting λcI857Kan phage (obtained from Ry Young). Following the infection, recombinants with 933W immunity were obtained as plaque formers on a lawn formed by K124 (a λ lysogen). The centers of turbid plaques were picked and streaked on an LB-plus-kanamycin plate for colonies. Putative lysogens were shown to have 933W immunity and to be refractory to mitomycin C treatment. The K37 derivative containing the λimm933WcI(Ind−) Kanr prophage was designated K10693.
(vi) Construction of λimm933WcI(Ind−) ts.
The starting strain used in the isolation of the cI temperature-sensitive (ts) mutation, K10710, is a derivative of K37 that is resistant to λ and has the mutS::amp allele (13) and a λimm933WcI(Ind−) Kanr prophage. The mutS allele was included to increase the mutation rate, and λ resistance was included to eliminate reinfection by released phage. The Ind− mutation largely eliminates spontaneous induction of the prophage. K10710 was grown in LB to mid-log phase, and bacteria were sedimented, resuspended in an equal volume of LB, and incubated at 42°C with shaking for 4 h. The bacteria were treated with CHCl3 and sedimented. The supernatant was plated on a K37 lawn and incubated overnight at 32°C for plaques. The centers of turbid plaques were picked for colonies by streaking onto LB-plus-kanamycin plates, which were incubated at either 32 or 42°C. Two of five tested colonies grew at 32°C and not at 42°C. Further testing showed that the prophages in these bacteria were induced at high temperature. DNA sequencing identified a codon change, causing a Thr-to-Ile change, at codon 170 as well as the Ind− mutation at codon 178. The K37 lysogen with the λimm933WcI(Ind−) ts prophage was designated K10717.
Construction of a plasmid with the cI gene.
The 933W cI gene was PCR amplified from strain K9675 using primers 5′-CCATGGTTCAGAATGAAAAAGTGC-3′ and 5′-CTCGAGCGAACTTTTCAG-CCACTCCCT-3′. The isolated PCR product was digested with NcoI and XhoI and inserted into the pET-33b(+) vector (Novagen) downstream of the T7 promoter by using the appropriate sites, yielding a fusion of the cI gene in frame with the coding sequence for a His6 tag. DNA sequencing confirmed the integrity of the insert. The constructed vector was then transformed into BL21(DE3), generating strain K10786. In vivo tests showed that the C-terminal His tag has no effect on immunity to superinfection.
Purification of rCI protein.
Recombinant CI (rCI) repressor protein fused to a His6 tag at its carboxy terminus was isolated from a culture of strain K10786 by using the Qiagen (Valencia, Calif.) protocol for isolating His6-tagged proteins under native conditions.
Gel mobility shift assays.
Radioactive DNA probes for OR and OL were generated by labeling the 5′ end of the sense primer with [γ-32P]ATP and subjecting the respective probe to PCR amplification with an unlabeled antisense primer, using DNA from a vir phage lysate or strain K9675 as the template (55). The primers used in the PCR amplification of the ∼250-bp OR probe are the same as those used for the mutagenesis of OR in the isolation of vir mutants. The primers used in the PCR amplification of the ∼220-bp OL probe were 5′-CATAAGCCCTTCTGATGACAA-3′ and 5′-ATAGACCTCCTG-ATCAACTTT-3′. The PCR products were isolated by ethanol precipitation (54). Gel mobility shift assays (55) were performed using increasing concentrations of purified rCI.
Isolation of RNA.
RNA used in the identification of the 933W PR and PL promoters was purified from extracts derived from infection of K37 with λimm933W (55). RNA used in the identification of the 933W PRM promoter was isolated from a culture of K9675 grown to mid-log phase. RNA was extracted by the Trizol method (65).
Primer extension analyses.
The 933W PR, PL, and PRM promoters were identified by primer extension analyses (65). The primers used to PCR amplify the ∼250-bp DNA fragment generated as the template for the sequencing reactions used in the identification of PR were 5′-ACAAGCCTTCGCA-ACTTCAG-3′ and 5′-CGCTAACTCCACAAGCCTTC-3′. The former primer was then used for the sequencing reactions and amplification of cDNA. The primers used to PCR amplify the ∼240-bp DNA fragment generated as the template for the sequencing reactions used in the identification of PL were 5′-CTTTGTCAGCGCCGTAGATTC-3′ and 5′-TG-TAACTCGCTAAACCATGC-3′. The former primer was then used for the sequencing reactions and amplification of cDNA. The primers used to PCR amplify the ∼190-bp DNA fragment generated as template for the sequencing reactions used in the identification of PRM were 5′-TGCGCTAGCCGCTGGGCGAA-3′ and 5′-ACAAGC-CTTCGCAACTT-CAG-3′. The former primer was then used for the sequencing reactions and amplification of cDNA. DNA sequencing was conducted using the Thermosequenase kit (USB) with an [α-33P]dNTP mixture (Amersham).
Toxin assay.
Soluble protein was obtained from cultures of K10595 and K10599 and subsequently analyzed for Stx2 levels by an enzyme-linked immunosorbent assay (ELISA) (59).
RESULTS
Isolation of virulent mutants.
Putative repressor-binding repeats in the 933W OR region were identified previously by sequence analysis (43) (Fig. 1B). We used the consensus sequence derived from these repeats to identify two similar sequences in the region upstream of N that we concluded were putative OL1 and OL2 repeats (Fig. 1); one of these sites was also identified as a possible operator repeat by Fattah et al. (11). Based on the approach that originally identified the λ operators (reviewed in references 19 and 47), we used mutations to demonstrate that these putative operator repeats function as repressor-binding sites in vivo. Virulent (vir) phage mutants grow in the presence of their cognate repressor because of nucleotide changes in their operators that significantly lower the affinity of the repressor for the operators, permitting transcription from the associated promoters in the presence of repressor. The classical λvir mutants were selected as variants that form plaques on a lawn formed by a λ lysogen (23). Later studies demonstrated that two mutations in OR and one mutation in OL, or one mutation in OR and two mutations in OL, are necessary and sufficient for λ virulence (12).
To isolate virulent mutants in 933W, a hybrid phage was constructed between λ and 933W, designated λimm933W. This construction was required because phage 933W is not stable in lysates, probably because of its large genome (41, 43). λimm933W was useful for these experiments because it has the operators, cI gene, and immediately surrounding regions from 933W, while the rest of the genome derived from λ. Hence, the genome size is close to that of λ, resulting in a more stable phage particle. Additionally, λimm933W has the att site of λ, permitting λ functions to excise the prophage from the E. coli chromosome (see below). PCR analysis showed that λimm933W contains a portion of the 933W genome extending from the N gene to the genes encoding replication functions (unpublished data).
Virulent mutants of λimm933W were generated in a two-step process. In planning the first step, we assumed that early-gene expression in phage 933W is analogous to that observed in λ. The N protein is the only gene product required for lytic growth encoded in the λ PL operon (reviewed in reference 15). We assumed that the same is true for 933W. Accordingly, a λimm933W mutant with base changes in OR, decreasing repressor affinity and permitting transcription from PR in the presence of repressor, should be able to grow lytically in a 933W lysogen when the N protein is supplied in trans. Therefore, for the first step in isolating λimm933Wvir mutants, DNA fragments containing the putative OR region that were synthesized under error-prone conditions were introduced into the λimm933W lysogen by electroporation. The recipient lysogen, K9838, has two contiguous prophages, λimm933W, the phage to be made virulent, and a defective λcI857, the supplier of recombination functions (64). In addition, the N protein (functional for 933W) was constitutively expressed from a plasmid, eliminating the need for a mutation(s) in OL, to allow λimm933W, containing mutations in OR, to grow in the presence of repressor. Recombinant phages with mutations in OR that reduce repressor binding sufficiently to allow for growth in the presence of repressor, referred to as partially vir mutants, were selected for their ability to form plaques on a lawn formed by K9835, a λimm933W lysogen carrying a plasmid that constitutively expresses an N protein that functions with 933W. However, because they have an intact OL, these partial vir mutants do not grow in a λimm933W lysogen, K9680, in which N is not supplied in trans. In the second step, fully virulent derivatives were isolated from partially virulent mutants by selecting spontaneous OL mutants as plaque formers on a lawn formed by a λimm933W lysogen (strain K9680).
DNA sequencing showed that the two λimm933Wvir mutants we isolated in independent experiments have three mutations in OR, located in OR2 and OR1, in combination with one mutation in OL, in either OL1 or OL2. The λimm933Wvir mutants isolated and the mutations in their operator regions are listed in Table 2 and illustrated in Fig. 1B. Both λimm933Wvir mutants contain changes in bp -3 of OR2 (Table 2; Fig. 1B). To determine if mutation of this position, when combined with a mutation in OL, is sufficient to confer virulence in λimm933W, we constructed a λimm933W partially vir mutant with a TA-to-AT change at bp -3 of OR2. This mutation was made using a single-stranded DNA oligonucleotide containing the base change (see Materials and Methods). The partially vir recombinant and the subsequent fully vir mutant containing this base change were isolated by using essentially the same steps used to generate the original λimm933Wvir mutants. The fully vir mutant containing the TA-to-AT bp change, λimm933WvirT-3A, in addition to a mutation in OL, has added mutations in both OR1 and OR2 (Table 2; Fig. 1B) that were not in the DNA oligonucleotide used in constructing the mutant. Therefore, we cannot determine if mutation of bp -3 in OR2, in combination with a mutation in OL, is sufficient to confer virulence. Despite several attempts by both direct and random mutagenesis, we never obtained λimm933Wvir mutants with fewer than three mutations in OR (data not shown).
TABLE 2.
λimm933Wvir mutants and mutations present in their respective operator repeats
| Phage | Mutation(s)a in:
|
|||
|---|---|---|---|---|
| OL1 | OL2 | OR1 | OR2 | |
| λimm933Wvir-5 | None | TA to CG at bp −3 | AT to GC at bp -6, AT to GC at bp -3 | TA to CG at bp -3 |
| λimm933Wvir-6 | TA to CG at bp 4 | None | TA to GC at bp 6, AT to GC at bp -6 | TA to AT at bp -3 |
| λimm933WvirT-3Ab | TA to CG at bp 4 | None | AT to CG at bp -6 | TA to AT at bp -3, CG to TA at bp 5 |
Nucleotides in operator repeats are numbered as shown in Fig. 1C.
λimm933Wvir phage mutant that contains a directed mutation of bp -3 in OR2.
In contrast to all other lambdoid phages analyzed, our studies indicate that three mutations in OR and one mutation in OL may be required for λimm933W virulence. It should be noted that the selection we used to isolate λimm933Wvir mutants differs from the way in which the classical λvir mutants were obtained (23). This may explain why the virulent mutants we obtained had more mutations in OR. The same selection strategy used here, however, was used in the isolation of H-19B virulent mutants, and those mutants had only one mutation in OR (55). Nonetheless, we emphasize that the results of our study do not rule out the possibility that fewer than three mutations in OR, with a mutation in OL, are necessary for virulence in 933W.
In vitro binding assays with repressor protein.
To confirm that the regions identified by sequence analyses and mutagenesis bind repressor in vitro, gel mobility shift assays were conducted with a recombinant form of the 933W CI protein (rCI). This protein, purified to near homogeneity (data not shown), contains a His6 tag at its carboxy terminus. The rCI protein is functional since an E. coli derivative, K10786, in which the rCI protein is expressed from a plasmid, is specifically immune to infection with either 933W or λimm933W (data not shown). Hence, the His tag does not interfere with the biological activity of this repressor protein under these conditions.
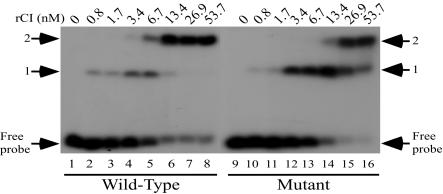
To examine repressor interactions with the OR repeats, gel mobility shift assays were conducted with increasing concentrations of rCI protein incubated with a ∼250-bp radiolabeled DNA probe that included the wild-type OR repeats. Three band shifts were observed when the OR probe was incubated with increasing concentrations of rCI (Fig. 2, lanes 1 to 8). These results suggest that there are three repressor-binding repeats in the right operator region. Whereas the first shift of the OR DNA probe occurred with an rCI concentration of ∼0.8 nM (Fig. 2, lane 2), the second shift of the probe required ca. fourfold more protein (∼3.4 nM) (lane 4). Incubation of the DNA probe with the same rCI concentration (∼3.4 nM) resulted in incomplete shifting to the third position (lane 4). A nearly complete shift of the DNA probe to the third position was evident when the probe was incubated with ∼13.4 nM rCI (lane 6), and complete shifting of the probe was observed with ∼53.7 nM rCI (lane 8).
FIG. 2.
Analysis of repressor interactions with the OR region DNA probes by gel mobility shift assays. Radiolabeled DNA probes containing the wild-type (lanes 1 to 8) or λimm933Wvir-6 mutant (lanes 9 to 16) OR sequences were incubated alone (lanes 1 and 9) or with increasing concentrations of rCI. The approximate concentrations of rCI incubated with the respective OR DNA probes are shown above the lanes.
Gel mobility shift assays showed that the rCI protein has decreased affinity for the OR DNA probe containing the nucleotide changes of the λimm933Wvir-6 mutant (Fig. 1B; Table 2). This decrease in rCI affinity for this probe is particularly noticeable in the second and third shifts of the probe containing the mutant sequence (Fig. 2, lanes 9 to 16). In comparison to the binding efficiencies evident when rCI was incubated with the probe containing the wild-type sequence, ca. fourfold more protein (∼13.4 nM) was required to shift the probe with the mutant sequence to the second position (lane 14). Whereas ∼53.7 nM rCI was sufficient to shift all of the bound probe with wild-type sequences, the same concentration of rCI did not shift the probe with mutant sequences to the third position (lanes 8 and 16).
To examine repressor interactions with the OL repeats, gel mobility shift assays were conducted with rCI protein and a ∼220-bp radiolabeled DNA probe that included the wild-type OL repeats. Two band shifts were observed when rCI was incubated with the OL probe (Fig. 3, lanes 1 to 8). This result is consistent with the presence of two repressor-binding repeats in the left operator region rather than the three found in the OL of λ and other lambdoid phages (45). The first shift was observed with ∼0.8 nM rCI (Fig. 3, lane 2). The second shift was detected when the DNA probe was incubated with ∼6.7 nM rCI (lane 5). These findings agree with those of Koudelka et al. (28), who also found that there are two repressor-binding sites in OL by gel shift analyses and DNase I footprinting.
FIG. 3.
Analysis of repressor interactions with the OL region DNA probes by gel mobility shift assays. Radiolabeled DNA probes containing the wild-type (lanes 1 to 8) or λimm933Wvir-6 mutant (lanes 9 to 16) OL sequences were incubated alone (lanes 1 and 9) or with increasing concentrations of rCI. The approximate concentrations of rCI incubated with the respective OL DNA probes are shown above the lanes.
Gel mobility shift assays conducted with rCI and a DNA probe matching the λimm933Wvir-6 OL region (Table 2; Fig. 1B) failed to result in two shifts of the probe at the same concentrations of protein required for shifting of the probe with wild-type OL sequences (Fig. 3, lanes 9 to 16). Whereas the bound DNA probe containing wild-type sequence was entirely shifted at rCI protein concentrations between ∼13.4 and ∼26.9 nM (lanes 6 and 7), the DNA probe containing the mutant sequence was not entirely shifted with ∼53.7 nM rCI (lane 16). These results show that, as with the mutant OR probe, rCI has decreased affinity for the OL probe containing the mutations present in OL of λimm933Wvir-6.
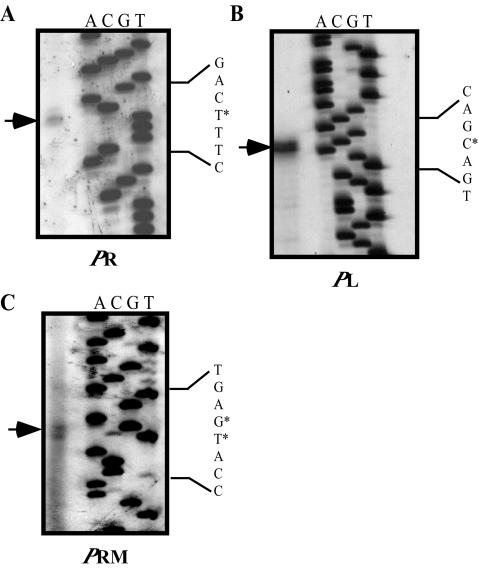
Identification of PR, PL, and PRM.
Evidence for the location of the 933W early promoters PR and PL was obtained by using primer extension to identify likely transcription start sites. RNAs used to identify the PL and PR transcription start sites were isolated from E. coli strain K37 infected with phage λimm933W. Based on the identification of a putative start site for the PR transcript (Fig. 4A), −10 and −35 sequences were assigned by comparing the upstream sequences to E. coli consensus promoter sequences (53) (Fig. 1B). As predicted by sequence analysis (43), we found that the 933W PR promoter spans all of OR2 and overlaps the left end of OR1 by 1 nucleotide (Fig. 1B). Based on the identification of a putative start site for the PL transcript (Fig. 4B), the −10 and −35 sequences were assigned according to promoter consensus sequences (53) (Fig. 1B). The putative 933W PL promoter elements overlap the two operator repeats in the OL region (Fig. 1B). In λ, binding of repressor to λ OL1 and λ OL2 is sufficient to repress PL. In 933W, the position of the PL promoter elements, relative to OL1 and OL2 (Fig. 1B), indicates that repression of PL is achieved much the same way as it is in λ.
FIG. 4.
Primer extension analyses determining the putative transcription start sites of phage early promoters PR (A), PL (B), and repressor maintenance promoter PRM (C). Sequencing reactions are shown to the right. An asterisk designates the putative start site of transcription. Assignment of the putative −10 and −35 promoter elements is shown in Fig. 1B.
Primer extension was used to identify the likely PRM transcription start site by using RNAs extracted from a culture of the 933W lysogen K9675 (Fig. 4C). The primer extension yielded a doublet, indicating that transcription can probably start at either of two adjacent nucleotides. Although transcription generally initiates at an A residue (53), our data indicate that, under these conditions, transcription probably starts at relatively equal efficiencies at the A and C residues upstream of the −10 sequence of PRM (Fig. 4C). We note that the PRM transcripts of both λ and HK022 initiate at the A of the AUG start codon of cI (2, 46). If the same is true for 933W, then transcription probably starts at the A residue, which is the A of the AUG start codon of cI in this case as well, at a greater frequency. As predicted by sequence analysis (43), we found that the 933W PRM −35 sequence overlaps OR2 and the −10 sequence flanks the right end of OR3 (Fig. 1B).
Construction of noninducible 933W and λimm933W prophages.
To extend our studies of the repressor control of 933W Stx expression, we constructed a derivative of 933W with a mutation in the cI gene that presumably blocks autocleavage of the repressor protein. The repressor proteins of lambdoid phages, as well as other autocleavable proteins, consist of an amino-terminal DNA-binding domain and a carboxy-terminal dimerization domain, connected by a linker called the hinge region (45). Little and colleagues showed for the λ and LexA repressors that RecA-mediated autocleavage occurs when a conserved Ala-Gly bond, positioned in the hinge region of the protein, is hydrolyzed by a nucleophilic attack of a conserved Ser-Lys catalytic dyad (56). Mutations changing either the conserved Ser or Lys residues in the λ repressor protein abrogate this autocleavage mechanism (31). The analogous Ser and Lys codons were identified in the 933W cI gene (J. W. Little, personal communication), and this information was used to construct a noninducible 933W prophage; Lys codon 178 of the 933W cI gene was changed to an Asn codon.
K10595, a lysogen with a 933W prophage that encodes a CI protein with the K178N change, produced very few infectious virions even when grown in the presence of the inducing agent mitomycin C (data not shown). Furthermore, spontaneous release of phage from K10693, a lysogen containing the hybrid phage λimm933W with the K178N cI mutation, was extremely low, less than 2.4 × 10−7 per lysogen. This was significantly lower than the spontaneous release observed with the lysogen carrying the wild-type isogenic λimm933W prophage, 2.7 × 10−3 per lysogen. Hence, the cI K178N mutant allele has the characteristics of a classic Ind− mutation and the prophages with the cIK178N change were designated λimm933WcI(Ind−) and 933WcI(Ind−). The few phages released from the λimm933WcI(Ind−) lysogen have spontaneous mutations that appear by plaque morphology to inactivate the repressor (data not shown).
Assessing the role of CI autocleavage on Stx production.
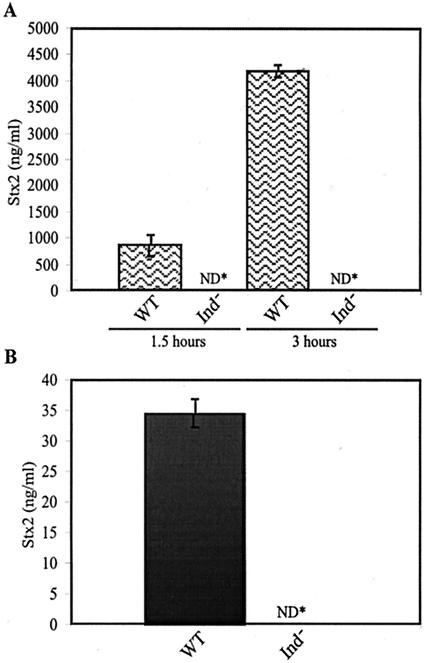
To assess the role of prophage induction in the production of Stx2 by a 933W lysogen, we compared toxin levels produced by strains K10599 and K10595, containing the wild-type or the cI(Ind−) 933W prophages, respectively, cultured in the presence or absence of mitomycin C. The essential idea underlying this approach is that if Stx expression depends on induction of the 933W prophage, then a lysogen with a 933WcI(Ind−) prophage should produce extremely low levels of Stx. An ELISA using anti-Stx2 antiserum was used to measure Stx levels. The levels of Stx2 in total protein obtained from sonicated cells and culture supernatant from a culture of K10595, the strain with the 933WcI(Ind−) prophage, were negligible when cultured in the presence or absence of the inducing agent mitomycin C (Fig. 5). In contrast, K10599, the strain with the 933W wild-type prophage, produced ∼25 to 35 ng of Stx per ml when bacteria were cultured in the absence of an inducing agent (Fig. 5B). The Stx levels increased by more than two orders of magnitude when K10599 was cultured in the presence of mitomycin C (Fig. 5A). These data demonstrate that production of Stx2 requires transcription initiating at the repressor-regulated phage early promoters. Removal of repression allows entry into lytic growth, a process abrogated by the K178N mutation in the 933W cI gene, which presumably results in a protein resistant to autocleavage. By showing that an Ind− mutation in the cI gene eliminates Stx production, these results provide the most direct evidence that prophage induction is required for toxin production by 933W lysogens (16, 60).
FIG. 5.
Amount of Stx in total cultures of the 933W lysogens K10599, containing a prophage encoding the wild-type repressor (WT), and K10595, containing a prophage encoding the noncleavable form of repressor (Ind−), as determined by ELISA using Stx antisera. Stx production was quantified from whole-cell sonicated lysates and culture supernatants obtained from cultures grown in the presence (A) and absence (B) of the prophage inducing agent mitomycin C. Stx production was quantified from cultures grown in the presence of mitomycin C at 1.5 and 3 h following the addition of the inducing agent. The total protein concentrations analyzed for Stx were 0.5 μg/ml (A) and 40 μg/ml (B). Samples were analyzed in triplicate, and the results are representative of four independent experiments. ND*, not detectable.
Isolation of a temperature-inducible prophage.
The isolation of the 933WcI(Ind−1) prophage provided a useful progenitor for constructing a 933WcIts mutant. Mutants of this class not only are valuable tools in studying the physiology of phages but also provide the most direct genetic proof that the involved gene encodes the repressor. A cIts prophage maintains repression at low temperature but, because the CI protein is unstable at higher temperatures, loses repression at high temperature.
Use of the cI(Ind−) mutant specifically enabled us to circumvent a major problem in isolating a cIts derivative of 933W. The background of phage released by a wild-type lysogen is quite high, making it difficult to isolate cIts derivatives from a lysogen carrying a prophage with a wild-type cI gene by mutagenesis and subsequent shift to high temperature. Because a prophage with a cI(Ind−) allele has such a low background of phage release, it can be used to search directly for ts mutants. We used the λimm933WcI(Ind−) Kanr hybrid phage as the progenitor in the search for a cIts mutation because the hybrid phage is far more stable than 933W. Details of the isolation of the temperature-inducible prophage, designated λimm933WcI(Ind−) ts, are outlined in Materials and Methods. DNA sequence analysis showed that this mutant had a single nucleotide change, in addition to the ind change, in the cI gene, encoding a Thr-to-Ile substitution at codon 169. As shown in Fig. 6, when a lysogen carrying the λimm933WcI(Ind−) ts prophage was grown at 32°C and then shifted to 42°C, the culture was lysed. The control lysogen carrying the λimm933WcIind prophage does not show any lysis following the temperature shift.
FIG. 6.
Thermal-induction of the cI(Ind−) ts mutant. The optical density of cultures of K10693 [λimm933WcI(Ind−1)] (⧫), and K10717 [λimm933WcI(Ind− ts)] (○), is shown. Lysogens were grown at 42°C following a shift of log phase bacteria from 32°C. Host cell lysis, represented as a decrease in the culture optical density, is used as a qualitative measurement of prophage induction.
DISCUSSION
The repressor-operator interactions of 933W are of particular importance because a number of studies have provided compelling evidence that significant levels of Stx production and release result from the induction of the prophage that carries the stx2 genes (16, 35, 60). Moreover, the relationship between prophage induction and Stx production and release has clinical relevance, since treatment of STEC-infected patients with antibiotics that induce prophages can exacerbate the course of the disease (1, 62).
Previous studies have led to the idea that there is a relationship between prophage induction and Stx expression in the case of the stx2 phages 933W and φ361 (16, 60). Although there is a promoter associated with the stx2 genes, transcription of these genes occurs primarily from Q-modified transcription initiating at the upstream phage late promoter PR′ (60). Q modification is required for transcription to transcend terminators located between PR′ and the stx genes (Fig. 1A). Q expression, in turn, requires transcription initiating at the early PR promoter. Since PR is under CI repressor control, expression of Q and, in turn, Stx from the prophage requires induction. Because loss of repression leads to prophage replication, induction also leads to a significant increase in the copy number of the stx genes. Hence, the repressor-operator interactions of these phages serve as primary regulators of Stx2 expression.
In this paper we report the isolation of three virulent (vir) mutants of λimm933W. These are designated vir because, even though they have the operator/promoter region (immunity region) of 933W, they can grow lytically in a 933W lysogen. Each of these mutants was shown to have base changes in its putative operator regions (Table 2). Moreover, by gel shift analyses we observed that purified repressor, rCI, exhibits decreased affinity for a DNA probe containing operator sequences matching that of a vir mutant, λimm933Wvir-6, relative to its affinity for a DNA probe containing wild-type operator sequences (Fig. 2 and 3). Thus, we can assume that in vivo λimm933Wvir mutants can grow in the presence of repressor because their operators contain base changes that lower the affinity of repressor binding, permitting expression from the early promoters PR and PL, which drive lytic growth. These results provide definitive evidence that the putative operators identified by sequence analyses (11, 43) are the sites required for repression of phage 933W.
Unlike the classical λvir mutant (21), which has two mutations in OR, the λimm933Wvir mutants we isolated have three mutations in OR, but like the classical λvir mutant, they have one mutation in OL. To assess the possibility that three mutations in 933W OR are required for virulence, we constructed a partially virulent mutant by site-directed mutagenesis. This mutant was then used to isolate a spontaneous fully virulent mutant. To generate the mutation, we employed a template that had the OR2 bp-3 mutation, the mutation shared by two isolated λimm933Wvir mutants. Again, when a fully virulent mutant was obtained, in addition to a mutation in OL, three mutations were identified in OR. These results suggest that virulence in 933W may differ from that seen with other lambdoid phages, which have either one or two mutations in OR. Like λ, P22 and HK022 require two mutations in OR for virulence (4, 44). In contrast, phages H-19B, 434, and 21 require only one mutation in OR for virulence (44, 55, 61).
Gel shift analyses conducted with purified 933W repressor, rCI, and a DNA probe with the sequence of the OR region were consistent with the existence of three binding repeats in the 933W OR (Fig. 2), as predicted by sequence analyses (43). It has been observed that in λ, binding of the repressor to λ OR2 is nearly concurrent with binding of repressor to λ OR1, due to cooperative interactions between the protein dimers bound to the two sites (45). We observed that ca. fourfold more rCI is required to shift the probe containing the 933W OR region to the second position (Fig. 2). Assuming that the first shift results from rCI binding to OR1 and the second shift results from rCI binding to both OR1 and OR2, it appears that binding of repressor to these sites may not be influenced by cooperativity, at least not under these in vitro conditions. In contrast, the rCI levels required to shift the probe to the third position in gel mobility shift assays were not significantly greater than the levels of rCI sufficient to shift the probe to the second position. A more detailed discussion of rCI binding to the OR repeats is included in reference 28.
Gel shift analyses using rCI incubated with a DNA probe with the sequence of the OL region indicate that there are only two repressor-binding sites in OL (Fig. 3). Our results are qualitatively consistent with those of Koudelka et al. (28), who used gel shift analyses and DNase I footprinting to examine repressor binding to OL. They detected repressor binding to only two sites in OL as well. Bacteriophage 933W is the only phage in the lambdoid family, of which we are aware, that contains only two repressor-binding repeats in OL.
We note that although the results of our gel mobility shift assays are qualitatively similar to the findings of Koudelka et al. (28), approximately 10-fold more rCI protein was required to shift the DNA probes in our assays (Fig. 2 and 3) (see the accompanying paper [28]). We offer three possible ways in which these quantitative differences can be explained. First, inefficient radiolabeling of our DNA probes might have resulted in an excess of unlabeled DNA probes that competed with radiolabeled DNA for binding with rCI, thus requiring more rCI to shift the radiolabeled probe. Second, since the rCI protein concentration reflects total protein, both active and inactive, it is possible that the percentage of active protein in our rCI preparation was lower than that of inactive protein. This would result in more total protein required to shift the DNA probes. Finally, the difference could be due to our use of a His6-tagged protein while Koudelka et al. (28) used the natural protein. Although the C-terminal His6 tag does not qualitatively affect repressor activity, it may quantitatively influence the binding activity of rCI.
A role for the third binding repeat in the λ OL region was identified by Dodd et al. (8). They found that binding of repressor to λ OL3 enhances the binding of repressor to λ OR3, which, in turn, leads to repression of transcription initiating at PRM. In λ, the repressor tetramers bound to OL1 and OL2 establish octamers with the repressor tetramers bound at OR1 and OR2 by looping out the intervening region (8, 49). Measurements of λ repressor binding to its respective OR sequences independent of OL sequences indicated that the concentrations of repressor necessary for binding OR3 are very high (25). Although repressor binding to OR3 turns off transcription from PRM, the high levels required for binding at OR3 made it difficult to see how OR3 contributes to regulation of repressor expression under physiological conditions in which the repressor levels are not high enough (34). However, the studies by Dodd et al. (9) demonstrate that interactions between OL1 · OL2 and OR1 · OR2 align the dimer bound to OL3 in position to interact with the dimer bound to OR3 so that repressor binding at OR3 is stabilized. In this way, the dimers bound to OL3 contribute to the effective repression of PRM by the CI protein. By controlling repressor expression from PRM, OL3 probably serves to help maintain repressor at proper levels to ensure that the prophage will be induced under appropriate conditions. Because 933W does not have an OL3, this model of PRM repression appears not to be applicable. The way in which 933W compensates physiologically for this lack of OL3 is an open question, which takes on added significance when considering the role of induction in the expression of stx from the 933W genome.
A recent study indicates that H-19B and 933W induce more readily than do the non-Stx-encoding lambdoid phages tested (33). Moreover, it has been suggested that this enhanced induction may have been selected to facilitate Stx production and/or release. Although H-19B and 933W appear to have similar spontaneous induction frequencies, their immunity regions differ in both structure and function. It seems plausible that these two phages have acquired independent mechanisms that make them more readily inducible. Because 933W does not possess OL3, it is likely not to regulate transcription initiating at PRM by precisely the same mechanisms as λ and presumably other lambdoid phages. However, it remains to be determined how phage 933W regulates transcription initiating at PRM. Phage H-19B repressor interactions with its respective operator repeats differ from what is observed in λ (55). Additionally, H-19B requires only one mutation in OR and one mutation in OL for virulence, suggesting the possibility of weak binding at OR. Due to the medical relevance of prophage induction to the progression of disease associated with STEC infection, examination of the immunity regions and, more specifically, the repressor-operator interactions of more Stx-encoding phages is warranted. In particular, it will be important to determine if high rates of spontaneous induction are a common attribute of phages that encode Stx.
Acknowledgments
This work was supported by Public Health Service grant AI11459-10. J.S.T. was supported in part by NIH training grant T32-GM08353.
Vic DiRita is thanked for helpful suggestions in preparing the manuscript. John Little is thanked for pointing out the codon change resulting in the ind mutation. Ry Young is thanked for supplying the λcI857Kanr phage. Gerald Koudelka is thanked for sharing data.
REFERENCES
- 1.Butler, T., M. R. Islam, M. A. Azad, and P. K. Jones. 1987. Risk factors for development of hemolytic uremic syndrome during shigellosis. J. Pediatr. 110:894-897. [DOI] [PubMed] [Google Scholar]
- 2.Cam, K. M., J. Oberto, and R. A. Weisberg. 1991. The early promoters of bacteriophage HK022: contrasts and similarities to other lambdoid phages. J. Bacteriol. 173:734-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell, A., and D. Botstein. 1983. Evolution of lambdoid phages, p. 365-380. In R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg (ed.), Lambda II. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 4.Carlson, N. G., and J. W. Little. 1993. Highly cooperative DNA binding by the coliphage HK022 repressor. J. Mol. Biol. 230:1108-1130. [DOI] [PubMed] [Google Scholar]
- 5.Cha, R. S., H. Zarbl, P. Keohavong, and W. G. Thilly. 1992. Mismatch amplification mutation assay (MAMA): application to the c-H-ras gene. PCR Methods Appl. 2:14-20. [DOI] [PubMed] [Google Scholar]
- 6.Court, D. L., S. Swaminathan, D. Yu, H. Wilson, T. Baker, M. Bubunenko, J. Sawitzke, and S. K. Sharan. 2003. Mini-lambda: a tractable system for chromosome and BAC engineering. Gene 315:63-69. [DOI] [PubMed] [Google Scholar]
- 7.Datz, M., C. Janetzki-Mittmann, S. Franke, F. Gunzer, H. Schmidt, and H. Karch. 1996. Analysis of the enterohemorrhagic Escherichia coli O157 DNA region containing lambdoid phage gene p and Shiga-like toxin structural genes. Appl. Environ. Microbiol. 62:791-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dodd, I. B., A. J. Perkins, D. Tsemitsidis, and J. B. Egan. 2001. Octamerization of lambda CI repressor is needed for effective repression of P(RM) and efficient switching from lysogeny. Genes Dev. 15:3013-3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dodd, I. B., K. E. Shearwin, A. J. Perkins, T. Burr, A. Hochschild, and J. B. Egan. 2004. Cooperativity in long-range gene regulation by the lambda C1 repressor. Genes Dev. 18:344-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellis, H. M., D. Yu, T. DiTizio, and D. L. Court. 2001. High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc. Natl. Acad. Sci. USA 98:6742-6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fattah, K. R., S. Mizutani, F. J. Fattah, A. Matsushiro, and Y. Sugino. 2000. A comparative study of the immunity region of lambdoid phages including Shiga-toxin-converting phages: molecular basis for cross immunity. Genes Genet. Syst. 75:223-232. [DOI] [PubMed] [Google Scholar]
- 12.Flashman, S. L. 1978. Mutational analysis of the operators of bacteriophage lambda. Mol. Gen. Genet. 166:61-73. [DOI] [PubMed] [Google Scholar]
- 13.Fowler, R. G., G. E. Degnen, and E. C. Cox. 1974. Mutational specificity of a conditional Escherichia coli mutator, mutD5. Mol. Gen. Genet. 133:179-191. [DOI] [PubMed] [Google Scholar]
- 14.Friedman, D. I., and D. L. Court. 2001. Bacteriophage lambda: alive and well and still doing its thing. Curr. Opin. Microbiol. 4:201-207. [DOI] [PubMed] [Google Scholar]
- 15.Friedman, D. I., and M. Gottesman. 1983. Lytic mode of lambda development, p. 21-51. In R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg (ed.), Lambda II. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 16.Fuchs, S., I. Muhldorfer, A. Donohue-Rolfe, M. Kerenyi, L. Emody, R. Alexiev, P. Nenkov, and J. Hacker. 1999. Influence of RecA on in vivo virulence and Shiga toxin 2 production in Escherichia coli pathogens. Microb. Pathog. 27:13-23. [DOI] [PubMed] [Google Scholar]
- 17.Gottesman, M. E., and M. B. Yarmolinsky. 1968. Integration-negative mutants of bacteriophage lambda. J. Mol. Biol. 31:487-505. [DOI] [PubMed] [Google Scholar]
- 18.Guarneros, G., and H. Echols. 1970. New mutants of bacteriophage lambda with a specific defect in excision from the host chromosome. J. Mol. Biol. 47:565-574. [DOI] [PubMed] [Google Scholar]
- 19.Gussin, G. N., A. D. Johnson, C. O. Pabo, and R. T. Sauer. 1983. Repressor and Cro protein: structure, function, and role in lysogenization, p. 93-121. In R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg (ed.), Lambda II. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 20.Hendrix, R. W. 2002. Bacteriophages: evolution of the majority. Theor. Popul. Biol. 61:471-480. [DOI] [PubMed] [Google Scholar]
- 21.Hopkins, N., and M. Ptashne. 1971. Genetics of virulence, p. 571-574. In A. D. Hershey (ed.), The bacteriophage lambda. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 22.Huang, A., S. de Grandis, J. Friesen, M. Karmali, M. Petric, R. Congi, and J. L. Brunton. 1986. Cloning and expression of the genes specifying Shiga-like toxin production in Escherichia coli H19. J. Bacteriol. 166:375-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacob, F., and E. L. Wollman. 1954. Etude genetique d'un bacteriophage tempere d'Escherichia coli 1 L systeme genetique du bacteriophage lambda. Ann. Inst. Pasteur (Paris) 85:653-673. [PubMed] [Google Scholar]
- 24.Johansen, B. K., Y. Wasteson, P. E. Granum, and S. Brynestad. 2001. Mosaic structure of Shiga-toxin-2-encoding phages isolated from Escherichia coli O157:H7 indicates frequent gene exchange between lambdoid phage genomes. Microbiology 147:1929-1936. [DOI] [PubMed] [Google Scholar]
- 25.Johnson, A. D., B. J. Meyer, and M. Ptashne. 1979. Interactions between DNA-bound repressors govern regulation by the lambda phage repressor. Proc. Natl. Acad. Sci. USA 76:5061-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karch, H., M. Bielaszewska, M. Bitzan, and H. Schmidt. 1999. Epidemiology and diagnosis of Shiga toxin-producing Escherichia coli infections. Diagn. Microbiol. Infect. Dis. 34:229-243. [DOI] [PubMed] [Google Scholar]
- 27.Karch, H., H. Schmidt, C. Janetzki-Mittmann, J. Scheef, and M. Kroger. 1999. Shiga toxins even when different are encoded at identical positions in the genomes of related temperate bacteriophages. Mol. Gen. Genet. 262:600-607. [DOI] [PubMed] [Google Scholar]
- 28.Koudelka, A. P., L. A. Hufnagle, and G. B. Koudelka. 2004. Purification and characterization of the repressor of the Shiga toxin-encoding bacteriophage 933W: DNA binding, gene regulation, and autocleavage. J. Bacteriol. 186:7659-7669. [DOI] [PMC free article] [PubMed]
- 29.Koudelka, G. B. 2000. Cooperativity: action at a distance in a classic system. Curr. Biol. 10:R704-R707. [DOI] [PubMed] [Google Scholar]
- 30.Li, X. T., N. Costantino, L. Y. Lu, D. P. Liu, R. M. Watt, K. S. Cheah, D. L. Court, and J. D. Huang. 2003. Identification of factors influencing strand bias in oligonucleotide-mediated recombination in Escherichia coli. Nucleic Acids Res. 31:6674-6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin, L. L., and J. W. Little. 1989. Autodigestion and RecA-dependent cleavage of Ind− mutant LexA proteins. J. Mol. Biol. 210:439-452. [DOI] [PubMed] [Google Scholar]
- 32.Little, J. W. 1995. The SOS regulatory system, p. 453-479. In E. C. C. Linn and A. S. Lynch (ed.), Regulation of gene expression in Escherichia coli. R.G. Landis, Georgetown, Tex.
- 33.Livny, J., and D. I. Friedman. 2004. Characterizing spontaneous induction of Stx encoding phages using a selectable reporter system. Mol. Microbiol. 51:1691-1704. [DOI] [PubMed] [Google Scholar]
- 34.Maurer, R., B. Meyer, and M. Ptashne. 1980. Gene regulation at the right operator (OR) bacteriophage lambda. I. OR3 and autogenous negative control by repressor. J. Mol. Biol. 139:147-161. [DOI] [PubMed] [Google Scholar]
- 35.Muhldorfer, I., J. Hacker, G. T. Keusch, D. W. Acheson, H. Tschape, A. V. Kane, A. Ritter, T. Olschlager, and A. Donohue-Rolfe. 1996. Regulation of the Shiga-like toxin II operon in Escherichia coli. Infect. Immun. 64:495-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neely, M. N., and D. I. Friedman. 1998. Arrangement and functional identification of genes in the regulatory region of lambdoid phage H-19B, a carrier of a Shiga-like toxin. Gene 223:105-113. [DOI] [PubMed] [Google Scholar]
- 37.Neely, M. N., and D. I. Friedman. 1998. Functional and genetic analysis of regulatory regions of coliphage H-19B: location of Shiga-like toxin and lysis genes suggest a role for phage functions in toxin release. Mol. Microbiol. 28:1255-1267. [DOI] [PubMed] [Google Scholar]
- 38.O'Brien, A. D., and R. K. Holmes. 1987. Shiga and Shiga-like toxins. Microbiol. Rev. 51:206-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Brien, A. D., and J. B. Kaper. 1998. Shiga toxin-producing Escherichia coli: yesterday, today, and tomorrow, p. 1-11. In A. D. O'Brien and J. B. Kaper (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. ASM Press, Washington, D.C.
- 40.O'Brien, A. D., J. W. Newland, S. F. Miller, R. K. Holmes, H. W. Smith, and S. B. Formal. 1984. Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science 226:694-696. [DOI] [PubMed] [Google Scholar]
- 41.Parkinson, J. S., and R. J. Huskey. 1971. Deletion mutants of bacteriophage lambda. I. Isolation and initial characterization. J. Mol. Biol. 56:369-384. [DOI] [PubMed] [Google Scholar]
- 42.Paton, J. C., and A. W. Paton. 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11:450-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plunkett, G., III, D. J. Rose, T. J. Durfee, and F. R. Blattner. 1999. Sequence of Shiga toxin 2 phage 933W from Escherichia coli O157:H7: Shiga toxin as a phage late-gene product. J. Bacteriol. 181:1767-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poteete, A. R., M. Ptashne, M. Ballivet, and H. Eisen. 1980. Operator sequences of bacteriophages P22 and 21. J. Mol. Biol. 137:81-91. [DOI] [PubMed] [Google Scholar]
- 45.Ptashne, M. 1992. A genetic switch. Cell Press and Blackwell Publications, Cambridge, Mass.
- 46.Ptashne, M., K. Backman, M. Z. Humayun, A. Jeffrey, R. Maurer, B. Meyer, and R. T. Sauer. 1976. Autoregulation and function of a repressor in bacteriophage lambda. Science 194:156-161. [DOI] [PubMed] [Google Scholar]
- 47.Ptashne, M., A. Jeffrey, A. D. Johnson, R. Maurer, B. J. Meyer, C. O. Pabo, T. M. Roberts, and R. T. Sauer. 1980. How the lambda repressor and cro work. Cell 19:1-11. [DOI] [PubMed] [Google Scholar]
- 48.Reisbig, R., S. Olsnes, and K. Eiklid. 1981. The cytotoxic activity of Shigella toxin. Evidence for catalytic inactivation of the 60 S ribosomal subunit. J. Biol. Chem. 256:8739-8744. [PubMed] [Google Scholar]
- 49.Revet, B., B. von Wilcken-Bergmann, H. Bessert, A. Barker, and B. Muller-Hill. 1999. Four dimers of lambda repressor bound to two suitably spaced pairs of lambda operators form octamers and DNA loops over large distances. Curr. Biol. 9:151-154. [DOI] [PubMed] [Google Scholar]
- 50.Riley, L. W., R. S. Remis, S. D. Helgerson, H. B. McGee, J. G. Wells, B. R. Davis, R. J. Hebert, E. S. Olcott, L. M. Johnson, N. T. Hargrett, P. A. Blake, and M. L. Cohen. 1983. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 308:681-685. [DOI] [PubMed] [Google Scholar]
- 51.Ritchie, J. M., P. L. Wagner, D. W. Acheson, and M. K. Waldor. 2003. Comparison of Shiga toxin production by hemolytic-uremic syndrome-associated and bovine-associated Shiga toxin-producing Escherichia coli isolates. Appl. Environ. Microbiol. 69:1059-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roberts, J. W., W. Yarnell, E. Bartlett, J. Guo, M. Marr, D. C. Ko, H. Sun, and C. W. Roberts. 1998. Antitermination by bacteriophage lambda Q protein. Cold Spring Harbor Symp. Quant. Biol. 63:319-325. [DOI] [PubMed] [Google Scholar]
- 53.Rosenberg, M., and D. Court. 1979. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu. Rev. Genet. 13:319-353. [DOI] [PubMed] [Google Scholar]
- 54.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 55.Shi, T., and D. I. Friedman. 2001. The operator-early promoter regions of Shiga-toxin bearing phage H-19B. Mol. Microbiol. 41:585-599. [DOI] [PubMed] [Google Scholar]
- 56.Slilaty, S. N., and J. W. Little. 1987. Lysine-156 and serine-119 are required for LexA repressor cleavage: a possible mechanism. Proc. Natl. Acad. Sci. USA 84:3987-3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tyler, J. S., and D. I. Friedman. 2004. Characterization of a eukaryotic-like tyrosine protein kinase expressed by the Shiga toxin-encoding bacteriophage 933W. J. Bacteriol. 186:3472-3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Unkmeir, A., and H. Schmidt. 2000. Structural analysis of phage-borne stx genes and their flanking sequences in shiga toxin-producing Escherichia coli and Shigella dysenteriae type 1 strains. Infect. Immun. 68:4856-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wagner, P. L., J. Livny, M. N. Neely, D. W. Acheson, D. I. Friedman, and M. K. Waldor. 2002. Bacteriophage control of Shiga toxin 1 production and release by Escherichia coli. Mol. Microbiol. 44:957-970. [DOI] [PubMed] [Google Scholar]
- 60.Wagner, P. L., M. N. Neely, X. Zhang, D. W. Acheson, M. K. Waldor, and D. I. Friedman. 2001. Role for a phage promoter in Shiga toxin 2 expression from a pathogenic Escherichia coli strain. J. Bacteriol. 183:2081-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wharton, R. P., E. L. Brown, and M. Ptashne. 1984. Substituting an alpha-helix switches the sequence-specific DNA interactions of a repressor. Cell 38:361-369. [DOI] [PubMed] [Google Scholar]
- 62.Wong, C. S., S. Jelacic, R. L. Habeeb, S. L. Watkins, and P. I. Tarr. 2000. The risk of the hemolytic-uremic syndrome after antibiotic treatment of Escherichia coli O157:H7 infections. N. Engl. J. Med. 342:1930-1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu, H., E. I. Petersen, S. B. Petersen, and M. R. el-Gewely. 1999. Random mutagenesis libraries: optimization and simplification by PCR. BioTechniques 27:1102-1104, 1106, 1108. [DOI] [PubMed] [Google Scholar]
- 64.Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu, R. R., and V. J. DiRita. 1999. Analysis of an autoregulatory loop controlling ToxT, cholera toxin, and toxin-coregulated pilus production in Vibrio cholerae. J. Bacteriol. 181:2584-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]