Abstract
Fibrotic disorders are associated with tissue accumulation of fibroblasts. We recently showed that caveolin (Cav)-1 gene suppression by a profibrotic cytokine, transforming growth factor (TGF)-β1, contributes to fibroblast proliferation and apoptosis resistance. Cav-1 has been shown to be constitutively suppressed in idiopathic pulmonary fibrosis (IPF), but mechanisms for this suppression are incompletely understood. We hypothesized that epigenetic processes contribute to Cav-1 down-regulation in IPF lung fibroblasts, and after fibrogenic stimuli. Cav-1 expression levels, DNA methylation status, and histone modifications associated with the Cav-1 promoter were examined by PCR, Western blots, pyrosequencing, or chromatin immunoprecipitation assays in IPF lung fibroblasts, normal fibroblasts after TGF-β1 stimulation, or in murine lung fibroblasts after bleomycin injury. Methylation-specific PCR demonstrated methylated and unmethylated Cav-1 DNA copies in all groups. Despite significant changes in Cav-1 expression, no changes in DNA methylation were observed in CpG islands or CpG island shores of the Cav-1 promoter by pyrosequencing of lung fibroblasts from IPF lungs, in response to TGF-β1, or after bleomycin-induced murine lung injury, when compared with respective controls. In contrast, the association of Cav-1 promoter with the active histone modification mark, H3 lysine 4 trimethylation, correlated with Cav-1 down-regulation in activated/fibrotic lung fibroblasts. Our data indicate that Cav-1 gene silencing in lung fibroblasts is actively regulated by epigenetic mechanisms that involve histone modifications, in particular H3 lysine 4 trimethylation, whereas DNA methylation does not appear to be a primary mechanism. These findings support therapeutic strategies that target histone modifications to restore Cav-1 expression in fibroblasts participating in pathogenic tissue remodeling.
Keywords: histone modification, DNA methylation, caveolin-1, lung fibroblasts, fibrosis
Clinical Relevance
The mechanisms for caveolin-1 suppression in idiopathic pulmonary fibrosis and by transforming growth factor-β1 are enigmatic. Our study demonstrates the involvement of histone modification of this gene, whereas DNA methylation does not appear to be a primary mechanism. These findings support therapeutic strategies to target histone modifications to augment caveolin-1 expression in diseases, such as idiopathic pulmonary fibrosis, with suppressed levels of this gene.
Caveolin (Cav)-1 is a scaffolding protein component of caveolae in plasma membranes (1). Caveolae participate in multiple cellular functions, including membrane trafficking, endocytosis, and signal transduction (1). Cav-1 is expressed in diverse cell types, such as fibroblasts and endothelial cells (2). It is one of the mostly extensively characterized Cavs of the three identified Cav family members. Cav-1 may function as a tumor suppressor, as well as an oncogene, depending on the cellular microenvironment (1).
Recent studies have implicated a critical role for Cav-1 in various fibrotic diseases (3). Fibrosis results from dysregulated tissue repair, during which fibroblasts differentiate into myofibroblasts and acquire apoptosis resistance, leading to persistent tissue remodeling and scarring that ends in loss of tissue function (4). Down-regulation of Cav-1 has been reported in a number of fibrotic diseases, including idiopathic pulmonary fibrosis (IPF) (5), scleroderma (6), cardiac fibrosis (7), and dermal keloids (8). Cav-1–deficient mice demonstrate impaired wound healing and altered lung morphology that mimics fibrosis (9). Restoration of Cav-1, either with a Cav-1 peptide (6, 8) or adenoviral gene transfer (10), showed decreases in fibrotic changes in cells/tissues, supporting antifibrotic effects of Cav-1.
Transforming growth factor (TGF)-β1 is a key mediator of fibrogenesis in diverse tissues and organs (11). TGF-β1 is a critical inducer of myofibroblast differentiation, and it promotes an apoptosis-resistant phenotype through multiple pathways, including p38 mitogen-activated protein kinase (MAPK) (12). TGF-β1 can down-regulate Cav-1 gene/protein in fibroblasts (5, 7, 8, 13), resulting in a hyperproliferative and apoptosis-resistant fibroblast phenotype (14, 15).
In human malignancies in which Cav-1 expression is down-regulated, including breast cancer (16), colon cancer (17), and ovarian cancer (18), DNA methylation at the Cav-1 gene promoter region has been reported to be a mechanism for constitutive Cav-1 down-regulation. Other epigenetic mechanisms, such as histone modifications, have been suggested by observations that the histone deacetylase inhibitor, trichostatin A, up-regulates Cav-1 expression (18). In a recent study in colon cancer, acetylation of histone H3 and H4 have been shown to correlate with Cav-1 expression (17). However, the epigenetic regulation of Cav-1 by other active histone modification marks, such as histone H3 lysine 4 trimethylation (H3K4Me3), has not been explored.
We previously reported that TGF-β1 down-regulates Cav-1 in lung fibroblasts (13). However, whether this TGF-β1–inducible expression and the constitutive low expression of Cav-1 in IPF fibroblasts involves epigenetic regulation is not known. In this study, we investigated the role of DNA methylation and histone modifications in the constitutive suppression of Cav-1 in primary lung fibroblasts from patients with IPF, as well as TGF-β1–induced down-regulation of Cav-1 and in an animal model of lung fibrosis.
Materials and Methods
See detailed protocols in the online supplement.
Cell Culture and Treatments
Human primary non-IPF control or IPF lung fibroblasts were derived from deidentified tissues from the University of Alabama at Birmingham (UAB) Tissue Procurement Facility, which is approved by the UAB Institutional Review Board. The diagnosis of IPF was made by a multidisciplinary approach according to the American Thoracic Society/European Respiratory Society guidelines (19). Demographic characteristics of the subjects are listed in Table E1 in the online supplement. Three fibroblast cell lines were randomly selected from subjects with IPF with constitutively low protein levels of Cav-1 (defined by ≥50% reduction in mean densitometric ratios of Cav-1:β-actin compared with non-IPF control fibroblasts). IMR-90 lung fibroblasts were from Coriell Institute for Medical Research (Camden, NJ), and were used before passage 5. Cells were in 1% FBS medium overnight before TGF-β1 treatment. To inhibit the p38 MAPK pathway, cells were treated with 10 μM SB203580 (Cell Signaling, Beverly, MA) for 2 hours before TGF-β1 treatment. TGF-β1 (R&D Systems, Minneapolis, MN) was added at 2 ng/ml for 24 hours.
DNA/RNA/Protein/Nuclear Extraction and Real-Time RT-PCR
Allprep (Qiagen, Valencia, CA) or EpiQuick Nuclear extraction kits (Epigentek, Brooklyn, NY) were used. Real-time RT-PCR was performed in triplicate and normalized to 18S or β-actin using the ΔΔ cycle threshold method (20). All primers are detailed in Table 1.
Table 1.
PCR Primer Sequences
| Name | Sequence |
|---|---|
| Cav1 (human; ENSG 00,000,105,974) | |
| RT-PCR | F: 5′-GAGCTGAGCGAGAAGCAAGT-3′ |
| R: 5′-TCCCTTCTGGTTCTGCAATC-3′ | |
| PCR for ChIP | F: 5′-GGCATAACCTGTTGGCATAAA-3′ |
| R: 5′-CTCCCAAACGCTTCGAAATAAG-3′ | |
| MSP | |
| Methylated primers | MF: 5′-TTATTTCGAAGCGTTTGGGAG-3′ |
| MR:5′-AACACTCGTTTACATCTAATCG-3′ | |
| Unmethylated primers | UF: 5′-TTATTTTGAAGTGTTTGGGAG-3′ |
| UR: 5′-AACACTCATTTACATCTAATCA-3′ | |
| Pyrosequencing | |
| Set 1 | F1: 5′-AGAGTAGGGAAAATGTTGTTTTAGG-3′ |
| R1: 5′-ATCTAAACACATCCCCAAAATTCT-3′ | |
| Sequencing primer 1 | 5′-AGGTTTAAAATAATTTGTTTAAGTA-3′ |
| Set 2: | F2: 5′-AGTAGGGAAAATGTTGTTTTAGG-3′ |
| R2: 5′-ATTTACCCCCAAACATACTAAC-3′ | |
| Sequencing primer 2 | 5′-GTTAGAATTTTGGGGATG -3′ |
| Set 3 | F3: 5′-TGTAAATATGGTATAATTTGTTGGTATAAA-3′ |
| R3: 5′-CCTTTAAATACATTTCACATCTAAACTTT-3′ | |
| Sequencing primer 3 | 5′-ACCTATAATATACTTTTAATAACAC-3′ |
| Set 4 | F4: 5′-GGAAGGGTTGGGATAGTTTT-3′ |
| R4: 5′-ACCCTCTATTACTAAAATCAATAAACTCAT-3′ | |
| Sequencing primer 4 | 5′-GTTAGTGTATTTTTTTTTAGTATTG-3′ |
| Cav1 (mouse; ENSMUSG 00,000,007,655) | |
| RT-PCR | F: 5′-GCGACCCCAAGCATCTCAACG-3′ |
| R: 5′-GGGTCACACACAGCCAAGCTCAA-3′ | |
| PCR for ChIP | F: 5′-TCCTTCCTCCCTCACAAGTA-3′ |
| R: 5′-AAGCTGAATCGTAGCCCTATTT-3′ | |
| Pyrosequencing | F1: 5′-AGGTTTGGAGGAGAGAAGGAATAT-3′ |
| R1: 5′-ACCTAACCCTACAACTTTTACTAACTTCA-3′ | |
| Sequencing primer 1 | 5′-GGAGAGAAGGAATATAGAG-3′ |
| F2: 5′-AGGAGGAGGAAAGGGAATTTTAGT-3′ | |
| R2: 5′-AAACAAAAATCAAAAATCAACCTAACTAT-3′ | |
| Sequencing primer 2 | 5′-GGTAGGAGTTTTTTGTTTGGTA-3′ |
| β-actin (human) | |
| RT-PCR | F:5′-TGC TAT CCA GGC TGT GCT AT-3′ |
| R: 5′-AGT CCA TCA CGA TGC CAG T-3′ | |
| β-actin (mouse) | |
| RT-PCR | F: 5′-AGT GTG ACG TTG ACA TCC GT-3′ |
| R: 5′-TGC TAG GAG CCA GAG CAG TA-3′ |
Definition of abbreviations: Cav1, caveolin-1; ChIP, chromatin immunoprecipitation; F, forward; M, methylated; MSP, methylation-specific PCR; PCR, polymerase chain reaction; R, reverse; U, unmethylated.
Antibodies and Immunoblotting
Anti–Cav-1 (no. 610406) was from BD Biosciences (San Jose, CA), β-tubulin (no. 2128) was from Cell Signaling, and anti-H3K4Me3 (no. 39159) was from Active Motif (Carlsbad, CA). Western blots were performed as previously reported (21).
DNA Methylation Changes by Methylation-Specific PCR and Pyrosequencing
Methylated and unmethylated primers for Cav-1 methylation-specific PCR (MSP) were derived from published studies (22). Quantitative methylation was performed by pyrosequencing (23).
Chromatin Immunoprecipitation Assays
Chromatin immunoprecipitation (ChIP) assays were performed per manufacturer’s protocol (Epigentek), with minor modifications (24). ChIPed-DNA was amplified by real-time PCR with primers delineated Table 1. Results were normalized to input DNA.
Animal Model of Pulmonary Fibrosis, Primary Murine Lung Fibroblast Isolation, and Immunohistochemistry
All animal studies were performed in accordance with UAB Institutional Animal Care and Use Committee–approved protocols. Healthy C57BL mice (6–8 wk old) were used. A single dose of normal saline or bleomycin sulfate (3 U/kg body weight) was instilled intratracheally, as previously described (25). Mice were killed on Days 5, 7, 12, and 28 after bleomycin injury. Lungs were prepared for histology and fibroblast primary culture (25). Fibroblasts were collected at passages 1–3 for DNA, RNA, whole-cell lysate, nuclear extract, and ChIP assays. PCR primers are listed in Table 1. Immunohistochemistry images were obtained with a Nikon TE2000U microscope equipped with a QiCam Fast Cooled high-resolution charge-coupled device camera (Nikon Inc., Melville, NY) with MetaMorph software (v.6.2r4; Universal Imaging, West Chester, PA). A semiquantitative Ashcroft scale, ranging from 0 to 8, was used to grade severity of lung fibrosis after bleomycin injury (26).
Statistical Analysis
Data are expressed as means (±SD). Student’s paired t test was used to compare two groups; one-way ANOVA was used for comparisons involving three or more groups. P values of 0.05 or less were considered statistically significant.
Results
Cav-1 DNA Methylation Status
Previous studies have reported decreased Cav-1 in IPF lung fibroblasts (5). We observed heterogeneity in the expression of Cav-1 in primary IPF lung fibroblasts, with 11 out of 15 samples showing a down-regulation in Cav-1 (data not shown), which is consistent with previous reports of fibroblast heterogeneity in IPF (27). We selected 3 of these 11 samples for the studies reported here (see Materials and Methods for detailed information). DNA methyltransferase (DNMT) 1 and DNMT3a were increased after treatment with TGF-β1 in non-IPF control lung fibroblasts (Figure E1; we were unable to detect DNMT3b). We sought to explore whether both constitutive Cav-1 suppression (IPF versus non-IPF controls) and TGF-β1–induced Cav-1 down-regulation (in IMR-90 fibroblasts) are mediated by DNA methylation.
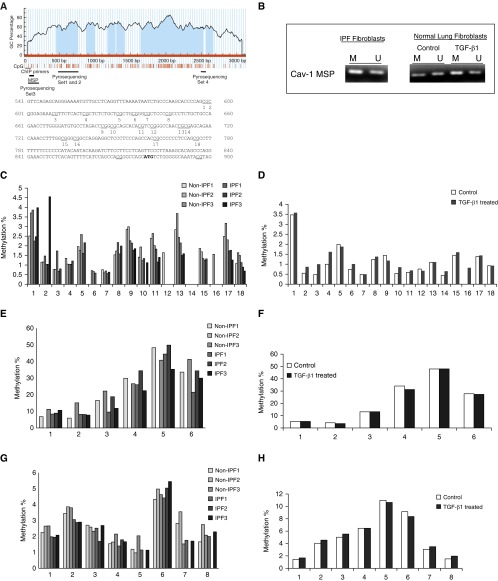
DNA methylation status at CpG island of Cav-1 gene.
First, we confirmed that Cav-1 levels are reduced in IPF fibroblasts, and after TGF-β1 stimulation in normal lung fibroblasts (Figure E1). MethPrimer (28) demonstrated the promoter region of human Cav-1 is CpG rich. (Figure 1A). Using MSP, we found that Cav-1 gene has methylated and unmethylated DNA copies in IPF fibroblasts and in normal lung fibroblasts stimulated with/without TGF-β1 (Figure 1B). Because MSP does not quantify the amount of DNA methylation of this gene, we further performed quantitative pyrosequencing. A total of 18 CpG sites in the defined CpG island (CGI) at the promoter region (ENSG00000105974, 596–780bp [Figure 1A], indicated by pyrosequencing set 1 and 2) were examined. Relatively low percentages (from undetectable to ∼4%) of DNA methylation were found in this region, without significant differences between IPF fibroblasts and non-IPF control fibroblasts (Figure 1C, n = 3 in each group). Similarly, both untreated and TGF-β1–treated lung fibroblasts demonstrated a similar pattern of Cav-1 DNA methylation at the CGI of the Cav-1 gene (from undetectable to ∼3.5%, in each group; Figure 1D).
Figure 1.
Human caveolin-1 (Cav-1) DNA methylation status. (A) Human Cav-1 genome browser view (28) and distribution of CpG sites. The red vertical lines under the figure mark each CpG site; the dark solid line represents approximate location of pryrosequencing primer sets, methylation-specific PCR (MSP) primers, and chromatin immunoprecipitation (ChIP) primer sets, which are all marked under the vertical lines of the CpG sites. The 18 CpG sites examined by pyrosequencing (sets 1 and 2; see C and D) are marked out and numbered accordingly. GC, guanine and cytosine. (B) MSP with bisulfite-modified DNA using specific primers that can distinguish methylated (M) and unmethylated (U) Cav-1 DNA copies, respectively, in idiopathic pulmonary fibrosis (IPF) fibroblasts and in normal lung fibroblasts treated without (control) or with transforming growth factor (TGF)-β1 (2 ng/ml) for 24 hours. (C and D) Pyrosequencing of CpG island (CGI) region of Cav-1 promoter region, covering the 18 CpG sites that are marked in A (region marked as pyrosequencing sets 1 and 2). The primers and probe are listed in Table 1. Bar graphs represent the methylation percentage at each CpG site in non-IPF versus IPF fibroblasts (C) or in normal IMR-90 lung fibroblasts treated without (control) or with TGF-β1 at 2 ng/ml for 24 hours (D). (E and F) Pyrosequencing of 5′ shore of CGI region of Cav-1 promoter, covering six CpG sites (the region indicated in A marked as pyrosequencing set 3). The primers and probe are listed in Table 1. Bar graphs represent the methylation percentage at each CpG site in non-IPF versus IPF fibroblasts (E) or in normal IMR-90 lung fibroblasts treated without (control) or with TGF-β1 at 2 ng/ml for 24 hours (F). (G and H) Pyrosequencing of CGI region of Cav-1 promoter region at the 3′ shore of CGI, covering eight CpG sites (the region indicated in A marked with pyrosequencing set 4). The primers and probes are listed in Table 1. Bar graphs represent the methylation percentage at each CpG site in non-IPF versus IPF fibroblasts (G) or in normal IMR-90 lung fibroblasts treated without (control) or with TGF-β1 at 2 ng/ml for 24 hours (H).
DNA methylation status at CGI shores of Cav-1 gene.
Because studies indicate that CGI shore methylation regulates Cav-1 expression in breast cancer (16), we measured the DNA methylation status of CpG sites further upstream at the 5′ shore as well as the 3′ shore downstream of the CGI. DNA methylation percentages were higher in the 5′ shore region than the CGI (ranging from ∼6 to ∼50%; Figures 1E and 1F). Despite the inability to obtain readings with this sequencing primer set in one of the non-IPF samples (non-IPF #2; Figure 1E), we did not detect any significant differences between IPF and non-IPF controls in the DNA methylation status within this region (pyrosequencing set 3; Figure 1A). Similarly, no changes in DNA methylation were detected in response to TGF-β1 treatment (Figure 1F). In contrast to the 5′ shore, DNA methylation levels are relatively low within the 3′ shore of the CGI region (ranging from <1 to ∼10%; Figures 1G and 1H). In concordance with the 5′ shore, no obvious differences in DNA methylation status were noted in comparisons of non-IPF versus IPF and untreated versus TGF-β1–treated lung fibroblasts (Figures 1G and 1H). Our data show that there are methylated and unmethylated DNA copies in the Cav-1 gene promoter. However, the DNA methylation status at CGI and CGI shores within the Cav-1 promoter region are similar in IPF and non-IPF control lung fibroblasts, and do not change in response to TGF-β1 treatment in non-IPF lung fibroblasts.
The Association of Active Histone Mark H3K4Me3 with the Cav-1 Promoter Region
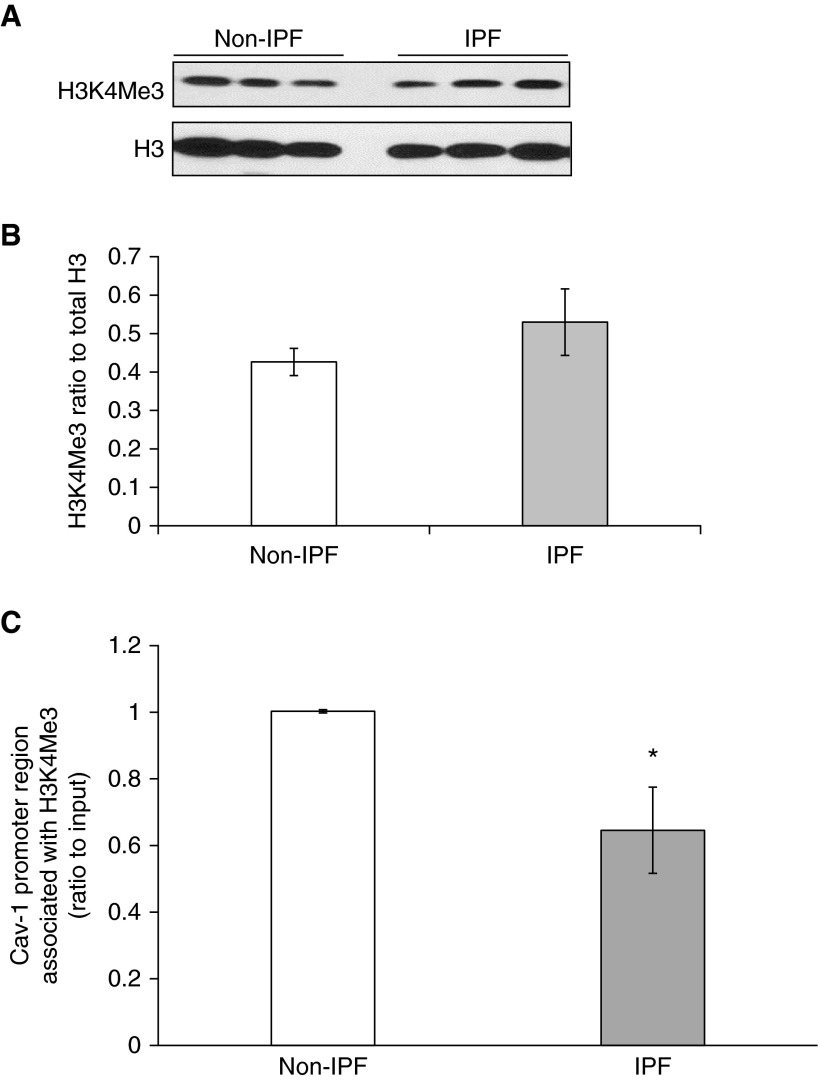
In addition to DNA methylation, histone modifications participate in epigenetic regulation of gene expression (29). We examined whether histone modifications are involved in the down-regulation of Cav-1 in IPF fibroblasts. Previous reports suggest that H3K4Me3 is an active histone mark, and it is important in regulation of tumor suppressor genes that include Cav-1 (30, 31). The global level of H3K4Me3 was measured in IPF fibroblasts, and then the association of H3K4Me3 with the Cav-1 promoter region was examined using ChIP assays. There were no significant differences of H3K4Me3 global levels between IPF versus non-IPF fibroblasts (Figures 2A and 2B). However, we observed significantly decreased association of Cav-1 with H3K4Me3 in IPF primary fibroblasts compared with non-IPF control cells (Figure 2C), corresponding to the constitutively low Cav-1 in these IPF fibroblasts.
Figure 2.
Histone modification H3 lysine 4 trimethylation (H3K4Me3) is associated with Cav-1 promoter region in lung fibroblasts. (A) Representative immunoblot of H3K4Me3 and total H3 in non-IPF and IPF lung fibroblasts; H3 was the loading control. (B) Densitometric analyses of the Western blots in A, after normalization to H3; bar represents mean (±SD) from the average of the samples (n = 3) of non-IPF and IPF. (C) The quantitative ChIP assays were performed to analyze the association of Cav-1 with H3K4Me3. Bars represent the relative levels of polymerase chain reaction (PCR) product of Cav-1 promoter region association with H3K4Me3; the regions of the PCR primers are marked out in Figure 1A. The results were normalized to input DNA and expressed as fold changes relative to non-IPF control. These assays were repeated in three different IPF and non-IPF cell lines. Bar graphs represent mean (±SD) from the average of three replicates. *P < 0.05, IPF versus non-IPF control.
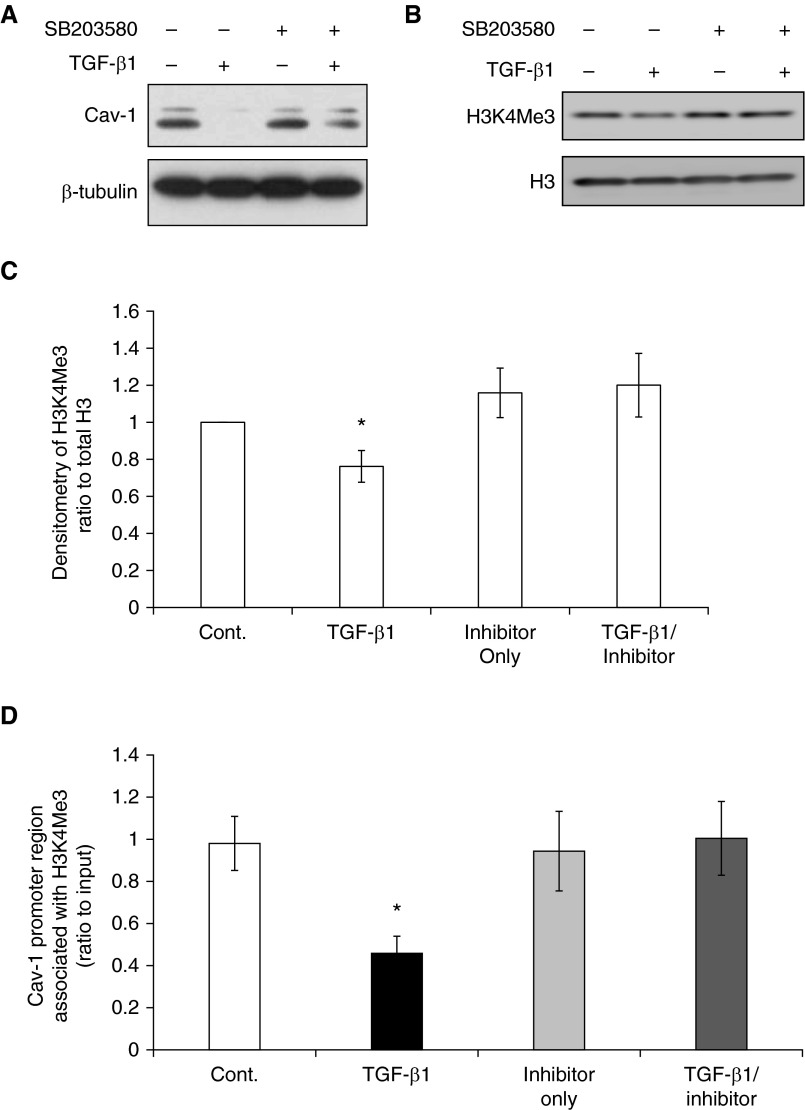
When normal lung fibroblasts (IMR-90) were treated with TGF-β1, we observed suppression of Cav-1 (Figure 3A), decreased global level of H3K4Me3 relative to total H3, and decreased association of H3K4Me3 with the Cav-1 promoter (Figure 3). We previously demonstrated that the p38 MAPK pathway mediates Cav-1 down-regulation by TGF-β (13). To determine if this pathway contributes to epigenetic regulation of Cav-1, the effects of p38 MAPK inhibition on H3K4Me3 histone modification were tested. Blocking p38 MAPK activity with the p38 MAPK inhibitor, SB203580, prevented TGF-β1–induced Cav-1 down-regulation and abrogated the decrease in H3K4Me3 levels (Figures 3B and 3C) in parallel with decreased association of this histone mark with the Cav-1 promoter (Figure 3D). These data suggest that the H3K4Me3 histone modification is likely involved in both the constitutive down-regulation of Cav-1 in IPF fibroblasts, and the TGF-β1–mediated suppression of Cav-1.
Figure 3.
The down-regulation of Cav-1 in normal lung fibroblasts by TGF-β1 is associated with histone modification H3K4Me3. (A) Representative immunoblots of the expression of Cav-1 in IMR-90 fibroblasts after treatment without or with TGF-β1 (2 ng/ml), p38 inhibitor SB203580 only (10 μM) for 24 hours, and SB203580 for 2 hours followed by 24 hours of TGF-β1 at 2 ng/ml; β-tubulin was used as loading control. See main text for detailed protocol. (B) Representative immunoblots of H3K4Me3 global level with nuclear extract with the same treatment as listed in A. Loading control was total H3. (C) Densitometric analyses of H3K4Me3 in B after being normalized to H3. (D) ChIP assays to examine the association of Cav-1 with H3K4Me3, in untreated control (Cont.), TGF-β1 treated for 24 hours, p38 inhibitor SB203580 only for 24 hours, and 2 hours of SB203580 followed by 24 hours of TGF-β1 treatment (TGF-β1/inhibitor). Bars represent the relative levels of PCR product of Cav-1 promoter region associated with H3K4Me3 (primers are marked in Figure 1A). The results are normalized to input DNA and expressed as fold changes relative to non-IPF control. Bars represent mean (±SD) from the average of at least three replicate experiments. *P < 0.05, TGF-β1 versus untreated control (Cont.).
Cav-1 Expression Is Transiently Down-Regulated in Response to Lung Injury in Mice
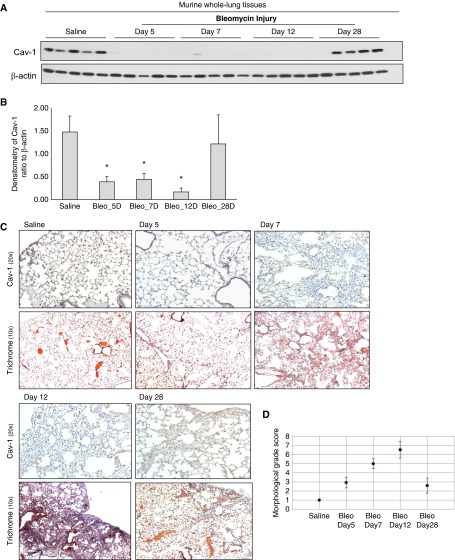
Changes in Cav-1 expression were examined at varying time points after bleomycin-induced lung injury in mice, as previously described (25). There is histopathological evidence that fibrosis develops in this model from Days 7 to 10, and peaks around Day 14 after injury (32). Cav-1 expression was examined on Day 5 after injury (when inflammation is relatively more active), as well as on Day 7 (when fibrosis is developing), Day 12 (near peak fibrosis), and Day 28 (during the resolving phase) (25). Cav-1 protein expression was markedly decreased in whole-lung lysates at Days 5, 7, and 12, with recovery on Day 28 after lung injury (Figures 4A and 4B, n = 5 in each group; one of the samples on Day 28 did not show recovery).
Figure 4.
Cav-1 expression in murine whole-lung tissues at different time points after bleomycin injury. (A) Expression of Cav-1 in murine lung tissues in 6- to 8-week-old C57BL mice, either treated with saline as control or with bleomycin at Days 5, 7, 12, and 28 after the injury. Whole-lung lysate was subjected to Western blots (each group, n = 5); loading control was β-actin. (B) Densitometric analyses of the Western blots in A. Results are ratio of Cav-1 to β-actin (the loading control). Each bar is the average of the ratio of all the samples (n = 5) in its group, representing mean (±SD). *P < 0.05, compared with saline control group. Bleo, bleomycin. (C) Paraffin sections of lung tissues from mice with saline or bleomycin injury at different time points were subjected to immunohistochemistry with an antibody that recognizes Cav-1 (brown; pictures were taken at 20×). Paraffin sections of lung from control (saline) or bleomycin-treated mice at different time points were stained with trichromatic (10×). (D) Semiquantitative Ashcroft score of lung sections at different time points after bleomycin injury (26).
Immunohistochemistry confirmed that Cav-1 expression was decreased in the lungs of injured mice on Day 5 and remained low at Days 7 and 12, with recovery on Day 28 (Figure 4C), similar to the pattern shown by Western blots. The transient decrease in Cav-1 expression correlated with the total lung expression of collagen 1A1 by Western blot (Figures E2A and E2B), by trichrome staining (Figure 4C), and by the semiquantitative Ashcroft scale (Figure 4D). These observations support the concept of Cav-1 suppression during the fibrotic phase of lung injury, with the potential for reversibility in association with recovery of Cav-1 expression during resolution of fibrosis.
Cav-1 Gene Expression Is Suppressed in Isolated Primary Murine Lung Fibroblasts during Active Fibrogenesis, and Its Promoter Is Associated with the Histone Mark, H3K4Me3
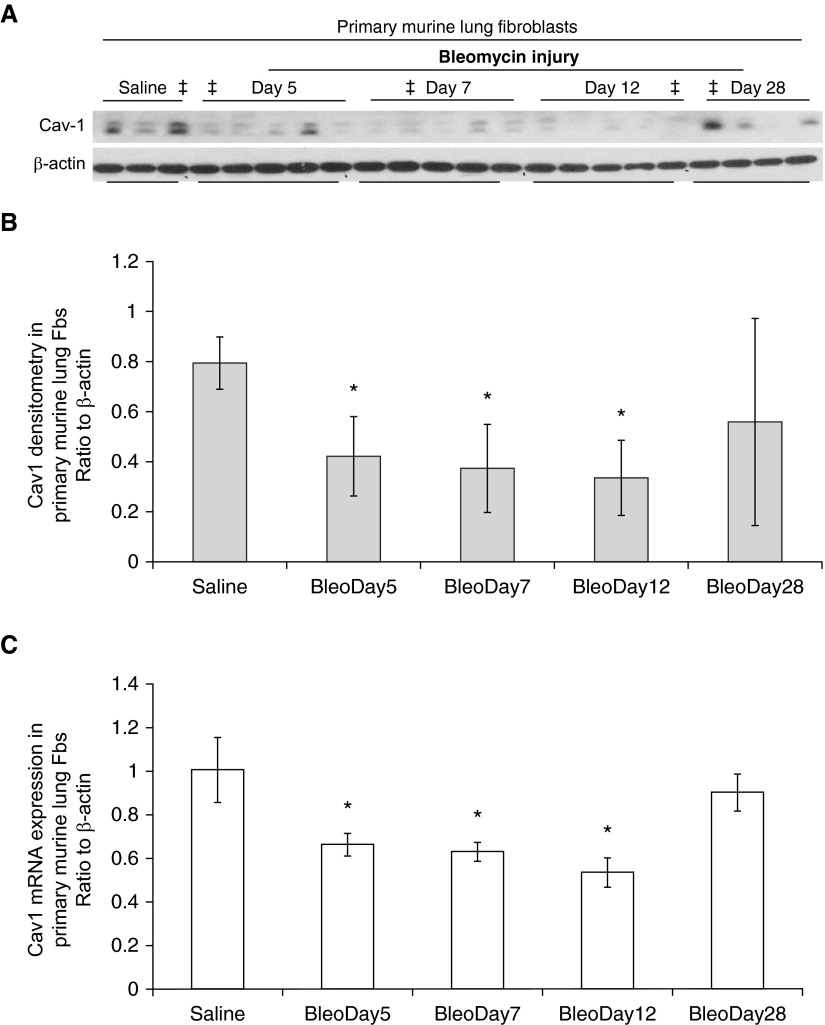
Primary murine lung fibroblasts were obtained after bleomycin injury, at the same time as we collected whole-lung homogenates. Cav-1 expression was measured in these cells (Figure 4). Despite more heterogeneity, primary murine lung fibroblasts showed down-regulation in Cav-1 paralleled that in whole-lung lysate analyses (Figures 5A and 5B). This in vitro finding in isolated fibroblasts for up to three passages in ex vivo cell culture after bleomycin injury supports a role for epigenetic regulation of this gene during the injury repair process.
Figure 5.
Expression of Cav-1 in primary murine lung fibroblasts (Fbs). Primary murine lung fibroblasts were obtained from mice treated with saline and at different time points after bleomycin injury. The fibroblasts were harvested under passage 3 for whole-cell lysate or mRNA. (A) Whole-cell lysates from primary lung fibroblasts were subjected to Western blots with Cav-1; β-actin was the loading control. (B) Densitometric analyses of the Western blots of Cav-1 in A, after normalization to β-actin. Bar graphs represent mean (±SD) from the average of the n from each group shown in A. *P < 0.05, each group was compared with saline group. (C) Primary lung fibroblasts of one cell line from each time point group were used to check the mRNA expression of Cav-1 by real-time RT-PCR, ratio to β-actin (the subjects were marked with ‡ in A, and the correlation of the same cell line’s DNA methylation status and histone association with Cav-1 promoter region are examined in Figures 6B and 6E, respectively). Bar graphs represent mean (±SD) from the average of at least three repeats of independent experiments. *P < 0.05, each group was compared with saline group.
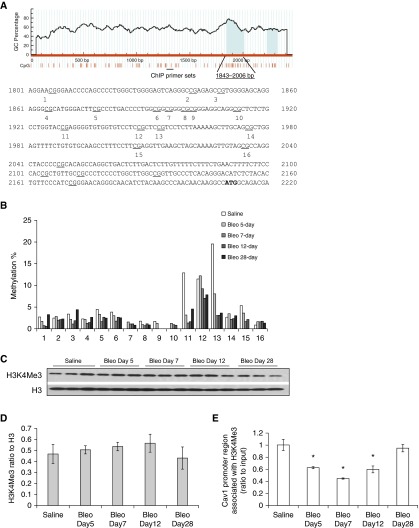
The concurrent relationships between Cav-1 mRNA expression and DNA methylation status, and the association of active histone mark H3K4Me3 with the Cav-1 promoter region from one cell line at each of the time points were examined (Figure 5A, the cell line marked with ‡). Cav-1 mRNA is significantly decreased on Days 5, 7, and 12, but recovered by Day 28 (Figure 5C). We identified CGI by MethPrimer (28) in the mouse Cav-1 gene promoter (ENSMUSG00000007655; Figure 6A). DNA methylation changes at these CpG sites of the CGI were similarly varied in the murine lung fibroblasts at different time points after bleomycin injury (Figure 6B). DNA methylation rates were low at most of these CpG sites, ranging from 0 to ∼5% with few exceptions. Some sites, such as CpG site number 12, showed relatively higher methylation percentage compared with others, but there was no statistically significant difference throughout the whole timecourse (Figure 6B and Figure E3). The relatively higher methylation rate in control mouse lung fibroblasts at CpG sites 11 and 13 (with ∼12 or ∼20%, respectively) are likely due to individual differences (see Figure E3, the average of three different murine cell lines at different time points). These findings suggest that DNA methylation is not responsible for regulation of Cav-1 gene expression level changes in the murine model of bleomycin injury.
Figure 6.
DNA methylation status and histone modification association of Cav-1 in murine lung fibroblasts at different time points after bleomycin injury. (A) Murine Cav-1 genome browser view (28) and distribution of CpG sites. The red vertical lines under the figure marked each CpG site; the approximate location of pryrosequencing regions are marked out and the 16 measured CpG sites numbered accordingly. The ChIP primer sets are marked under the vertical lines of the CpG sites. (B) Cav-1 DNA methylation percentage at the CGI near promoter region from the murine fibroblasts; the cell mRNA levels correspond to values given in Figure 5C. (C) Representative immunoblots of the global level of H3K4Me3 with nuclear extract at different time points, as marked in murine primary lung fibroblasts; total H3 was the loading control. (D) Densitometric analyses of the Western blots in C of H3K4Me3, after normalization to total H3. Bar graphs represent mean (±SD) from the average of each group (n = 3) from C. (E) The quantitative ChIP assays, showing the relative levels of Cav-1 promoter region association with H3K4Me3, the region of the PCR primers are marked out in A. The same set of cells as shown in B (DNA methylation) and Figure 5 (protein and mRNA levels expression). The results were normalized to input DNA and expressed as fold changes relative to saline control. Bar graphs represent mean (±SD) from the average of at least three independent experiments; *P < 0.05, each group compared with saline control.
To examine the association of Cav-1 with active histone mark H3K4Me3, global H3K4Me3 levels were analyzed and then ChIP assays were performed in isolated murine lung fibroblasts at different time points after lung injury. H3K4Me3 levels in these cells did not change significantly (Figures 6C and 6D). However, the binding of H3K4Me3 with the Cav-1 promoter region was significantly diminished during the time before and around the peak of fibrosis (Days 5, 7, and 12 after bleomycin injury), whereas the binding recovered during the resolution phase, on Day 28 after injury (Figure 6E). These data support a role for histone modifications involving H3K4Me3 in the epigenetic control of Cav-1 gene expression in primary murine lung fibroblasts after lung injury.
Discussion
This study investigated epigenetic changes that regulate Cav-1 gene expression in lung fibroblasts after injury. We confirmed that Cav-1 expression is down-regulated in the majority of IPF lung fibroblasts, and the suppression of Cav-1 in response to TGF-β1 (5, 13). Our data show that low constitutive expression of Cav-1 in IPF lung fibroblasts, as well as down-regulation of this protein by TGF-β1, is due to Cav-1 gene silencing by diminished binding of the active histone mark, H3K4Me3, with its promoter. Moreover, our data indicate that this regulatory process does not appear to be due to changes in DNA methylation. These findings were replicated in murine lung fibroblasts during the active (versus resolving) phase of fibrosis after bleomycin-induced lung injury. These data demonstrate a critical role for histone modifications in Cav-1 epigenetic silencing during active fibrosis in an experimental animal model and among ex vivo IPF lung fibroblasts.
CGIs are CpG-rich regions in vertebrate genomes (33). CGIs are generally associated with the promoter region and characterized by a transcriptionally permissive chromatin state (34). Hypermethylation of CGI promoters usually result in stable transcriptional repression; in addition, the location of methylation is critical to gene transcription (35). For instance, DNA methylation can block transcriptional initiation when it is in the immediate vicinity of the transcriptional start site, but when DNA methylation is within the gene body, then transcription elongation is stimulated (33). Recent studies have shown that DNA methylation of non-CGI promoter regions can also silence genes (36). Differential DNA methylation at CGI shores, which are up to 2 kb away from the CGI, and represents sites of relatively low CpG density (34, 36), has been shown to correlate well with gene expression in cancers (34, 37). Alterations of DNA methylation are known to be involved in IPF (38); however, the relationships of specific gene methylation patterns to IPF pathogenesis are not well understood. We investigated whether the Cav-1 DNA methylation status at the promoter region of CGI, as well as in CGI shores, is associated with Cav-1 expression in lung fibroblasts in the context of a fibrogenic stimulus.
Our data demonstrate that DNA methylation level is low at CGIs and remains at similar levels, despite the constitutively low expression of Cav-1 in IPF fibroblasts in comparison to non-IPF control fibroblasts, after TGF-β1 treatment of normal lung fibroblasts, and in response to bleomycin lung injury. Although the DNA methylation percentage is highest at the CGI 5′ shore of the Cav-1 promoter region, no obvious changes were observed at either the 5′ or 3′ CGI shores, despite the observed changes in Cav-1 expression. This is consistent with a previous study of Cav-1 methylation status in breast cancer cells (16). However, in that study, the Cav-1 CGI shores (both 5′ and 3′) were found to be associated with Cav-1 expression (16). It should be noted that technical limitations prevented us from examining all CpG sites within the CGI and CGI shores. The discrepant findings may also be due to the use of different cell types and diseases (breast cancer cells versus lung fibroblasts). Nonetheless, our data provide support to the notion that the down-regulation of Cav-1 in fibrogenic lung fibroblasts is not likely primarily regulated by DNA methylation.
We used a murine model of bleomycin-induced lung fibrosis (25, 39) to corroborate findings in our in vitro cell models. Down-regulation of Cav-1 was similar in lung fibroblasts after lung injury in both in vitro and in vivo models. The mediator(s) responsible for the down-regulation of Cav-1 in the animal model remains unclear. Several growth factors/cytokines, including TGF-β1, known to down-regulate Cav-1 are induced during bleomycin lung fibrosis (40). TGF-β1 mRNA and protein expression are increased in myofibroblasts as early as Day 3 after bleomycin-induced pulmonary fibrosis in the rat (41).
The mouse Cav-1 gene sequence is different from that of humans, particularly at the promoter region of CGIs. It is possible that the murine Cav-1 gene expression may be regulated by DNA methylation at the CGI shores; testing this would require techniques other than pyrosequencing to cover longer sequences of DNA.
Recent studies suggest that DNA methylation is probably not a primary mechanism for gene silencing (42, 43); instead, rapid changes in chromatin structure are regulated by histone modifications. We observed the enrichment of active histone mark, H3K4Me3, with Cav-1 promoter region in the cells expressing high levels of Cav-1. Our data indicate that, after injury or TGF-β1 treatment, regardless of the global level of H3K4Me3, the specific association of this active mark with the Cav-1 promoter changes. We do not exclude the possibility that other histone marks may also participate in the epigenetic regulation of Cav-1, including possible posttranslation modifications. For example, in MCF-7 breast cancer cells with low Cav-1 expression, the Cav-1 gene was enriched with H3K4Me2 at the promoter region (16); H3K4Me2 is usually present on poised inactive genes, whereas H3K4Me3 is concomitant with active transcription (44). In another study of colon cancer cells, it was reported that Cav-1 is up-regulated by prolonged hyperacetylation of the promoter region and enriched for histone H3 and H4 acetylation (17). We observed a general correlation between H3K9Ac and Cav-1 expression in human lung fibroblasts; however, these data were not replicated in the animal model of lung fibrosis (data not shown). The association of H3K4Me3 with Cav-1 is the most consistent in the models we tested. Additional studies are required to examine the association of other histone marks with Cav-1 gene expression. The relationship between H3K4Me3 levels and gene expression is complex, and is influenced by several factors that include other histone modifications and DNA methylation (45). The global H3K4Me3 levels should not be used to predict Cav-1 expression in our model.
In addition to DNA methylation and histone modifications, micro-RNAs may also participate in epigenetic regulation of Cav-1 gene expression (46–49). It is possible that micro-RNAs may coordinately regulate Cav-1 in concert with H3K4Me3 histone modification.
In summary, we show that Cav-1 gene expression is associated with the H3K4Me3 histone mark in lung fibroblasts from patients with IPF and in response to TGF-β1, as well as in murine lung fibroblasts after fibrogenic lung injury; in contrast, DNA methylation was not identified to be a primary mechanism for this effect. Based on the relatively small sample size and the potential for heterogeneity, it is important to recognize the influence of multiple factors, such as age, sex, and smoking status in regulating Cav-1 levels. Together, these findings support a role for therapeutic strategies that target histone modifications in preference to DNA methylation to augment Cav-1 expression in diseases characterized by pathologic suppression of this gene.
Footnotes
This work was supported by American Thoracic Society Foundation/Coalition for Pulmonary Fibrosis and the Pulmonary Fibrosis Foundation Research Program 2011 (Y.Y.S.), and by National Institutes of Health grants HL127203 (S.K.H.) and P01 HL114470 and VA BX003056 (V.J.T.).
Author Contributions: Y.Y.S. and V.J.T. conceived, designed, and coordinated the study, analyzed the data, and wrote the manuscript; H.L. and A.M.S. performed the experiments; S.R.D. and S.K.H analyzed the data and assisted with manuscript preparation.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2016-0034OC on August 25, 2016
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Quest AF, Gutierrez-Pajares JL, Torres VA. Caveolin-1: an ambiguous partner in cell signalling and cancer. J Cell Mol Med. 2008;12:1130–1150. doi: 10.1111/j.1582-4934.2008.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parat MO. The biology of caveolae: achievements and perspectives. Int Rev Cell Mol Biol. 2009;273:117–162. doi: 10.1016/S1937-6448(08)01804-2. [DOI] [PubMed] [Google Scholar]
- 3.Gvaramia D, Blaauboer ME, Hanemaaijer R, Everts V. Role of caveolin-1 in fibrotic diseases. Matrix Biol. 2013;32:307–315. doi: 10.1016/j.matbio.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 4.Thannickal VJ, Toews GB, White ES, Lynch JP, III, Martinez FJ. Mechanisms of pulmonary fibrosis. Annu Rev Med. 2004;55:395–417. doi: 10.1146/annurev.med.55.091902.103810. [DOI] [PubMed] [Google Scholar]
- 5.Wang XM, Zhang Y, Kim HP, Zhou Z, Feghali-Bostwick CA, Liu F, Ifedigbo E, Xu X, Oury TD, Kaminski N, et al. Caveolin-1: a critical regulator of lung fibrosis in idiopathic pulmonary fibrosis. J Exp Med. 2006;203:2895–2906. doi: 10.1084/jem.20061536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tourkina E, Richard M, Gööz P, Bonner M, Pannu J, Harley R, Bernatchez PN, Sessa WC, Silver RM, Hoffman S. Antifibrotic properties of caveolin-1 scaffolding domain in vitro and in vivo. Am J Physiol Lung Cell Mol Physiol. 2008;294:L843–L861. doi: 10.1152/ajplung.00295.2007. [DOI] [PubMed] [Google Scholar]
- 7.Miyasato SK, Loeffler J, Shohet R, Zhang J, Lindsey M, Le Saux CJ. Caveolin-1 modulates TGF-β1 signaling in cardiac remodeling. Matrix Biol. 2011;30:318–329. doi: 10.1016/j.matbio.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang GY, Yu Q, Cheng T, Liao T, Nie CL, Wang AY, Zheng X, Xie XG, Albers AE, Gao WY. Role of caveolin-1 in the pathogenesis of tissue fibrosis by keloid-derived fibroblasts in vitro. Br J Dermatol. 2011;164:623–627. doi: 10.1111/j.1365-2133.2010.10111.x. [DOI] [PubMed] [Google Scholar]
- 9.Drab M, Verkade P, Elger M, Kasper M, Lohn M, Lauterbach B, Menne J, Lindschau C, Mende F, Luft FC, et al. Loss of caveolae, vascular dysfunction, and pulmonary defects in caveolin-1 gene-disrupted mice. Science. 2001;293:2449–2452. doi: 10.1126/science.1062688. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, Maciejewski BS, Drouillard D, Santos M, Hokenson MA, Hawwa RL, Huang Z, Sanchez-Esteban J. A role for caveolin-1 in mechanotransduction of fetal type II epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2010;298:L775–L783. doi: 10.1152/ajplung.00327.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darby IA, Hewitson TD. Fibroblast differentiation in wound healing and fibrosis. Int Rev Cytol. 2007;257:143–179. doi: 10.1016/S0074-7696(07)57004-X. [DOI] [PubMed] [Google Scholar]
- 12.Horowitz JC, Lee DY, Waghray M, Keshamouni VG, Thomas PE, Zhang H, Cui Z, Thannickal VJ. Activation of the pro-survival phosphatidylinositol 3-kinase/AKT pathway by transforming growth factor-beta1 in mesenchymal cells is mediated by p38 MAPK–dependent induction of an autocrine growth factor. J Biol Chem. 2004;279:1359–1367. doi: 10.1074/jbc.M306248200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanders YY, Cui Z, Le Saux CJ, Horowitz JC, Rangarajan S, Kurundkar A, Antony VB, Thannickal VJ. SMAD-independent down-regulation of caveolin-1 by TGF-β: effects on proliferation and survival of myofibroblasts. PLoS One. 2015;10:e0116995. doi: 10.1371/journal.pone.0116995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xia H, Khalil W, Kahm J, Jessurun J, Kleidon J, Henke CA. Pathologic caveolin-1 regulation of PTEN in idiopathic pulmonary fibrosis. Am J Pathol. 2010;176:2626–2637. doi: 10.2353/ajpath.2010.091117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cerezo A, Guadamillas MC, Goetz JG, Sánchez-Perales S, Klein E, Assoian RK, del Pozo MA. The absence of caveolin-1 increases proliferation and anchorage-independent growth by a Rac-dependent, Erk-independent mechanism. Mol Cell Biol. 2009;29:5046–5059. doi: 10.1128/MCB.00315-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rao X, Evans J, Chae H, Pilrose J, Kim S, Yan P, Huang RL, Lai HC, Lin H, Liu Y, et al. CpG island shore methylation regulates caveolin-1 expression in breast cancer. Oncogene. 2013;32:4519–4528. doi: 10.1038/onc.2012.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dasgupta N, Kumar Thakur B, Ta A, Das S. Caveolin-1 is transcribed from a hypermethylated promoter to mediate colonocyte differentiation and apoptosis. Exp Cell Res. 2015;334:323–336. doi: 10.1016/j.yexcr.2015.03.020. [DOI] [PubMed] [Google Scholar]
- 18.Wiechen K, Diatchenko L, Agoulnik A, Scharff KM, Schober H, Arlt K, Zhumabayeva B, Siebert PD, Dietel M, Schäfer R, et al. Caveolin-1 is down-regulated in human ovarian carcinoma and acts as a candidate tumor suppressor gene. Am J Pathol. 2001;159:1635–1643. doi: 10.1016/S0002-9440(10)63010-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raghu G, Collard HR, Egan JJ, Martinez FJ, Behr J, Brown KK, Colby TV, Cordier JF, Flaherty KR, Lasky JA, et al. ATS/ERS/JRS/ALAT Committee on Idiopathic Pulmonary Fibrosis. An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. Am J Respir Crit Care Med. 2011;183:788–824. doi: 10.1164/rccm.2009-040GL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanders YY, Pardo A, Selman M, Nuovo GJ, Tollefsbol TO, Siegal GP, Hagood JS. Thy-1 promoter hypermethylation: a novel epigenetic pathogenic mechanism in pulmonary fibrosis. Am J Respir Cell Mol Biol. 2008;39:610–618. doi: 10.1165/rcmb.2007-0322OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanders YY, Tollefsbol TO, Varisco BM, Hagood JS. Epigenetic regulation of thy-1 by histone deacetylase inhibitor in rat lung fibroblasts. Am J Respir Cell Mol Biol. 2011;45:16–23. doi: 10.1165/rcmb.2010-0154OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sunaga N, Miyajima K, Suzuki M, Sato M, White MA, Ramirez RD, Shay JW, Gazdar AF, Minna JD. Different roles for caveolin-1 in the development of non–small cell lung cancer versus small cell lung cancer. Cancer Res. 2004;64:4277–4285. doi: 10.1158/0008-5472.CAN-03-3941. [DOI] [PubMed] [Google Scholar]
- 23.Huang SK, Fisher AS, Scruggs AM, White ES, Hogaboam CM, Richardson BC, Peters-Golden M. Hypermethylation of PTGER2 confers prostaglandin E2 resistance in fibrotic fibroblasts from humans and mice. Am J Pathol. 2010;177:2245–2255. doi: 10.2353/ajpath.2010.100446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanders YY, Hagood JS, Liu H, Zhang W, Ambalavanan N, Thannickal VJ. Histone deacetylase inhibition promotes fibroblast apoptosis and ameliorates pulmonary fibrosis in mice. Eur Respir J. 2014;43:1448–1458. doi: 10.1183/09031936.00095113. [DOI] [PubMed] [Google Scholar]
- 25.Hecker L, Logsdon NJ, Kurundkar D, Kurundkar A, Bernard K, Hock T, Meldrum E, Sanders YY, Thannickal VJ. Reversal of persistent fibrosis in aging by targeting Nox4–Nrf2 redox imbalance. Sci Transl Med. 2014;6:231ra47. doi: 10.1126/scitranslmed.3008182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ashcroft T, Simpson JM, Timbrell V. Simple method of estimating severity of pulmonary fibrosis on a numerical scale. J Clin Pathol. 1988;41:467–470. doi: 10.1136/jcp.41.4.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DePianto DJ, Chandriani S, Abbas AR, Jia G, N’Diaye EN, Caplazi P, Kauder SE, Biswas S, Karnik SK, Ha C, et al. Heterogeneous gene expression signatures correspond to distinct lung pathologies and biomarkers of disease severity in idiopathic pulmonary fibrosis. Thorax. 2015;70:48–56. doi: 10.1136/thoraxjnl-2013-204596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 29.Ben-Porath I, Cedar H. Epigenetic crosstalk. Mol Cell. 2001;8:933–935. doi: 10.1016/s1097-2765(01)00399-9. [DOI] [PubMed] [Google Scholar]
- 30.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 31.Yamane K, Tateishi K, Klose RJ, Fang J, Fabrizio LA, Erdjument-Bromage H, Taylor-Papadimitriou J, Tempst P, Zhang Y. PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol Cell. 2007;25:801–812. doi: 10.1016/j.molcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 32.Chaudhary NI, Schnapp A, Park JE. Pharmacologic differentiation of inflammation and fibrosis in the rat bleomycin model. Am J Respir Crit Care Med. 2006;173:769–776. doi: 10.1164/rccm.200505-717OC. [DOI] [PubMed] [Google Scholar]
- 33.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 34.Illingworth RS, Bird AP. CpG islands—‘a rough guide’. FEBS Lett. 2009;583:1713–1720. doi: 10.1016/j.febslet.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 35.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 36.Han H, Cortez CC, Yang X, Nichols PW, Jones PA, Liang G. DNA methylation directly silences genes with non-CpG island promoters and establishes a nucleosome occupied promoter. Hum Mol Genet. 2011;20:4299–4310. doi: 10.1093/hmg/ddr356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanders YY, Ambalavanan N, Halloran B, Zhang X, Liu H, Crossman DK, Bray M, Zhang K, Thannickal VJ, Hagood JS. Altered DNA methylation profile in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2012;186:525–535. doi: 10.1164/rccm.201201-0077OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moeller A, Ask K, Warburton D, Gauldie J, Kolb M. The bleomycin animal model: a useful tool to investigate treatment options for idiopathic pulmonary fibrosis? Int J Biochem Cell Biol. 2008;40:362–382. doi: 10.1016/j.biocel.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moore BB, Hogaboam CM. Murine models of pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. 2008;294:L152–L160. doi: 10.1152/ajplung.00313.2007. [DOI] [PubMed] [Google Scholar]
- 41.Zhang K, Flanders KC, Phan SH. Cellular localization of transforming growth factor-beta expression in bleomycin-induced pulmonary fibrosis. Am J Pathol. 1995;147:352–361. [PMC free article] [PubMed] [Google Scholar]
- 42.Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.You JS, Kelly TK, De Carvalho DD, Taberlay PC, Liang G, Jones PA. OCT4 establishes and maintains nucleosome-depleted regions that provide additional layers of epigenetic regulation of its target genes. Proc Natl Acad Sci USA. 2011;108:14497–14502. doi: 10.1073/pnas.1111309108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sims RJ, III, Reinberg D. Histone H3 Lys 4 methylation: caught in a bind? Genes Dev. 2006;20:2779–2786. doi: 10.1101/gad.1468206. [DOI] [PubMed] [Google Scholar]
- 45.Okitsu CY, Hsieh CL. DNA methylation dictates histone H3K4 methylation. Mol Cell Biol. 2007;27:2746–2757. doi: 10.1128/MCB.02291-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang S, Liu X, Li X, Sun S, Sun F, Fan B, Zhao S. MicroRNA-124 reduces caveolar density by targeting caveolin-1 in porcine kidney epithelial PK15 cells. Mol Cell Biochem. 2013;384:213–219. doi: 10.1007/s11010-013-1800-x. [DOI] [PubMed] [Google Scholar]
- 47.Miao L, Xiong X, Lin Y, Cheng Y, Lu J, Zhang J, Cheng N. miR-203 inhibits tumor cell migration and invasion via caveolin-1 in pancreatic cancer cells. Oncol Lett. 2014;7:658–662. doi: 10.3892/ol.2014.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lino Cardenas CL, Henaoui IS, Courcot E, Roderburg C, Cauffiez C, Aubert S, Copin MC, Wallaert B, Glowacki F, Dewaeles E, et al. miR-199a-5p is upregulated during fibrogenic response to tissue injury and mediates TGFbeta-induced lung fibroblast activation by targeting caveolin-1. PLoS Genet. 2013;9:e1003291. doi: 10.1371/journal.pgen.1003291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin DH, Yue P, Pan C, Sun P, Wang WH. MicroRNA 802 stimulates ROMK channels by suppressing caveolin-1. J Am Soc Nephrol. 2011;22:1087–1098. doi: 10.1681/ASN.2010090927. [DOI] [PMC free article] [PubMed] [Google Scholar]