Abstract
Airway epithelial CD55 down-regulation occurs in several hypoxia-associated pulmonary diseases, but the mechanism is unknown. Using in vivo and in vitro assays of pharmacologic inhibition and gene silencing, the current study investigated the role of hypoxia-inducible factor (HIF)-1α in regulating airway epithelial CD55 expression. Hypoxia down-regulated CD55 expression on small-airway epithelial cells in vitro, and in murine lungs in vivo; the latter was associated with local complement activation. Treatment with pharmacologic inhibition or silencing of HIF-1α during hypoxia-recovered CD55 expression in small-airway epithelial cells. HIF-1α overexpression or blockade, in vitro or in vivo, down-regulated CD55 expression. Collectively, these data show a key role for HIF-1α in regulating the expression of CD55 on airway epithelium.
Keywords: hypoxia, complement regulatory proteins, lung
Clinical Relevance
Previous studies have reported that down-regulation of CD55, a complement regulatory protein, expressed on airway epithelium was associated with local complement activation in idiopathic pulmonary fibrosis and obliterative bronchiolitis. However, the molecular mechanisms mediating this airway epithelial CD55 down-regulation remain unknown. The data show a central role for hypoxia-inducible factor (HIF)-1α in mediating down-regulation of airway epithelial–derived CD55. These data suggest that local complement activation in hypoxia-associated pulmonary diseases may, in part, be mediated by HIF-1α–dependent down-regulation of airway epithelial CD55. This report provides insight on a potential therapeutic intervention for attenuating complement-mediated injuries in hypoxic pulmonary diseases by preserving airway epithelial CD55 expression.
Hypoxia is a complicating factor in several pulmonary diseases, such as obliterative bronchiolitis (OB) and idiopathic pulmonary fibrosis (IPF) (1, 2). Physiological changes involved in both of these pulmonary diseases are driven by complex interplay between innate and adaptive immunity, along with many other hypoxia-mediated molecular mechanisms of the immune system. Our group reported novel roles for complement activation in OB and IPF, due to complement-mediated cytotoxicity on airway epithelium (3–5). These findings correlated with airway epithelial CD55 down-regulation, a key complement regulatory protein (CRP) in OB and IPF (3, 5).
CD55 is a glycosylphosphatidylinositol-anchored protein present on various cell types, including murine and human airway epithelial membranes (6), and functions by regulating complement activation via dissociation of complement C3 and C5 convertases (6). Accordingly, airway epithelial CD55 could protect against lung diseases associated with complement activation. However, mechanisms regulating CD55 in pulmonary pathologies have not been defined.
Because CD55 is down-regulated on cell membranes of transplanted airway epithelium and in pulmonary fibrosis (3, 5), we hypothesized that tissue hypoxia, a condition common to both diseases, may mediate CD55 down-regulation. In particular, we hypothesize that hypoxia-inducible factor (HIF)-1α, a transcription factor induced potently by hypoxia (7), regulates airway epithelial CD55 expression, as indicated by small-airway epithelial cells (SAECs) and murine lungs. Data in the current study show a crucial role for HIF-1α in regulating CD55 expression on airway epithelium in vitro and in vivo.
Materials and Methods
Animals
C57BL/10 male mice (Harlan, Indianapolis, IN) were housed in Laboratory Animal Resource Center at Indiana University School of Medicine (Indianapolis, IN), according to institutional guidelines. Studies were approved by Indiana University School of Medicine Institutional Care and Use Committee (3).
Cells
SAECs were from Lonza Clonetics (Walkersville, MD).
Hypoxia
Mice were placed in a custom-made hypobaric hypoxia chamber (10% O2) for 24 hours, as previously reported (8). SAECs were incubated in 1% O2 hypoxic chamber (The Baker Co., Ruskinn Technology Ltd., Sanford, ME) for various time points (8). Mice and SAECs were exposed to 21% O2 for normoxia (9).
HIF-1α Inhibition
HIF-1α was pharmacologically inhibited by pretreating SAECs with 100 nM chetomin and vehicle control (Sigma-Aldrich, St. Louis, MO), or by intratracheal instillation of mice with 1 mg/ml chetomin and vehicle control. Additional details are available in the online supplement.
RNA Interference
SAECs were transfected with 50 nM of human HIF-1α small interference RNA (siRNA; M-004018-05-0005; Dharmacon Technologies, Pittsburgh, PA), or control siRNA (D-001600-01-05; Dharmacon Technologies). Mice were intratracheally instilled with 50 μg of mouse HIF-1α siRNA (L-040638-00-0005; Dharmacon Technologies), or control siRNA. Additional details are available in the online supplement.
HIF-1α Stabilization
SAECs were treated with 1 μM dimethyloxaloylglycine (DMOG; Sigma-Aldrich) and vehicle control. Mice were intratracheally instilled with vehicle control or 1 mg/ml DMOG. Additional details are available in the online supplement.
Adenoviral Transduction
Normoxic SAECs were transduced with 50 plaque-forming units per cell AdCA5 or AdLACZ vectors (10).
Immunoblot
Cytoplasmic/Nuclear Extraction (Thermo Fisher Scientific, Rockford, IL) was used to detect cytoplasmic CD55 and nuclear HIF-1α in SAECs and mouse lung homogenates. Antibodies for immunoblots included: CD55 (sc-9156 [H-319]; Santa Cruz Biotechnology, Santa Cruz, CA); HIF-1α (cat. no. AF1935; R&D Systems, Minneapolis, MN), vinculin (ab18058; Abcam, Cambridge, MA), lamin β1 (ab16048; Abcam), and β-actin (ab8224; Abcam). Immunoblot analysis was performed as previously described (5).
Real-Time Polymerase Chain Reaction
Total RNA and cDNA were prepared as previously described (11). Taqman mastermix and primers (Applied Biosystems Life Technologies, Grand Island, NY) were used. Mouse primers included: CD55 Mm00438377_m1, HIF-1α Mm00468869_m1, and β-actin Mm00607939_s1. The human primers included: CD55 Hs00892618_m1, HIF-1α Hs00153153_m1, and β-actin Hs01060665_g1 (3). Relative gene expression was completed as previously described (3).
Immunohistochemistry
CD55 staining was achieved by 1:100 dilution of rabbit anti-human CD55 (sc-31208, Santa Cruz Biotechnology) as previously described (3, 5). The antigen retrieval was done by 1M EDTA in a pressure cooker. The secondary antibody was donkey-anti-goat (1:100) for 60 min from Jackson Immunology/Research Labs. For HIF-1α staining, HIF-1α antibody at 1:100 dilution (NB.100-479; Novus Biologicals, Littleton, CO) was used. Dako’s DAKO Envision+ Rabbit was used for HIF-1a (Dako North America Inc., Carpinteria, CA). Additional details are available in the online supplement. Images were captured with Olympus BX41 microscope and DP12 camera (Olympus Imaging America Inc., Melville, NY).
C3a ELISA
Optical density was analyzed at 450 nm on Spectra Max Plus Softmax Pro 3.1.2 software (Molecular Devices, Sunnyvale, CA) for C3a Mouse ELISA (MBS703819; MyBioSource Inc., San Diego, CA).
Statistical Analysis
One-way ANOVA (Bonferroni), two-tailed paired t test, and unpaired t test were performed using GraphPad Prism 4 (GraphPad Prism Software, San Diego, CA). Data represent mean (±SEM). Statistical significance was set at P < 0.05.
Results
Hypoxia Induces HIF-1α and Down-Regulates CD55 Expression
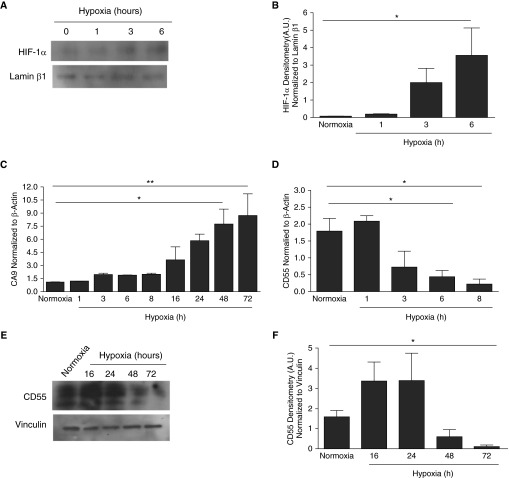
Hypoxia is the most potent inducer of HIF-1α in several tissues, but its induction in normal human SAECs has not been reported. Hypoxia induced a time-dependent induction of HIF-1α in SAECs (Figures 1A and 1B, P < 0.05). Data showing concurrent induction of carbonic anhydrase IX (CA9), an HIF-1α–dependent gene (12, 13), demonstrated that hypoxia-induced HIF-1α was active transcriptionally (Figure 1C). Notably, CD55 transcripts were down-regulated within 6 hours of hypoxia in SAECs (Figure 1D, P < 0.05), which is the same time point at which HIF-1α was induced. However, CD55 protein down-regulation occurred at 72 hours of hypoxia (Figures 1E and 1F, P < 0.05).
Figure 1.
Hypoxia induces hypoxia-inducible factor (HIF)-1α in small-airway epithelial cells (SAECs) and down-regulates CD55. SAECs were exposed to hypoxia (1% O2) at times shown. Normoxic (21% O2) cells were controls. (A) Hypoxic SAECs were probed for HIF-1α. Lamin β1 served as an internal loading control. (B) Densitometry analysis indicated significant up-regulation of HIF-1α by 6 hours of hypoxia versus normoxic untreated (0 hour controls) conditions. Data are representative of means (±SEM); n = 5; *P < 0.05 versus normoxia; one-way ANOVA with Bonferroni posttest. (C) CA9 transcripts increased with hypoxia exposure compared with normoxia. Data represent means (±SEM); n = 3; *P < 0.05 or **P < 0.01 versus normoxia; one-way ANOVA with Bonferroni posttest. (D) CD55 transcripts were significantly down-regulated by 6 hours of hypoxia compared with the controlled normoxic conditions. Values represent means (±SEM); n = 3; *P < 0.05 versus normoxia; one-way ANOVA with Bonferroni posttest. (E) Hypoxic SAECs were probed for CD55. Vinculin was the internal loading control. (F) Densitometry analysis shows significant down-regulation of CD55 by 72 hours of hypoxic versus normoxic conditions. Data represent means (±SEM); n = 3; *P < 0.05 versus normoxia; one-way ANOVA with Bonferroni posttest. A.U., absorbance units.
Inhibition of HIF-1α Restores CD55 Expression in Hypoxic Human SAECs
In preliminary studies, SAECs from multiple donors were studied to determine the time course of hypoxia-induced CD55 down-regulation. Whereas Figures 1E and 1F depict CD55 down-regulation at 72 hours of hypoxia, in subsequent studies, hypoxia-induced down-regulation of CD55 protein expression was maximal at 24 hours due to donor-dependent variations in SAECs from multiple donors used in these experiments. Therefore, the 24-hour time point was used in all subsequent experiments. To determine the linkage between HIF-1α expression and CD55 down-regulation, HIF-1α was pharmacologically inhibited or silenced through the use of chetomin or HIF-1α siRNA, respectively. Vehicle treatment had no effect on hypoxia-induced down-regulation of CD55 (see Figure E1A in the online supplement). Chetomin treatment significantly recovered CD55 transcript expression as compared with 6-hour hypoxia or vehicle controls (Figure E1A, P < 0.001). Similar to prior reports (14), preliminary studies confirmed that 100 nM was the optimal dosing for chetomin-mediated effects (Figure E2A). Chetomin restored CD55 protein expression at 24 hours of hypoxia as compared with hypoxic vehicle controls (Figures E1B and E1C, P < 0.05). To confirm the HIF-1α–specific effects of chetomin, we assessed transcription of HIF-1α–dependent gene, CA9 (12, 13). CA9 transcripts were down-regulated in chetomin-treated, 6-hour hypoxic SAECs compared with 6-hour hypoxic SAECs in the presence of vehicle control, as well as in normoxic conditions (Figure E1D, P < 0.05).
Gene silencing via siRNA was used to assess the role of HIF-1α on CD55 expression in hypoxic conditions. Control experiments confirmed the efficacy of siRNA to block HIF-1α expression (Figures 2A and 2B, P < 0.05). To further confirm HIF-1α silencing and its transcriptional activity, CA9 transcripts were also assessed. CA9 expression was down-regulated in hypoxic SAECs transfected with HIF-1α siRNA compared with 6-hour hypoxic SAECs transfected with or without control siRNA and control normoxic cells (Figure 2C, P < 0.01). CD55 transcripts were restored within 6 hours of hypoxia in cells silenced with HIF-1α siRNA compared with normoxic cells or cells transfected with the control siRNA in hypoxic conditions (Figure 2D, P < 0.05). Notably, silencing HIF-1α also recovered CD55 protein expression in hypoxic SAECs compared with normoxic control cells or hypoxic cells transfected with control siRNA (Figures 2E and 2F, P < 0.05).
Figure 2.
Silencing HIF-1α in hypoxic SAECs recovers CD55. (A) HIF-1α is silenced in 6-hour hypoxic SAECs transfected with 50 nM HIF-1α small interfering RNA (siRNA) versus 6-hour hypoxic SAECs with control siRNA (50 nM) or in normoxia. Lamin β1 was the loading control. (B) HIF-1α was repressed in 24-hour hypoxic SAECs transfected with HIF-1α siRNA versus control siRNA or normoxic levels. Data represent means (±SEM); n = 3; *P < 0.05 versus normoxia or control siRNA in 24 hours of hypoxia; one-way ANOVA with Bonferroni posttest. (C) CA9 transcripts were repressed in HIF-1α siRNA–transfected 6-hour hypoxic SAECs versus 6-hour hypoxic SAECs with or without control siRNA. Values represent means (±SEM); n = 3; **P < 0.01 versus nomoxia, 6 hours of hypoxia, or control siRNA; one-way ANOVA with Bonferroni posttest. (D) CD55 transcripts were recovered in HIF-1α siRNA–transfected 6-hour hypoxic SAECs versus 6-hour hypoxic SAECs in the presence or absence of control siRNA. Values represent means (±SEM); n = 3; *P < 0.05 versus nomoxia, 6 hours of hypoxia, or control siRNA; one-way ANOVA with Bonferroni posttest. (E) CD55 was assessed in normoxic, 24-hour hypoxic HIF-1α siRNA, or control siRNA–transfected SAECs. Vinculin was the loading control. (F) CD55 was recovered in 24-hour HIF-1α siRNA SAECs versus control siRNA or normoxic levels. Values represent means (±SEM); n = 3; *P < 0.05 versus normoxia, 6 hours of hypoxia, or control siRNA; one-way ANOVA with Bonferroni posttest.
Induction of HIF-1α in Normoxia Down-Regulates CD55 Expression In Vitro
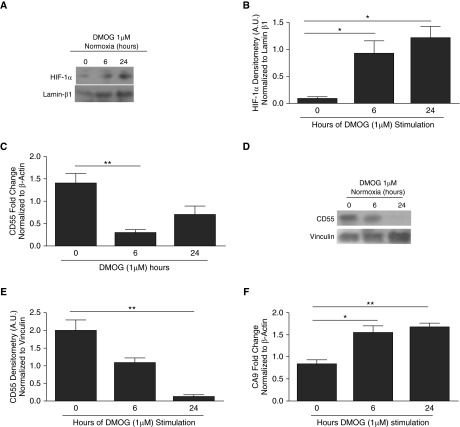
We investigated if HIF-1α induction under normoxic conditions was sufficient to suppress CD55 expression. HIF-1α was stabilized in normoxic SAECs by treating SAECs with DMOG, a prolyl hydroxylase (PHD) inhibitor. Preliminary studies confirmed the optimal dosing and timing to observe for DMOG-mediated effects (Figure E3A). Control experiments confirmed that DMOG stabilized HIF-1α protein expression in SAECs under normoxic conditions at 6 hours and increased significantly by 6–24 hours (Figures 3A and 3B, P < 0.05). Similar results were observed in SAECs with no DMOG treatment (0 h) and vehicle control (data not shown). Therefore, only 0-hour DMOG treatment results were used to compare the effects of DMOG. Consistent with the induction of HIF-1α, CD55 transcripts were down-regulated after 6 hours of DMOG treatment (Figure 3C, P < 0.01), and CD55 protein down-regulation was observed at 24 hours (Figures 3D and 3E, P < 0.01). Data showing that CA9 transcripts were significantly higher in DMOG-treated SAECs provide evidence that DMOG-induced effects are due to stabilizing HIF-1α (Figure 3F, P < 0.05, P < 0.01).
Figure 3.
Stabilization of HIF-1α in normoxia by dimethyloxaloylglycine (DMOG) results in CD55 down-regulation in SAECs. (A) DMOG-treated (1 μM) SAECs were probed for HIF-1α protein expression. Lamin β1 was used as internal loading control. (B) Densitometry analysis shows HIF-1α induction within 24-hour DMOG (1 μM)-treated SAECs compared with cells with no DMOG. Values represent means (±SEM); n = 3; *P < 0.05 versus nontreated cells (0 hour controls); one-way ANOVA with Bonferroni posttest. (C) CD55 transcripts were down-regulated in 6-hour DMOG (1 μM)-treated SAECs compared with the nontreated cells (0 hour controls). Data represent means (±SEM); n > 3; **P < 0.01 versus 0 hour (nontreated cells); one-way ANOVA with Bonferroni posttest. (D) CD55 was probed for in DMOG (1 μM)-treated SAECs. Lamin β1 was used as internal loading control. (E) Densitometry analysis shows CD55 down-regulation by 24 hours of DMOG (1 μM)-treated SAECs compared with the nontreated cells (0 hour controls). Values represent means (±SEM); n = 3; **P < 0.01 versus nontreated cells (0 hour controls); one-way ANOVA with Bonferroni posttest. (F) CA9 transcripts were up-regulated in DMOG (1 μM)-treated SAECs compared with nontreated cells. Data represent means (±SEM); n = 3; *P < 0.05, or **P < 0.01 versus nontreated cells (0 hour controls); one-way ANOVA with Bonferroni posttest.
To confirm further specificity of DMOG-induced effects, we silenced PHD2, a PHD enzyme with increased selectivity for HIF-1α ubiquitination and degradation in normoxia (15, 16). PHD2 siRNA transfection showed dose-dependent down-regulation of PHD2 transcripts in normoxic SAECs that correlated with decreased protein expression (Figures E4A–E4D, P < 0.05). However, no significant change in CD55 protein expression was observed in PHD2 siRNA–transfected normoxic SAECs versus control siRNA (Figures E4C and E4E, P < 0.05, P < 0.01). To confirm that silencing PHD2 results in increased stabilization of HIF-1α in normoxia, we assessed and quantified HIF-1α protein expression in normoxic SAECs that were transfected with 50–150 nM PHD2 siRNA (Figures E5A and E5B). HIF-1α expression was induced in normoxic SAECs that were transfected with 50–150 nM PHD2 siRNA compared with control siRNA (Figures E5A and E5B, P < 0.05, P < 0.01).
HIF-1α was also stabilized in normoxic SAECs by transduction with an AdCA5, an adenoviral vector, to express constitutively active HIF-1α (10). HIF-1α expression was significantly higher in AdCA5-transduced normoxic cells at 24 hours compared with HIF-1α basal levels expressed in normoxic SAECs transduced with AdLACZ (Figures E6A and E6B, P < 0.05). AdCA5-induced HIF-1α correlated with down-regulated CD55 expression in the same cells (Figures E6C and E6D, P < 0.05).
Hypoxia Down-Regulates Airway CD55 In Vivo
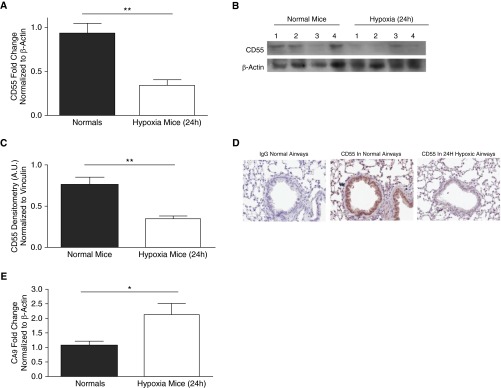
CD55 expression was assessed in the lungs of C57BL/6 mice after 24 hours of hypoxia (10% O2) in vivo. Analysis of whole-lung homogenates revealed that CD55 transcript and protein expression were down-regulated (Figures 4A–4C, P < 0.01). Immunohistochemistry (IHC) analysis revealed that this CD55 down-regulation occurred in 24-hour hypoxic mouse airway epithelium (Figure 4D). The expression of the HIF-1α–dependent gene, CA9, was also assessed to confirm the presence of HIF-1α and its activity. CA9 transcripts were increased in 24-hour hypoxic mouse lungs compared with lungs of normal mice, thus indicating hypoxia-induced HIF-1α activity (Figure 4E, P < 0.05).
Figure 4.
In vivo, hypoxia results in CD55 down-regulation in mouse lungs. (A) CD55 transcript levels were significantly down-regulated in 24-hour hypoxic mouse lungs compared with lungs of normal mice. Values represent means (±SEM); n > 6; **P < 0.01 versus normal mice; two-tailed unpaired t test. (B) CD55 expression was probed for CD55 in lung homogenates of 24-hour normal and hypoxic mice; β-actin was used as the loading control. (C) Densitometry analysis depicts statistically significant down-regulation of CD55 in 24-hour hypoxic mouse lungs compared with normal mice. Data represent means (±SEM); n = 4; **P < 0.01 versus normal mice; two-tailed unpaired t test. (D) Immunohistochemistry (IHC) analysis shows CD55 down-regulation in 24-hour hypoxic mouse airways compared with control normal mice. Original magnification: ×20. (E) HIF-1α–dependent gene, CA9, was transcriptionally induced in lungs of 24-hour hypoxic mice compared with normal mice. Values represent means (±SEM); n > 4; *P < 0.05 versus normal mice; two-tailed unpaired t test.
Down-Regulation of CD55 in Hypoxic Mouse Lungs Results in Local Complement Activation
Due to the CD55 down-regulation observed in hypoxic mouse lungs (Figure 4), we assessed local complement activation in bronchoalveolar lavage fluid (BALF) of these hypoxic mice as a functional correlate for the repression of this specific CRP. Interestingly, C3a levels were elevated in the BALF from these 24-hour hypoxic mice (Figure E7A, P < 0.05).
Silencing HIF-1α In Vivo during Hypoxia Recovers CD55 Airway Expression
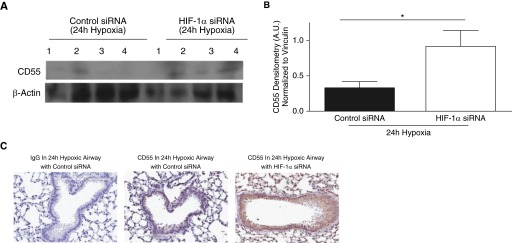
We next investigated if silencing HIF-1α expression in vivo could recover CD55 in hypoxic mouse lungs. After intratracheal instillation of HIF-1α siRNA or control siRNA, C57BL/10 mice were placed in hypoxic conditions (10% O2) for 72 hours. HIF-1α silencing was confirmed via densitometry analysis of HIF-1α in whole-lung homogenates (Figures E8A and E8B, P < 0.01). IHC analysis also showed a trend for HIF-1α down-regulation in hypoxic mouse airways (Figure E8C). CA9 transcription was assessed to confirm the effect of HIF-1α silencing. CA9 transcripts were elevated in 24-hour hypoxic mouse lungs compared with normal lungs (Figure E8D, P < 0.001), but down-regulated in 24-hour hypoxic mouse lungs treated with HIF-1α siRNA (Figure E8D, P < 0.05). Interestingly, CD55 expression was restored in hypoxic mouse lung homogenates with HIF-1α siRNA compared with mouse lung homogenates with control siRNA (Figures 5A and 5B, P < 0.05). IHC analysis shows recovery of CD55 expression in HIF-1α–silenced 24-hour hypoxic mouse airways (Figure E8C). CD55 recovery was also observed after 24 hours of hypoxia in mice that were instilled intratracheally with chetomin to inhibit HIF-1α (Figures E9A and E9B, P < 0.01). This was further confirmed by IHC analysis depicting CD55 recovery observed on airway epithelium of chetomin-instilled hypoxic mouse lungs compared with hypoxic mouse lungs with vehicle control, and the specificity of the drug was confirmed by assessing the CA9 transcripts (Figures E9C and E9D).
Figure 5.
CD55 expression recovered in hypoxic mouse lungs in which HIF-1α was silenced. (A) CD55 expression was probed for in lung homogenates of 24-hour hypoxic mice intratracheally instilled with HIF-1α siRNA (50 μg) or control siRNA (50 μg); β-actin was used as internal loading control. (B) Densitometry analysis indicated that CD55 was restored significantly in lung homogenates of 24-hour hypoxic mice intratracheally instilled with HIF-1α siRNA (50 μg) compared with control siRNA (50 μg). Data represent means (±SEM); n = 4; *P < 0.05 versus control siRNA; two-tailed unpaired t test. (C) IHC analysis show CD55 down-regulation in hypoxic mouse lungs in which HIF-1α is silenced compared with hypoxic mouse lungs with control siRNA. Original magnification: ×20.
Induction of HIF-1α In Vivo during Normoxia Down-Regulates Airway CD55 Expression
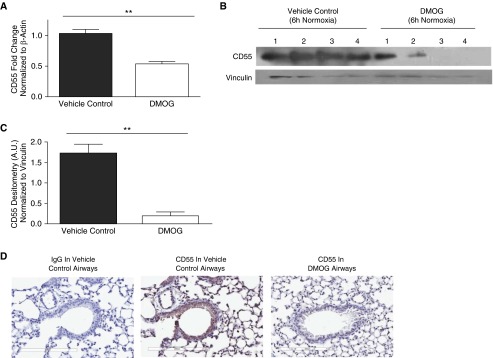
HIF-1α stabilization was achieved in vivo by intratracheal instillation of DMOG during normoxia. Compared with vehicle control, DMOG induced HIF-1α expression during normoxia (Figures E10A and E10B, P < 0.05). IHC analysis shows HIF-1α stabilization in normoxic mouse airways of mice instilled with DMOG versus vehicle control airways (Figure E10C). The specificity of this effect was confirmed by data showing increased CA9 in lungs of these mice (Figure E10D, P < 0.05). HIF-1α stabilization by DMOG in normoxic mouse lungs by 6 hours resulted in down-regulation of CD55 transcripts (Figure 6A, P < 0.01) and protein (Figures 6B and 6C, P < 0.01) compared with vehicle controls. Suppression of CD55 was further confirmed by IHC analysis of DMOG-instilled mouse airways in comparison to the controls (Figure 6D). Collectively, these results confirm that HIF-1α stabilization in the absence of hypoxia also mediates CD55 down-regulation.
Figure 6.
Induction of HIF-1α results in CD55 down-regulation. (A) CD55 transcript levels were significantly down-regulated in 6-hour lung homogenates of mice intratracheally instilled with DMOG (1 mg/ml) compared with vehicle controls. Data represent means (±SEM); n = 3; **P < 0.01 versus vehicle controls; two-tailed unpaired t test. (B) CD55 expression was probed in mouse lung homogenates 6 hours after intratracheal instillation with DMOG (1 mg/ml) or vehicle controls. Vinculin was used as internal loading control. (C) Densitometry analysis indicated significant CD55 down-regulation in lung homogenates intratracheally instilled with DMOG (1 mg/ml) or vehicle controls. Values represent means (±SEM); n = 4; **P < 0.01 versus vehicle controls; two-tailed unpaired t test. (D) IHC shows CD55 down-regulation within 6 hours after DMOG instillation versus vehicle control airways. Original magnification: ×20.
Discussion
Experimental findings highlighted in these studies demonstrate a role for HIF-1α in regulating airway epithelial CD55 expression. HIF-1α inhibition via chetomin or siRNA in hypoxic SAECs recovered CD55 expression and down-regulated the HIF-1α–dependent gene, CA9, in vitro and in vivo. In vivo data suggest that CD55 down-regulation in hypoxic mouse lungs correlated with increases in local complement activity, as shown by elevated C3a levels in BALF after 24 hours of hypoxia. In contrast, stabilizing HIF-1α in normoxic SAECs through AdCA5, or in vivo and in vitro through DMOG, resulted in CD55 down-regulation. Furthermore, stabilizing HIF-1α expression by silencing PHD2 during normoxia down-regulated CD55 expression.
In addition to CD55, other CRPs, such as CD46/CRRY or CD59, can regulate complement activation. However, our studies focused on CD55, due to its uniform expression on membranes of many cell types, including airway epithelial cells (17). CD55, a glycosylphosphatidylinositol-anchored protein, functions by preventing/dissociating C3 and C5 convertases, thereby inhibiting complement activation (18). In addition, CD55 has a role in protection from natural killer cell–mediated injury extravasation of neutrophils in epithelial cells, and is a receptor for certain viruses (17). Along with the role in innate immunity, CD55 also functions in adaptive immunity by having a costimulatory role in T cell activation and differentiation (19). Therefore, the observed CD55 down-regulation in some of these pulmonary pathologies (3, 5) may exacerbate lung injury through CD55 eliciting innate and adaptive immune responses.
These findings may be of particular interest for understanding the pathogenesis of pulmonary diseases, such as OB and IPF, where hypoxia occurs and down-regulation of CD55 is observed (1, 2). Therefore, it is possible that HIF-1α induced by airway hypoxia may contribute to the CD55 down-regulation observed in these diseases. It is plausible that regulating HIF-1α expression in these pulmonary diseases could preserve CD55 expression on airway epithelium, which could attenuate complement activation observed in OB (3) and IPF (5).
To the best of our knowledge, only two other studies report a link between CD55 and HIF-1α. Colgan and colleagues (20) reported that HIF-1α induced CD55 on human colorectal adenocarcinoma cells, and Collard and colleagues (21) reported a similar effect on hypoxic human umbilical vein endothelial cells. These data are in contrast to the current study, in which HIF-1α down-regulated CD55 on airway epithelium, and are likely accounted for by the use of primary cells in the current study rather than immortalized cell lines in prior reports (20–22). In addition, the current study uses lung cells, whereas the prior reports used nonlung cells to assess HIF-1α effects on CD55. These data could suggest tissue-specific effects of HIF-1α on CD55 expression. Factors, such as duration of hypoxic exposure and the concentration of oxygen used to induce hypoxia, may also contribute to hypoxia-dependent CD55 expression.
In normoxia, PHDs hydroxylate proline residues (p402 and p564) on HIF-1α, enabling E3 ubiquitin ligase, known as Von Hippel-Lindau, to ubiquitinate and degrade HIF-1α by 26S proteasome (23). In hypoxia, PHDs are inactivated, and HIF-1α is stabilized (23) and translocated to the nucleus to bind to its constitutively expressed subunit, HIF-1β. This heterodimer binds to hypoxia response elements on the DNA sequence and assembles with coactivators or corepressors for target gene activation or repression, respectively (24). This translocation process may take some time, which may explain the temporal lag between induction of HIF-1α and activation of CA9 (Figure 1). HIF-1α transcription is constitutive, but basal levels of HIF-1α in normoxia are observed due to the cyclical nature of its synthesis and degradation (23, 25).
Although other forms of PHD enzymes exist, PHD2 was silenced, due to its selectivity for HIF-1α among the other HIF isoforms, and its abundant expression in various cells (16). Silencing PHD2 resulted in up-regulated HIF-1α expression, which correlated with decreased CD55 transcript levels, but not CD55 protein during normoxia. This differential expression between CD55 transcripts and protein when PDH2 was silenced could be due to activity of other PHD isoforms compensating for the loss of PHD2. This question will be addressed in future studies.
Although data in the current study show that HIF-1α down-regulates CD55 expression, the exact molecular mechanisms of this process are not known. We speculate that transcriptional down-regulation may occur through epigenetic modifications of CD55 by recruitment of corepressors and histone-modifying complexes, as indicated by studies where HIF-1α resulted in transcriptional repression of other genes (26, 27). Some of the corepressors involved in HIF-1α–mediated gene repression are characterized as histone deacetylases (HDACs). Although the formation of the type of corepressor–HIF-1α complex depends on the stimulus and specificity of the cell or tissue, some of the corepressors known to bind HIF-1α in hypoxic conditions include HDAC1, HDAC2, HDAC3, HDAC4, HDAC5, HDAC7, and sirtuin-3 (28–32). However, it is likely that HIF-1α and HDAC interactions, under certain conditions, can also enhance transactivation of genes (32, 33). Therefore, it may be possible that corepressors with functional characteristics like methylase or demethylase activity may mediate CD55 down-regulation (34). Although other isoforms of HIFs exist, such as HIF-2α and HIF-3α, we focused on HIF-1α for our studies due to its ubiquitous expression and its strong induction during hypoxia (24). However, the current data do not exclude the possibility of other HIF isoforms in regulating CD55.
A potential limitation of our study involves the use of 10% oxygen for in vivo experiments, and 1% oxygen in vitro. Due to lethality associated with prolonged exposure, we were not able to conduct in vivo studies using 1% oxygen in vivo, although 10% oxygen is reported commonly for in vivo rodent studies (35, 36). The effects of various oxygen concentrations on CD55 expression in vitro will be assessed in future studies.
In summary, these studies highlight a role for HIF-1α in regulating CD55 on airway epithelium. It is intriguing to speculate that targeting the HIF-1α pathway could be a therapeutic intervention in lung diseases associated with complement activation.
Acknowledgments
Acknowledgments
AdLacZ and AdCA5 were given by Dr. Gregg Semenza from Johns Hopkins University (Baltimore, MD). All hypoxia studies were conducted in hypoxia chambers with the generous consent of Drs. Tim Lahm and Mircea Ivan at their laboratories at Indiana University School of Medicine (Indianapolis, IN).
Footnotes
This work was supported by National Institutes of Health (NIH) grants HL067177, HL096845, and P01AI084853 (D.S.W., Primary Investigator); P.H.P. was supported by NIH T32 AI060519 (J.S.B., Primary Investigator).
Author Contributions: Study concept and design—P.H.P. and D.S.W.; acquisition of samples and data—P.H.P., A.J.F., E.A.M., C.J.T., K.P.L., A.G., and G.E.S.; critical revision of manuscript—P.H.P., M.M., K.P., J.R., J.S.B., T.L., and D.S.W.; statistical analysis—P.H.P., K.R., and D.S.W.; obtained funding—J.R., J.S.B., and D.S.W.; full responsibility of study integrity and accuracy—P.H.P. and D.S.W.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2015-0237OC on August 5, 2016
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Wilkes DS. Airway hypoxia, bronchiolar artery revascularization, and obliterative bronchiolitis/bronchiolitis obliterans syndrome: are we there yet? Am J Respir Crit Care Med. 2010;182:136–137. doi: 10.1164/rccm.201004-0508ED. [DOI] [PubMed] [Google Scholar]
- 2.Bodempudi V, Hergert P, Smith K, Xia H, Herrera J, Peterson M, Khalil W, Kahm J, Bitterman PB, Henke CA. miR-210 promotes IPF fibroblast proliferation in response to hypoxia. Am J Physiol Lung Cell Mol Physiol. 2014;307:L283–L294. doi: 10.1152/ajplung.00069.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suzuki H, Lasbury ME, Fan L, Vittal R, Mickler EA, Benson HL, Shilling R, Wu Q, Weber DJ, Wagner SR, et al. Role of complement activation in obliterative bronchiolitis post-lung transplantation. J Immunol. 2013;191:4431–4439. doi: 10.4049/jimmunol.1202242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iwata T, Philipovskiy A, Fisher AJ, Presson RG, Jr, Chiyo M, Lee J, Mickler E, Smith GN, Petrache I, Brand DB, et al. Anti–type V collagen humoral immunity in lung transplant primary graft dysfunction. J Immunol. 2008;181:5738–5747. doi: 10.4049/jimmunol.181.8.5738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gu H, Mickler EA, Cummings OW, Sandusky GE, Weber DJ, Gracon A, Woodruff T, Wilkes DS, Vittal R. Crosstalk between TGF-β1 and complement activation augments epithelial injury in pulmonary fibrosis. FASEB J. 2014;28:4223–4234. doi: 10.1096/fj.13-247650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pandya PH, Wilkes DS. Complement system in lung disease. Am J Respir Cell Mol Biol. 2014;51:467–473. doi: 10.1165/rcmb.2013-0485TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilkes DS. Chronic lung allograft rejection and airway microvasculature: is HIF-1 the missing link? J Clin Invest. 2011;121:2155–2157. doi: 10.1172/JCI58329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lahm T, Albrecht M, Fisher AJ, Selej M, Patel NG, Brown JA, Justice MJ, Brown MB, Van Demark M, Trulock KM, et al. 17β-Estradiol attenuates hypoxic pulmonary hypertension via estrogen receptor-mediated effects. Am J Respir Crit Care Med. 2012;185:965–980. doi: 10.1164/rccm.201107-1293OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao X-J, Wang L, Shiva S, Tejero J, Wang J, Frizzell S, Gladwin MT. Mechanisms for cellular NO oxidation and nitrite formation in lung epithelial cells. Free Radic Biol Med. 2013;61:428–437. doi: 10.1016/j.freeradbiomed.2013.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarkar K, Fox-Talbot K, Steenbergen C, Bosch-Marcé M, Semenza GL. Adenoviral transfer of HIF-1α enhances vascular responses to critical limb ischemia in diabetic mice. Proc Natl Acad Sci U S A. 2009;106:18769–18774. doi: 10.1073/pnas.0910561106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan L, Benson HL, Vittal R, Mickler EA, Presson R, Fisher AJ, Cummings OW, Heidler KM, Keller MR, Burlingham WJ, et al. Neutralizing IL-17 prevents obliterative bronchiolitis in murine orthotopic lung transplantation. Am J Transplant. 2011;11:911–922. doi: 10.1111/j.1600-6143.2011.03482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giatromanolaki A, Koukourakis MI, Sivridis E, Pastorek J, Wykoff CC, Gatter KC, Harris AL. Expression of hypoxia-inducible carbonic anhydrase-9 relates to angiogenic pathways and independently to poor outcome in non–small cell lung cancer. Cancer Res. 2001;61:7992–7998. [PubMed] [Google Scholar]
- 13.McDonald PC, Dedhar S. Carbonic anhydrase IX (CAIX) as a mediator of hypoxia-induced stress response in cancer cells. Subcell Biochem. 2014;75:255–269. doi: 10.1007/978-94-007-7359-2_13. [DOI] [PubMed] [Google Scholar]
- 14.Yano K, Horinaka M, Yoshida T, Yasuda T, Taniguchi H, Goda AE, Wakada M, Yoshikawa S, Nakamura T, Kawauchi A, et al. Chetomin induces degradation of XIAP and enhances TRAIL sensitivity in urogenital cancer cells. Int J Oncol. 2011;38:365–374. doi: 10.3892/ijo.2010.874. [DOI] [PubMed] [Google Scholar]
- 15.Peng J, Lai ZG, Fang ZL, Xing S, Hui K, Hao C, Jin Q, Qi Z, Shen WJ, Dong QN, et al. Dimethyloxalylglycine prevents bone loss in ovariectomized C57BL/6J mice through enhanced angiogenesis and osteogenesis. PLoS One. 2014;9:e112744. doi: 10.1371/journal.pone.0112744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fong GH, Takeda K. Role and regulation of prolyl hydroxylase domain proteins. Cell Death Differ. 2008;15:635–641. doi: 10.1038/cdd.2008.10. [DOI] [PubMed] [Google Scholar]
- 17.Piccoli AK, Alegretti AP, Schneider L, Lora PS, Xavier RM. Expression of complement regulatory proteins CD55, CD59, CD35, and CD46 in rheumatoid arthritis. Rev Bras Reumatol. 2011;51:503–510. [PubMed] [Google Scholar]
- 18.Meri S. Complement activation in diseases presenting with thrombotic microangiopathy. Eur J Intern Med. 2013;24:496–502. doi: 10.1016/j.ejim.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 19.Sutavani RV, Bradley RG, Ramage JM, Jackson AM, Durrant LG, Spendlove I. CD55 costimulation induces differentiation of a discrete T regulatory type 1 cell population with a stable phenotype. J Immunol. 2013;191:5895–903. doi: 10.4049/jimmunol.1301458. [DOI] [PubMed] [Google Scholar]
- 20.Louis NA, Hamilton KE, Kong T, Colgan SP. HIF-dependent induction of apical CD55 coordinates epithelial clearance of neutrophils. FASEB J. 2005;19:950–959. doi: 10.1096/fj.04-3251com. [DOI] [PubMed] [Google Scholar]
- 21.Collard CD, Väkevä A, Büküsoglu C, Zünd G, Sperati CJ, Colgan SP, Stahl GL. Reoxygenation of hypoxic human umbilical vein endothelial cells activates the classic complement pathway. Circulation. 1997;96:326–333. doi: 10.1161/01.cir.96.1.326. [DOI] [PubMed] [Google Scholar]
- 22.Varsano S, Rashkovsky L, Shapiro H, Ophir D, Mark-Bentankur T. Human lung cancer cell lines express cell membrane complement inhibitory proteins and are extremely resistant to complement-mediated lysis; a comparison with normal human respiratory epithelium in vitro, and an insight into mechanism(s) of resistance. Clin Exp Immunol. 1998;113:173–182. doi: 10.1046/j.1365-2249.1998.00581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kong X, Lin Z, Liang D, Fath D, Sang N, Caro J. Histone deacetylase inhibitors induce VHL and ubiquitin-independent proteasomal degradation of hypoxia-inducible factor 1α. Mol Cell Biol. 2006;26:2019–2028. doi: 10.1128/MCB.26.6.2019-2028.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ke Q, Costa M. Hypoxia-inducible factor-1 (HIF-1) Mol Pharmacol. 2006;70:1469–1480. doi: 10.1124/mol.106.027029. [DOI] [PubMed] [Google Scholar]
- 25.Harris CL, Spiller OB, Morgan BP. Human and rodent decay-accelerating factors (CD55) are not species restricted in their complement-inhibiting activities. Immunology. 2000;100:462–470. doi: 10.1046/j.1365-2567.2000.00066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen KF, Lai YY, Sun HS, Tsai SJ. Transcriptional repression of human cad gene by hypoxia inducible factor-1α. Nucleic Acids Res. 2005;33:5190–5198. doi: 10.1093/nar/gki839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romney SJ, Newman BS, Thacker C, Leibold EA. HIF-1 regulates iron homeostasis in Caenorhabditis elegans by activation and inhibition of genes involved in iron uptake and storage. PLoS Genet. 2011;7:e1002394. doi: 10.1371/journal.pgen.1002394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Charron CE, Chou PC, Coutts DJ, Kumar V, To M, Akashi K, Pinhu L, Griffiths M, Adcock IM, Barnes PJ, et al. Hypoxia-inducible factor 1α induces corticosteroid-insensitive inflammation via reduction of histone deacetylase-2 transcription. J Biol Chem. 2009;284:36047–36054. doi: 10.1074/jbc.M109.025387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee KJ, Lee KY, Lee YM. Downregulation of a tumor suppressor RECK by hypoxia through recruitment of HDAC1 and HIF-1α to reverse HRE site in the promoter. Biochim Biophys Acta. 2010;1803:608–616. doi: 10.1016/j.bbamcr.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 30.He Q, Gao Z, Yin J, Zhang J, Yun Z, Ye J. Regulation of HIF-1α activity in adipose tissue by obesity-associated factors: adipogenesis, insulin, and hypoxia. Am J Physiol Endocrinol Metab. 2011;300:E877–E885. doi: 10.1152/ajpendo.00626.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barneda-Zahonero B, Parra M. Histone deacetylases and cancer. Mol Oncol. 2012;6:579–589. doi: 10.1016/j.molonc.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen L, Endler A, Shibasaki F. Hypoxia and angiogenesis: regulation of hypoxia-inducible factors via novel binding factors. Exp Mol Med. 2009;41:849–57. doi: 10.3858/emm.2009.41.12.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seo H-W, Kim E-J, Na H, Lee M-O. Transcriptional activation of hypoxia-inducible factor-1α by HDAC4 and HDAC5 involves differential recruitment of p300 and FIH-1. FEBS Lett. 2009;583:55–60. doi: 10.1016/j.febslet.2008.11.044. [DOI] [PubMed] [Google Scholar]
- 34.Zhou X, Sun H, Chen H, Zavadil J, Kluz T, Arita A, Costa M. Hypoxia induces trimethylated H3 lysine 4 by inhibition of JARID1A demethylase. Cancer Res. 2010;70:4214–4221. doi: 10.1158/0008-5472.CAN-09-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patel D, Alhawaj R, Wolin MS. Exposure of mice to chronic hypoxia attenuates pulmonary arterial contractile responses to acute hypoxia by increases in extracellular hydrogen peroxide. Am J Physiol Regul Integr Comp Physiol. 2014;307:R426–R433. doi: 10.1152/ajpregu.00257.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Døhlen G, Carlsen H, Blomhoff R, Thaulow E, Saugstad OD. Reoxygenation of hypoxic mice with 100% oxygen induces brain nuclear factor-κB. Pediatr Res. 2005;58:941–945. doi: 10.1203/01.PDR.0000182595.62545.EE. [DOI] [PubMed] [Google Scholar]