Abstract
RNA polymerase from mesophilic Deinococcus radiodurans displays the same cold sensitivity of promoter opening as RNA polymerase from the closely related thermophilic Thermus aquaticus. This suggests that, contrary to the accepted view, cold sensitivity of promoter opening by thermophilic RNA polymerases may not be a consequence of their thermostability.
Thermophilic enzymes are known to work at low temperatures much slower than their mesophilic counterparts. Such cold sensitivity is believed to be a penalty for their more rigid structure, which ensures high protein thermostability. The increased rigidity of thermophilic enzymes is supposed to reduce their conformational mobility at low temperatures and to block structural changes required for catalysis (5, 16). Accordingly, it is anticipated that comparative study of thermophilic enzymes may help to characterize conformational dynamics involved in catalysis.
Promoter opening by RNA polymerase (RNAP)—a crucial step of transcription involving DNA melting around the point of RNA initiation—seemed to conform to this paradigm. It was demonstrated long ago that RNAPs from moderately thermophilic Bacillus species opened promoters at higher temperatures than the RNAP from mesophilic Escherichia coli (13, 15); both the core and σ subunit were responsible for the observed cold sensitivity (12). Using chemical probes, it was shown that thermostable RNAP from Thermotoga maritima opened promoters through a series of isomerization events that were very similar to those observed with the E. coli enzyme but that occurred at higher temperatures (10). Recent studies of Thermus thermophilus (18) and Thermus aquaticus RNAPs (11) confirmed cold sensitivity of promoter opening by thermophilic RNAPs relative to that by E. coli RNAP. It was concluded that cold sensitivity of promoter opening by thermophilic RNAPs was a penalty for adaptation to high temperatures, resulting in more rigid protein structure (18). This conclusion was based, however, on a taciturn assumption that all mesophilic RNAPs were similar to E. coli RNAP, i.e., they were cold resistant. However, scrutiny of limited information available about RNAPs from mesophilic bacteria other than E. coli hinted that this may not be the case (8, 17). To clarify the issue, we compared RNAPs from T. aquaticus and Deinococcus radiodurans. T. aquaticus RNAP was chosen because X-ray structures of T. aquaticus and closely related T. thermophilus RNAPs were available. D. radiodurans RNAP was chosen because this bacterium is the closest mesophilic counterpart of T. aquaticus (7, 9). The strong phylogenetic relationship between T. aquaticus and D. radiodurans suggested that most differences between their RNAPs were likely to result from different temperature adaptations of these bacteria. Surprisingly, many characteristics ascribed previously to thermophilic RNAPs were also found in the mesophilic D. radiodurans enzyme. This demonstrated that comparison of thermophilic RNAPs with E. coli RNAP as the only representative of mesophilic enzymes indeed may lead to misleading conclusions.
We isolated E. coli and D. radiodurans cores from corresponding cells essentially as described previously (1, 3, 4). Recombinant T. aquaticus core and E. coli and T. aquaticus σ subunits were overproduced in E. coli cells containing appropriate plasmids (1, 11). The D. radiodurans rpoD gene was amplified from the genomic sequence (GenBank accession number NP_294640.1) and cloned into the pET28 plasmid. The recombinant protein was overproduced and purified by analogy with the T. aquaticus σA subunit. E. coli, T. aquaticus, and D. radiodurans holoenzymes were reconstituted by mixing core enzymes with a fivefold excess of σ subunit. We then compared promoter binding, open complex formation, elongation, and termination by these RNAPs. We found that T. aquaticus RNAP differed from E. coli RNAP in most assays used. It displayed reduced stability of promoter complexes at the optimal temperature, was more resistant to rifampin and more sensitive to streptolydigin than the E. coli enzyme, was less prone to abortive RNA synthesis, and showed specific differences in RNA termination (Table 1). As expected, D. radiodurans RNAP was thermosensitive and had a mesophilic temperature optimum. At the same time, the D. radiodurans enzyme behaved very similarly to T. aquaticus RNAP in most other transcription assays (Table 1 and data not shown).
TABLE 1.
Properties of E. coli, T. aquaticus, and D. radiodurans RNAPs
| Characteristic | RNAP
|
||
|---|---|---|---|
| E. coli | T. aquaticus | D. radio- durans | |
| Temperature optimuma (°C) | 37 | 60 | 37 |
| Residual activity (%) after heating for 10 min at 65°C | 1.5 | 74 | 2 |
| Ratio of activities (%) at 20 and 45°C | 53 | 0.25 | 0.6 |
| Open complex half-life timeb | >30 min | <20 s | <20 s |
| Sensitivity to rifampinc (μg/ml) | 0.1 | 100 | 100 |
| Sensitivity to streptolydiginc (μg/ml) | >10 | <0.1 | <0.1 |
Measured in a multiple-round transcription assay on a template containing T7 A1 promoter and λ tR2 terminator (14); the activity was quantified as the sum of runoff and terminated RNA products.
Measured in the transcription assay at the optimum temperature in the presence of 5 μg of heparin/ml.
Concentration of antibiotic required to inhibit 90% of the RNAP activity.
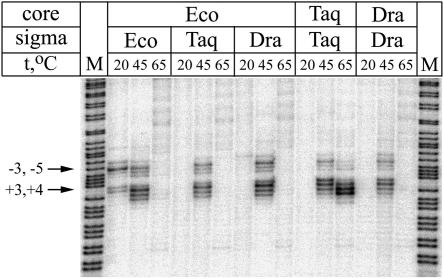
Most importantly, T. aquaticus and D. radiodurans RNAPs possessed a similar cold sensitivity of promoter opening in KMnO4 probing experiments that detect thymines in single-stranded but not in double-stranded DNA. We studied DNA melting by E. coli, T. aquaticus, and D. radiodurans RNAPs in lacUV5 promoter complexes at different temperatures using end-labeled DNA fragment as described elsewhere (2). In brief, holoenzyme of RNAP (100 nM core plus 500 nM σ) was incubated with the promoter DNA (10 nM) in transcription buffer (40 mM Tris-HCl [pH 7.9], 40 mM KCl, 10 mM MgCl2) for 10 min at the temperatures indicated and treated for 10 s with 5 mM KMnO4. The modified thymines were detected by piperidine strand cleavage (2). All three RNAPs melted DNA around the starting point of transcription at 45°C. However, only E. coli RNAP opened the promoter at 20°C (Fig. 1). At 65°C, only T. aquaticus RNAP melted the promoter, which conformed to its higher thermostability in comparison with the mesophilic RNAPs.
FIG. 1.
Permanganate footprints of E. coli (Eco), D. radiodurans (Dra), and T. aquaticus (Taq) RNAPs and hybrid holoenzymes on the nontemplate strand of lacUV5 promoter at 20, 45, and 65°C. Arrows with numbers indicate the positions of hyperreactive thymine residues relative to the starting point of transcription. M lanes are A+G cleavage markers.
To determine the role of the core and σ subunits in cold sensitivity of promoter opening, we performed KMnO4 footprinting experiments with hybrid core-σ holoenzymes. Unfortunately, the E. coli σ70 subunit did not form active holoenzymes with T. aquaticus or D. radiodurans core polymerases (data not shown) (11). However, holoenzymes containing E. coli core and T. aquaticus or D. radiodurans σA subunits were active and opened lacUV5 promoter at 45°C but not at 20°C (Fig. 1). Thus, cold sensitivity of promoter opening by T. aquaticus and D. radiodurans RNAPs is apparently determined primarily by their σ subunits. It should be emphasized that the KMnO4 probing experiments presented here assessed promoter opening under equilibrium conditions, while it remains possible that T. aquaticus and D. radiodurans RNAPs do possess some differences on intermediate stages of the open complex formation.
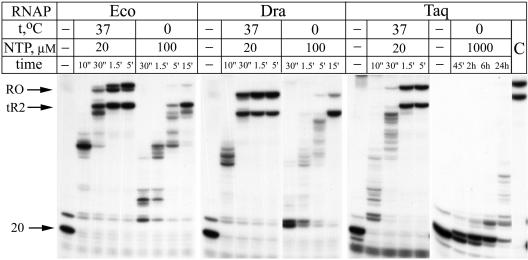
It was previously shown that thermophilic T. thermophilus and T. aquaticus RNAPs possess certain defects in RNA elongation compared with the E. coli enzyme (11, 18). To measure the rate of RNA synthesis by E. coli, T. aquaticus, and D. radiodurans RNAPs, we preformed stalled elongation complexes on a DNA template containing T7 A1 promoter and λ tR2 terminator (14) and followed RNA elongation at different temperatures. As is seen from Fig. 2, T. aquaticus and D. radiodurans enzymes reached the tR2 terminator at 37°C as fast as the E. coli enzyme. At 0°C, it took about 15 min to reach the terminator for the E. coli and D. radiodurans enzymes at 100 μM NTP, while the T. aquaticus enzyme failed to reach the terminator even after 24 h at 1 mM NTP. At lower substrate concentrations (100 μM NTP), T. aquaticus RNAP elongation was immeasurably slow (data not shown). Control experiments showed that the T. aquaticus RNAP elongation complex was not dissociated or irreversibly inactivated at 0°C and was fully active after transfer to 37°C (Fig. 2, lane C). The dramatic defect in T. aquaticus RNAP elongation observed at 0°C suggests that elongation requires conformational changes that are severely impeded at low temperature.
FIG. 2.
RNA elongation by E. coli (Eco), D. radiodurans (Dra), and T. aquaticus (Taq) RNAPs on the T7 A1 promoter fragment followed by λ tR2 terminator. The starting 20-mer RNA, runoff, and terminated transcripts are shown by arrows. Elongation was measured at different NTP concentrations at 37 and 0°C. The sample in lane C was incubated for 24 h at 0°C with 1 mM NTP and then transferred to 37°C for the additional 5 min.
Our data show that the dramatic cold sensitivity of elongation displayed by thermophilic T. aquaticus RNAP may be related to thermal adaptation. Since D. radiodurans RNAP had the same elongation rates as the E. coli enzyme at all temperatures tested, one can conclude that the cold sensitivity of RNA elongation is the only T. aquaticus RNAP “defect” which is correlated with its thermophily and thus could be regarded as the penalty for its thermostability. In contrast, other functional differences between T. aquaticus and E. coli RNAPs, including cold sensitivity of promoter opening by the thermophilic enzyme, may not be consequences of adaptation to high temperature. These differences may be a neutral character or reflect adaptation to some unknown condition common to T. aquaticus and D. radiodurans and some other bacteria. The high level of homology between T. aquaticus and D. radiodurans RNAPs implies that that the nature of the observed promoter opening defects is similar for both enzymes. At the same time, we also cannot exclude the possibility that these features arise from different sequence and structure determinants that have evolved independently in these bacteria. Ignorance on the adaptive role of the functional peculiarities of T. aquaticus and D. radiodurans RNAPs does not prevent use of these enzymes as natural mutants that have diverged from E. coli by evolutionary design. As was shown for a few other enzymes (6), such mutants may provide useful phenotypes for structural studies that cannot be obtained with standard genetic approaches.
Acknowledgments
This work was supported by NIH grant GM30717 to A.G. and by Russian Foundation for Basic Research grant 02-04-48525.
We thank Konstantin Severinov for plasmids, sharing unpublished information, and reading the manuscript. A.K. is grateful to N. Korzheva, L. Minakhin, V. Epshtein, and A. Mustaev for the continued help during the work.
REFERENCES
- 1.Borukhov, S., and A. Goldfarb. 1993. Recombinant Escherichia coli RNA polymerase: purification of individually overexpressed subunits and in vitro assembly. Protein Expr. Purif. 4:503-511. [DOI] [PubMed] [Google Scholar]
- 2.Brodolin, K., A. Mustaev, K. Severinov, and V. Nikiforov. 2000. Identification of RNA polymerase β′ subunit segment contacting the melted region of the lacUV5 promoter. J. Biol. Chem. 275:3661-3666. [DOI] [PubMed] [Google Scholar]
- 3.Burgess, R. R. 1969. A new method for the large scale purification of Escherichia coli deoxyribonucleic acid-dependent ribonucleic acid polymerase. J. Biol. Chem. 244:6160-6167. [PubMed] [Google Scholar]
- 4.Burgess, R. R., and J. J. Jendrisak. 1975. A procedure for the rapid, large-scale purification of Escherichia coli DNA-dependent RNA polymerase involving Polymin P precipitation and DNA-cellulose chromatography. Biochemistry 14:4634-4638. [DOI] [PubMed] [Google Scholar]
- 5.Fields, P. A. 2001. Protein function at thermal extremes: balancing stability and flexibility. Comp. Biochem. Physiol. A 129:417-431. [DOI] [PubMed] [Google Scholar]
- 6.Golding, G. B., and A. M. Dean. 1998. The structural basis of molecular adaptation. Mol. Biol. Evol. 15:355-369. [DOI] [PubMed] [Google Scholar]
- 7.Griffiths, E., and R. S. Gupta. 2004. Distinctive protein signatures provide molecular markers and evidence for the monophyletic nature of the Deinococcus-Thermus phylum. J. Bacteriol. 186:3097-3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Juang, Y. L., and J. D. Helmann. 1994. A promoter melting region in the primary sigma factor of Bacillus subtilis. Identification of functionally important aromatic amino acids. J. Mol. Biol. 235:1470-1488. [DOI] [PubMed] [Google Scholar]
- 9.Makarova, K. S., L. Aravind, Y. I. Wolf, R. L. Tatusov, K. W. Minton, E. V. Koonin, and M. J. Daly. 2001. Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol. Mol. Biol. Rev. 65:44-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meier, T., P. Schickor, A. Wedel, L. Cellai, and H. Heumann. 1995. In vitro transcription close to the melting point of DNA: analysis of Thermotoga maritima RNA polymerase-promoter complexes at 75 degrees C using chemical probes. Nucleic Acids Res. 23:988-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Minakhin, L., S. Nechaev, E. A. Campbell, and K. Severinov. 2001. Recombinant Thermus aquaticus RNA polymerase, a new tool for structure-based analysis of transcription. J. Bacteriol. 183:71-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nikiforov, V. G. 1971. Hybrid RNA polymerases formed from core enzymes and sigma factors of E. coli and thermophilic B. megaterium. FEBS Lett. 16:74-76. [DOI] [PubMed] [Google Scholar]
- 13.Nikiforov, V. G. 1970. Substrate dependent heterogeneity of initiation by RNA polymerase from thermophilic B. megaterium. FEBS Lett. 9:186-188. [DOI] [PubMed] [Google Scholar]
- 14.Nudler, E., M. Kashlev, V. Nikiforov, and A. Goldfarb. 1995. Coupling between transcription termination and RNA polymerase inchworming. Cell 81:351-357. [DOI] [PubMed] [Google Scholar]
- 15.Remold-O'Donnell, E., and W. Zillig. 1969. Purification and properties of DNA-dependent RNA-polymerase from Bacillus stearothermophilus. Eur. J. Biochem. 7:318-323. [DOI] [PubMed] [Google Scholar]
- 16.Vieille, C., and G. J. Zeikus. 2001. Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for thermostability. Microbiol. Mol. Biol. Rev. 65:1-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wiggs, J. L., J. W. Bush, and M. J. Chamberlin. 1979. Utilization of promoter and terminator sites on bacteriophage T7 DNA by RNA polymerases from a variety of bacterial orders. Cell 16:97-109. [DOI] [PubMed] [Google Scholar]
- 18.Xue, Y., B. P. Hogan, and D. A. Erie. 2000. Purification and initial characterization of RNA polymerase from Thermus thermophilus strain HB8. Biochemistry 39:14356-14362. [DOI] [PubMed] [Google Scholar]