Abstract
Previous studies have indicated that in adult smokers, a history of childhood pneumonia is associated with reduced lung function and chronic obstructive pulmonary disease. There have been few previous investigations using genome-wide association studies to investigate genetic predisposition to pneumonia. This study aims to identify the genetic variants associated with the development of pneumonia during childhood and over the course of the lifetime. Study subjects included current and former smokers with and without chronic obstructive pulmonary disease participating in the COPDGene Study. Pneumonia was defined by subject self-report, with childhood pneumonia categorized as having the first episode at <16 years. Genome-wide association studies for childhood pneumonia (843 cases, 9,091 control subjects) and lifetime pneumonia (3,766 cases, 5,659 control subjects) were performed separately in non-Hispanic whites and African Americans. Non-Hispanic white and African American populations were combined in the meta-analysis. Top genetic variants from childhood pneumonia were assessed in network analysis. No single-nucleotide polymorphisms reached genome-wide significance, although we identified potential regions of interest. In the childhood pneumonia analysis, this included variants in NGR1 (P = 6.3 × 10−8), PAK6 (P = 3.3 × 10−7), and near MATN1 (P = 2.8 × 10−7). In the lifetime pneumonia analysis, this included variants in LOC339862 (P = 8.7 × 10−7), RAPGEF2 (P = 8.4 × 10−7), PHACTR1 (P = 6.1 × 10−7), near PRR27 (P = 4.3 × 10−7), and near MCPH1 (P = 2.7 × 10−7). Network analysis of the genes associated with childhood pneumonia included top networks related to development, blood vessel morphogenesis, muscle contraction, WNT signaling, DNA damage, apoptosis, inflammation, and immune response (P ≤ 0.05). We have identified genes potentially associated with the risk of pneumonia. Further research will be required to confirm these associations and to determine biological mechanisms.
Clinical Trial Registration: NCT00608764
Keywords: pneumonia, genome-wide association study, pediatrics, genetic epidemiology, chronic obstructive pulmonary disease
Clinical Relevance
To the best of our knowledge, this study is the first to investigate the genetic factors that may be associated with pneumonia during childhood and across the lifetime. It may help identify children at risk of lung disease such as chronic obstructive pulmonary disease later in life. This could ultimately direct us toward a subtype of chronic obstructive pulmonary disease with early childhood origins and could help better prognosticate disease risk and treatment response.
Pneumonia is a common pediatric diagnosis, especially in young children, that poses a significant risk of respiratory disease later in life (1, 2). Pneumonia is less common in adults, although smokers are a subpopulation known to experience increased rates of pneumonia (3–5). Our previous investigations have demonstrated that pneumonia in childhood is a risk factor for chronic obstructive pulmonary disease (COPD), reduced lung function, and airway disease on chest computed tomography scans in adult smokers (6). This variability in the prevalence of childhood pneumonia and its association with an increased risk of lung disease later in life suggest some underlying genetic susceptibility.
Few previous investigations have used genome-wide association studies (GWAS) to investigate the genetic predisposition to pneumonia (7). The objective of this study was to identify the genetic susceptibility loci involved in the development of childhood pneumonia in adult smokers from the COPDGene study population. The primary analysis used GWAS to assess genetic associations with childhood pneumonia. We focused on childhood pneumonia because it may have an increased likelihood of being driven by genetic factors, as compared with pneumonia in adult smokers, which may be driven more by environmental factors such as smoking, an exposure known to be associated with pneumonia development (5). A secondary analysis used GWAS to assess the history of lifetime pneumonia to identify potential common genes and pathways influencing both childhood and adult pneumonias.
We hypothesized that this GWAS would identify susceptibility loci related to both childhood and lifetime pneumonia. Furthermore, we postulated that these loci were likely to show evidence of associations with genetic variants related to lung function, lung development, immune response, COPD, and asthma. Some of the results of these studies have been reported previously in the form of an abstract (8).
Materials and Methods
Subjects
We evaluated 10,192 current and former U.S. smokers with and without COPD from the COPDGene Study. The subjects were 45–80 years old, of non-Hispanic white (NHW) or African American (AA) race, with at least a 10 pack-year smoking history. Subjects were excluded for a history of lung disease other than COPD or asthma. COPDGene was approved by institutional review boards at each of the 21 clinical sites (9). All participants provided written informed consent. Study protocol, enrollment criteria, and data collection forms have been described previously and are available at www.copdgene.org (9, 10).
Data Collection
Pneumonia history was collected from responses to a modified American Thoracic Society Respiratory Epidemiology Questionnaire (10, 11). Genotyping was performed on DNA extracted from blood samples using HumanOmniExpress (Illumina, San Diego, CA). Standard quality control measures were completed on DNA and single nucleotide polymorphism (SNP) data, as described previously (9, 12, 13). Additional genotypes were imputed from 1,000 Genomes Phase I v3 reference panels (hg19) using minimac and MaCH (13–16). COPDGene datasets are available publicly (dbGaP accession number phs000179.v1.p1).
Case Identification
Pneumonia was defined by subject self-report, as described previously (6). The subjects were classified as having childhood pneumonia if their first episode was at <16 years of age or during childhood, and lifetime pneumonia if they had ever had pneumonia. Additional subjects were excluded from lifetime pneumonia analysis for not knowing whether they had history of pneumonia.
Statistical Analysis
Four independent GWAS were performed in subjects with childhood pneumonia and lifetime pneumonia using PLINK 1.90, with initial evaluations completed separately in NHW and AA populations (17). A logistic regression of SNPs on the basis of case-control status was performed, adjusted for sex and genetic ancestry (12). Lambda (λ) was calculated to assess the degree of genomic inflation after principal component adjustment (18). Genome-wide significance for SNPs was defined as P ≤ 5 × 10−8 (19, 20). Results from NHW and AA populations were combined in separate metaanalyses using a fixed-effect model weighted by inverse variance (17, 21). Sensitivity analyses were run, adjusting for pack-years of smoking and assessing subjects with multiple lifetime pneumonias.
In the individual population-based GWAS, SNPs with minor allele frequency (MAF) ≥5% were included for further analysis. Childhood pneumonia GWAS included 5,870,826 NHW SNPs and 8,151,810 in AAs. Lifetime pneumonia GWAS included 5,870,929 NHW SNPs and 8,150,616 in AAs. For meta-analysis, SNPs with MAF ≥1% in either population were included: 6,950,410 SNPs for childhood pneumonia and 6,950,930 for lifetime pneumonia. A lower MAF cutoff was used for meta-analysis to account for the more modest correlation of variants between ancestries (21, 22).
In single SNP analyses, annotation of variants to genes was performed using National Center for Biotechnology Information databases dbSNP/Gene, UCSC Genome Browser, and LocusZoom (23–26). For gene-based analysis, SNPs were mapped to genes with VEGAS2 using the Monte Carlo approach with simulations from a multivariate normal to estimate the null distribution and to generate gene-based P values (27–29). VEGAS2 software performs adjustment for multiple testing using a Bonferroni-corrected threshold of P < 2.0 × 10−6 (∼25,000 genes) (27, 28). The resulting VEGAS2 genes from childhood pneumonia analysis with P < 0.01 were included in the network analysis using Metacore (see online supplement) (30–32).
Results
The COPDGene Study consists of 10,192 current and former smokers. Subjects with both genotype and phenotype data available were included in GWAS analysis (see Figures E1 and E2 and Table E1 in the online supplement). GWAS QQ and Manhattan plots can be viewed in Figures E3–E8; the lambda values ranged from 0.89 to 1.01. No SNPs in either childhood pneumonia or lifetime pneumonia GWAS reached genome-wide significance (P ≤ 5 × 10−8). However, regions of interest were identified at near genome-wide significance, with SNPs at P < 1 × 10−6 (Tables 1 and 2). Top SNPs from the pneumonia GWAS were examined for overlap with known COPD variants; there was a nominal association of CHRNA3, cholinergic receptor nicotinic α 3 subunit, with lifetime pneumonia, which was not surprising in this cohort of smokers (13).
Table 1.
Top GWAS Variants Associated with Childhood and Lifetime Pneumonia*: Individual GWAS
| Individual GWAS | SNP | Nearest Gene | Locus | Rsq | Risk Allele | Frequency (%) | OR | P Value |
|---|---|---|---|---|---|---|---|---|
| NHW childhood pneumonia (n = 685 cases, 5,967 control subjects) | rs16833920 | MATN1† | 1p35.2 | 0.99 | G | 64.82 | 1.38 | 2.80E-07 |
| NHW lifetime pneumonia (n = 2,284 cases, 3,422 control subjects) | rs55762539 | MCPH1† | 8p23.1 | 0.78 | C | 42.45 | 1.22 | 8.28E-07 |
| AA lifetime pneumonia (n = 882 cases, 2,237 control subjects) | rs1107268 | LOC339862 | 3p24.3 | 0.95 | G | 25.32 | 1.38 | 8.67E-07 |
| rs140541031 | RAPGEF2 | 4q32.1 | 0.81 | A | 17.55 | 1.47 | 8.40E-07 | |
| rs34476983 | PHACTR1 | 6p24.1 | 0.86 | C | 87.12 | 1.66 | 6.06E-07 |
Definition of abbreviations: AA, African American; GWAS, genome-wide association study; NHW, non-Hispanic white; OR, odds ratio; Rsq, estimation of imputation quality; SNP, single-nucleotide polymorphism.
Includes the top SNP from each region where GWAS P value was ≤1 × 10−6.
SNP is near these genes, not in the genes.
Table 2.
Top GWAS Variants Associated with Childhood and Lifetime Pneumonia*: GWAS Meta-Analysis
| GWAS Meta-Analysis | SNP | Nearest Gene | Locus | NHW Rsq | AA Rsq | Risk Allele | NHW FRQ (%) | AA FRQ (%) | OR | P Value | I2 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Childhood pneumonia (n = 843 cases, 9,091 control subjects) | rs188808012 | NRG1 | 8p12 | 0.95 | 0.85 | T | 1.48 | 1.01 | 2.46 | 6.32E-08 | 65.44 |
| rs77554123 | PAK6 | 15q14.1 | 0.99 | 0.51 | A | 5.16 | 4.66 | 1.70 | 3.28E-07 | 37.09 | |
| Lifetime pneumonia (n = 3,766 cases, 5,659 control subjects) | rs56373127 | PRR27† | 4q13.3 | 0.93 | 0.93 | G | 41.59 | 25.29 | 1.18 | 4.34E-07 | 0 |
| rs11136992 | MCPH1† | 8p23.1 | 0.72 | 0.74 | T | 40.34 | 37.75 | 1.21 | 2.71E-07 | 15.36 |
Definition of abbreviations: AA, African American; FRQ, frequency; GWAS, genome-wide association study; I2, heterogeneity index; NHW, non-Hispanic white; OR, odds ratio; Rsq, estimation of imputation quality; SNP, single-nucleotide polymorphism.
Includes the top SNP from each region where GWAS P value was ≤1 × 10−6.
SNP is near these genes, not in the genes.
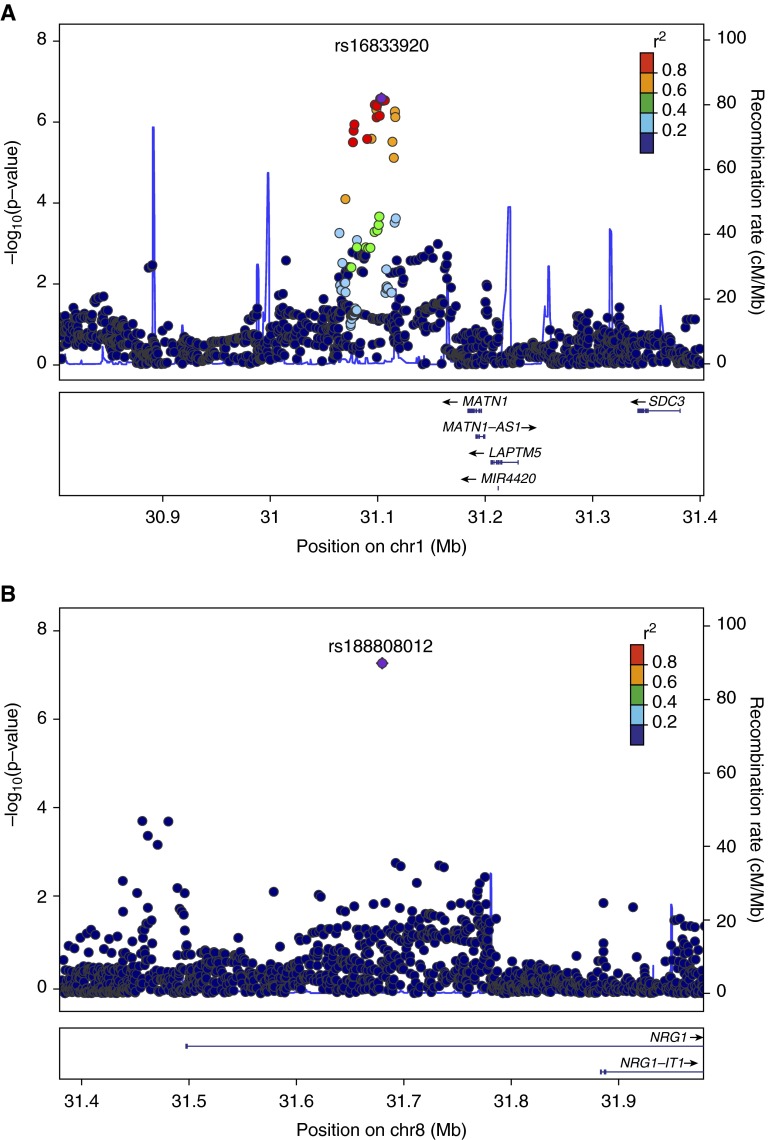
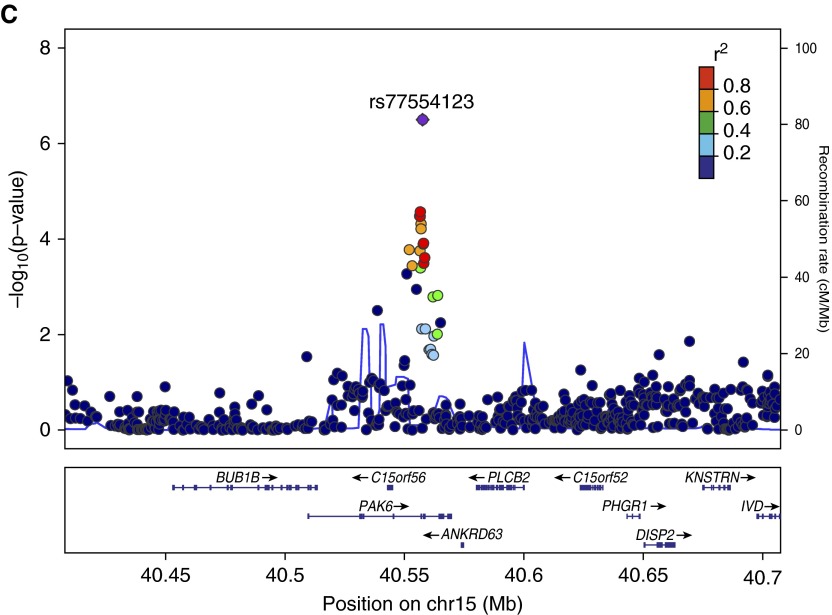
Childhood pneumonia GWAS included 6,652 NHW subjects with a case prevalence of 10.3% and 3,282 AA subjects with a prevalence of 4.8% (Figure E1 and Table E1). NHW analysis identified a potential region of interest near MATN1, matrilin 1, cartilage matrix protein (Tables 1 and 2 and Figure 1). The AA GWAS had no variants identified with P < 1.0 × 10−6. Meta-analysis on the combined NHW and AA populations included 9,934 subjects with a childhood pneumonia case prevalence of 8.5%. Meta-analysis identified variants of interest approaching genome-wide significance in NGR1, neuregulin 1 and PAK6, p21 protein (Cdc42/Rac)-activated kinase 6.
Figure 1.
Locus plots for top childhood pneumonia variants (Genome build hg19, linkage disequilibrium population 1,000 Genomes Nov 2012 EUR). Regional LocusZoom plots from (A) non-Hispanic whites, near MATN1, 1p35.2 (rs16833920), (B) meta-analysis NRG1, 8p12 (rs188808012), and (C) meta-analysis PAK6, 15q14 (rs77554123). Index single-nucleotide polymorphisms (SNPs) in purple and regional SNPs plotted in colors represent their degree of linkage disequilibrium with the index SNP, as measured by r2, the squared coefficient of correlation. The solid blue lines show the recombination rates. chr, chromosome.
Lifetime pneumonia GWAS included 6,306 NHW subjects with a case prevalence of 45.7% and 3,119 AA subjects with a prevalence of 28.3% (Figure E2 and Table E1). NHW analysis identified a potential region of interest near MCPH1, microcephalin 1 (Tables 1 and 2 and Figure E9). AA analysis identified variants of interest in uncharacterized LOC339862, RAPGEF2, Rap guanine nucleotide exchange factor (GEF) 2, and PHACTR1, phosphatase and actin regulator 1. Meta-analysis of the combined NHW and AA populations included 9,425 subjects with a lifetime pneumonia prevalence of 40.0%. Meta-analysis identified variants of interest approaching genome-wide significance near PRR27, proline rich 27 (also known as C4orf40), and near MCPH1.
Gene-based analysis of childhood pneumonia SNPs using VEGAS2 identified 585 genes in the NHW population and 1,141 genes in the AA population with gene-based P ≤ 0.01. Gene-based analysis of lifetime pneumonia SNPs found 732 genes in the NHW population and 1,159 genes in the AA population with gene-based P ≤ 0.01. Two genes in the childhood pneumonia analysis, RNF216P1 and TLE3, reached the Bonferroni-corrected threshold of P < 2.0 × 10−6 for significance in gene-based testing, and no genes reached this threshold in the lifetime pneumonia analysis; however, there were genes of interest with P values approaching the level of significance (Table 3) (27, 28). The top 10 genes for childhood pneumonia included EPAS1, endothelial PAS domain protein 1, ORAI1, ORAI calcium release-activated calcium modulator 1, TLE3, transducing-like enhancer of split 3, and for lifetime pneumonia, included uncharacterized LOC339862.
Table 3.
Top Genes in VEGAS2 Gene-Based Analyses of Childhood and Lifetime Pneumonia
| Childhood Pneumonia |
Lifetime Pneumonia |
|||||||
|---|---|---|---|---|---|---|---|---|
| Gene | Locus | nSNPs* | P Value | Gene | Locus | nSNPs* | P Value | |
| NHW | ANKRD63 | 15q15.1 | 209 | 5.00E-06 | PRICKLE2 | 3p14.1 | 557 | 1.00E-05 |
| C15orf56 | 15q15.1 | 177 | 1.00E-05 | PRICKLE2-AS3 | 3p14.1 | 300 | 1.20E-05 | |
| PLCB2 | 15q15 | 266 | 2.50E-05 | FAM184A | 6q22.31 | 646 | 4.20E-05 | |
| TAAR9 | 6q23.2 | 288 | 6.10E-05 | HPCA | 1p35-p34.2 | 115 | 5.00E-05 | |
| TAAR8 | 6q23.2 | 324 | 8.40E-05 | MCM9 | 6q22.31 | 365 | 5.30E-05 | |
| MORN3 | 12q24.31 | 165 | 0.000134 | TMEM54 | 1p35-p34 | 112 | 5.50E-05 | |
| STX7 | 6q23.1 | 364 | 0.000139 | HAND2-AS1 | 4q34.1 | 226 | 5.80E-05 | |
| ATXN7L1 | 7q22.3 | 805 | 0.000142 | FNDC5 | 1p35.1 | 104 | 5.90E-05 | |
| ORAI1 | 12q24.31 | 148 | 0.000158 | WFDC1 | 16q24.3 | 691 | 6.10E-05 | |
| EPAS1 | 2p21-p16 | 451 | 0.000163 | ASF1A | 6q22.31 | 238 | 7.80E-05 | |
| AA | RNF216P1 | 7p22.1 | 623 | 1.00E-06 | LARGE-AS1 | 22q12.3 | 476 | 1.60E-05 |
| TLE3 | 15q22 | 365 | 2.00E-06 | LOC339862† | 3p24.3 | 562 | 3.00E-05 | |
| RBAK | 7p22.1 | 636 | 3.00E-06 | VLDLR-AS1 | 9p24.2 | 949 | 7.70E-05 | |
| MMD2 | 7p22.1 | 792 | 4.00E-06 | NARS2 | 11q14.1 | 929 | 9.00E-05 | |
| RBAK-RBAKDN | 7p22.1 | 655 | 5.00E-06 | TMEM88B | 1p36.33 | 261 | 9.30E-05 | |
| RBAKDN | 7p22.1 | 527 | 7.00E-06 | MIR3688-1 | — | 222 | 0.000107 | |
| MIR629 | 15q23 | 217 | 7.00E-06 | FAM71E1 | 19q13.33 | 371 | 0.000113 | |
| ZNF862 | 7q36.1 | 497 | 1.70E-05 | VWA1 | 1p36.33 | 284 | 0.000114 | |
| HNRNPK | 9q21.32-q21.33 | 274 | 1.90E-05 | MIR3688-2 | — | 222 | 0.000119 | |
| HERC2P10 | 15q13.2 | 358 | 2.10E-05 | ANKRD65 | 1p36.33 | 295 | 0.000121 | |
Definition of abbreviations: AA, African Americans; GWAS, genome-wide association study; NHW, non-Hispanic whites; SNPs, single-nucleotide polymorphisms.
Number of SNPs that map to the gene.
An SNP in this gene was near genome-wide significance in the lifetime pneumonia AA GWAS (Table 1).
Network analysis of childhood pneumonia genes in Metacore, performed separately for NHW and AA populations, identified association with networks including development, blood vessel morphogenesis, muscle contraction, DNA damage, cytoskeleton, cell cycle, cell adhesion, WNT signaling, inflammation, immune response, and apoptosis (P ≤ 0.05) (Table 4). The top network in both NHW and AA populations was related to development, specifically blood vessel morphogenesis (Table 4 and Figure E10).
Table 4.
Top Networks for Childhood Pneumonia Genes
| Networks | Childhood Pneumonia Genes | Total Genes | P Value |
|---|---|---|---|
| NHW GWAS | |||
| Development | |||
| Blood vessel morphogenesis* | 13 | 228 | 0.004 |
| Muscle contraction | |||
| Muscle contraction* | 10 | 173 | 0.010 |
| Nitric oxide signaling in the cardiovascular system | 7 | 124 | 0.032 |
| DNA damage | |||
| Checkpoint | 8 | 124 | 0.011 |
| Core | 3 | 29 | 0.033 |
| Cytoskeleton | |||
| Intermediate filaments | 6 | 81 | 0.014 |
| Regulation of cytoskeleton rearrangement | 10 | 183 | 0.014 |
| Cell cycle | |||
| Meiosis | 7 | 106 | 0.015 |
| S phase | 8 | 149 | 0.030 |
| G1-S growth factor regulation | 9 | 195 | 0.050 |
| Cell adhesion | |||
| Leukocyte chemotaxis | 10 | 205 | 0.029 |
| Cell junctions | 8 | 162 | 0.045 |
| Signal transduction | |||
| WNT signaling* | 9 | 177 | 0.030 |
| Androgen receptor nuclear signaling | 7 | 126 | 0.035 |
| Cholecystokinin signaling | 6 | 106 | 0.045 |
| Inflammation | |||
| Kallikrein-kinin system | 9 | 185 | 0.038 |
| Immune response | |||
| Phagocytosis | 10 | 222 | 0.046 |
| Transport | |||
| Iron transport | 6 | 108 | 0.049 |
| Neurophysiological process | |||
| Taste signaling | 3 | 34 | 0.050 |
| AA GWAS | |||
| Development | |||
| Blood vessel morphogenesis* | 23 | 228 | 0.001 |
| Skeletal muscle development | 15 | 144 | 0.005 |
| Ossification and bone remodeling | 15 | 157 | 0.012 |
| Cartilage development | 8 | 66 | 0.016 |
| Cardiac development | |||
| Role of NADPH oxidase and ROS | 16 | 134 | 0.001 |
| BMP TGF-β signaling | 12 | 117 | 0.014 |
| Signaling: Wnt† β-catenin, Notch, VEGF, IP3, integrin | 13 | 150 | 0.037 |
| Muscle contraction | |||
| Muscle contraction* | 18 | 173 | 0.002 |
| Cell adhesion | |||
| Cadherins | 18 | 180 | 0.004 |
| Apoptosis | |||
| Apoptotic mitochondria | 9 | 77 | 0.014 |
| Regulation of metabolism | |||
| Bile acid regulation of lipid metabolism and negative FXR-dependent regulation of bile acids concentration | 8 | 71 | 0.025 |
| Transport | |||
| Calcium transport* | 16 | 192 | 0.030 |
| Reproduction | |||
| Male sex differentiation | 19 | 243 | 0.035 |
Definition of abbreviations: AA, African American; BMP, bone morphogenic protein; FXR, farnesoid X receptor; GWAS, genome-wide association study; IP3, inositol 1,4,5-trisphosphate; NADPH, nicotinamide adenine dinucleotide phosphate; NHW, non-Hispanic white; ROS, reactive oxygen species; TGF, transforming growth factor; VEGF, vascular endothelial growth factor.
Networks included are those with P ≤0.05.
Shared networks in both NHW and AA populations.
Sensitivity Analyses
To assess whether additional adjustment for pack-years would significantly affect the lifetime pneumonia GWAS results, we repeated GWAS analyses independently in NHW and AA populations with adjustment for pack-years of smoking. No SNPs reached genome-wide significance, and the resulting top SNPs were similar (Table E2).
To assess whether subjects who experience multiple pneumonias may have a stronger genetic association, we ran additional GWAS analyses independently in NHW and AA populations comparing subjects who reported more than one lifetime pneumonia with those with no history of pneumonia. No SNPs reached the level of genome-wide significance.
Discussion
We have presented results from, to the best of our knowledge, the first GWAS of all-cause pneumonia susceptibility during childhood and over the lifetime. In a meta-analysis of smokers from two ethnic groups, we have identified potential genes and chromosomal regions of interest. No SNPs reached genome-wide significance.
In young children in the United States, the prevalence of pneumonia annually is ∼3–5% (2). In this GWAS population of adult smokers, the prevalence of childhood pneumonia was 8.5%. This increased prevalence in the COPDGene population was possibly related to the fact that a significant number of these subjects had risk factors for developing childhood pneumonia, including childhood asthma and living with a smoker during childhood (6).
Variants of interest identified in the childhood pneumonia GWAS were found in NRG1, PAK6, and near MATN1, genes related to lung function, cardiovascular system growth and development, and repair processes. The top variant possibly associated with childhood pneumonia was in NRG1. NRG1 has been associated with lung function in a GWAS network analysis from the Framingham Heart Study (33). NRG1 plays a critical role in cardiovascular system growth and development, including angiogenesis, and surfactant synthesis in the fetal lung (23, 34, 35). NRG1 encodes neuregulin-1, a signaling protein that is part of the epidermal growth factor family, which is important in the coordination of epithelial repair processes (23, 26, 36). Expression of specific NRG1 isoforms in human airway epithelium has been found to induce the expression of MUC5AC and MUC5B protein, which are predominant mucins in asthma and COPD (37). NRG1 has also been found to be a contributor in the pathophysiology of acute lung injury (38). PAK6 encodes protein kinases that function in cytoskeleton rearrangement and apoptosis (23). MATN1 is a cartilage matrix protein expressed in skeletal and cartilage tissue during embryogenesis and throughout the lifespan, which is important to both development and repair processes (39).
Among the top NHW childhood pneumonia genes from the gene-based analysis were EPAS1, a key regulator gene in COPD, and ORAI1, which is involved in calcium channel conductance in the bronchial epithelium, smooth muscle physiology, and susceptibility to injury from cigarette smoke in both asthma and COPD (40–43). Among the top AA childhood pneumonia genes, TLE3 has been found in previous GWAS to be significantly associated with lung function as part of the THSD4-UACA-TLE3 locus in the Hutterites, a founder population from South Tyrol (44). TLE3 codes for a transcriptional corepressor protein that is in a family of proteins that participate in the Notch signaling pathway, which plays a role in development by affecting cell fate determination (23, 26).
Network analysis of the significant childhood pneumonia genes identified the same top network for both the NHW and the AA population, a development network related to blood vessel morphogenesis, which included EPAS1. A number of other networks related to development were present among the significant networks in the AA analysis. A network for muscle contraction was also a top finding for both the NHW and the AA population. In addition, there were shared networks in both populations for WNT signaling, which has been implicated in the pathogenesis of impaired lung function related to asthma and also to COPD (45, 46). Interestingly, out of 30 significant networks identified in the analysis, there was only one immune response network implicated, which is less than we would have predicted given that the cases were all subjects with childhood pneumonia.
Variants identified as possibly associated with lifetime pneumonia GWAS were found in RAPGEF2, PHACTR1, and near MCPH1, genes related to vascular functions, embryonic hematopoiesis, and damage response. RAPGEF2 has been found to be critical for murine embryonic hematopoiesis and to be differentially expressed in pulmonary arterial hypertension (47, 48). PHACTR1 plays a role in the reorganization and regulation of the actin cytoskeleton and appears to influence important vascular functions (49, 50). MCPH1 is a gene that encodes a DNA damage response protein (23).
Previous investigations of genetic susceptibility to pneumonia have largely been candidate gene studies. The only previous pneumonia GWAS in humans focused on sepsis caused by pneumonia and looked at the primary outcome of 28-day survival, not SNPs associated with the risk of developing pneumonia (7, 51). In this population, the combined prevalence of childhood pneumonia and childhood asthma was 1.6%. Although it would have been interesting to run GWAS looking at genetic determinants of the shared diagnoses, the power would be too limited for this analysis (Table E1).
Potential Limitations
We acknowledge that this investigation is limited by reliance on subject recall for classification of childhood pneumonia. Previous studies have shown that self-reported pneumonia diagnosis has a relatively good agreement with the medical record (52). We have demonstrated previously that recall bias did not play a significant role in the characterization of childhood pneumonia in COPDGene, and that 96.1% of subjects reporting childhood pneumonia indicated that they had pneumonia diagnosed by a medical provider (6).
The current study has limited power to detect significant variants at the strict genome-wide level. Although the GWAS meta-analysis includes 9,934 subjects, only 843 had childhood pneumonia and a significant proportion of these were in the NHW population. The childhood pneumonia meta-analysis was estimated to have 80% power to detect an SNP of MAF 0.2 with an odds ratio of 1.46 (53). GWAS can require populations of 10–20 times this size to detect a significant variants (54). Despite these limitations, there were a number of SNPs identified as approaching genome-wide significance and these may be involved in critical pathways related to lung growth and development, pulmonary function, and future respiratory disease susceptibility. To strengthen these findings, replication will be an important part of future investigations, followed by biologic validation of relevant genes.
Conclusions
Our childhood pneumonia GWAS in current and former adult smokers identified potential genes of interest related to lung growth and development, vascularization, lung function, and repair processes. Genes showing suggestive evidence of an association with childhood pneumonia have known relationships to acute lung injury, asthma, and COPD. Network analysis of childhood pneumonia genes implicated top networks related to development, particularly blood vessel morphogenesis, muscle contraction, and WNT signaling. These results could help explain a genetic predisposition for developing pneumonia during childhood and respiratory disease in adult smokers. Further exploration of the genetic susceptibility loci will be required to learn more about the pathways of disease association and to propose networks of disease susceptibility on the basis of these findings.
Acknowledgments
Acknowledgments
The authors thank the COPDGene Investigators.
COPDGene Investigators: Core Units Administrative Core: James Crapo, M.D. (PI); Edwin Silverman, M.D., Ph.D. (PI); Barry Make, M.D.; Elizabeth Regan, M.D., Ph.D. Genetic Analysis Core: Terri H. Beaty, Ph.D.; Nan Laird, Ph.D.; Christoph Lange, Ph.D.; Michael Cho, M.D.; Stephanie Santorico, Ph.D.; John Hokanson, M.P.H., Ph.D.; Dawn DeMeo, M.D., M.P.H.; Nadia Hansel, M.D., M.P.H.; Craig Hersh, M.D., M.P.H.; Peter Castaldi, M.D., M.Sc.; Merry-Lynn McDonald, Ph.D.; Emily Wan, M.D.; Megan Hardin, M.D.; Jacqueline Hetmanski, M.S.; Margaret Parker, M.S.; Marilyn Foreman, M.D.; Brian Hobbs, M.D.; Robert Busch, M.D.; Adel El-Boueiz, M.D.; Peter Castaldi, M.D.; Megan Hardin, M.D.; Dandi Qiao, Ph.D.; Elizabeth Regan, M.D.; Eitan Halper-Stromberg, Ferdouse Begum; Sungho Won; Sharon Lutz, Ph.D. Imaging Core: David A. Lynch, M.B.; Harvey O. Coxson, Ph.D.; MeiLan K. Han, M.D., M.S., M.D.; Eric A. Hoffman, Ph.D.; Stephen Humphries, M.S.; Francine L Jacobson, M.D.; Philip F. Judy, Ph.D.; Ella A. Kazerooni, M.D.; John D. Newell, Jr., M.D.; Elizabeth Regan, M.D.; James C. Ross, Ph.D.; Raul San Jose Estepar, Ph.D.; Berend C. Stoel, Ph.D.; Juerg Tschirren, Ph.D.; Eva van Rikxoort, Ph.D.; Bram van Ginneken, Ph.D.; George Washko, M.D.; Carla G. Wilson, M.S.; Mustafa Al Qaisi, M.D.; Teresa Gray; Alex Kluiber; Tanya Mann; Jered Sieren; Douglas Stinson; Joyce Schroeder, M.D.; Edwin Van Beek, M.D., Ph.D. PFT QA Core, Salt Lake City, UT: Robert Jensen, Ph.D. Data Coordinating Center and Biostatistics, National Jewish Health, Denver, CO: Douglas Everett, Ph.D.; Anna Faino, M.S.; Matt Strand, Ph.D.; Carla Wilson, M.S. Epidemiology Core, University of Colorado Anschutz Medical Campus, Aurora, CO: John E. Hokanson, M.P.H., Ph.D.; Gregory Kinney, M.P.H., Ph.D.; Sharon Lutz, Ph.D.; Kendra Young, Ph.D.; Katherine Pratte, M.S.P.H.; Lindsey Duca, M.S. COPDGene Investigators: Clinical Centers, Ann Arbor, VA: Jeffrey L. Curtis, M.D.; Carlos H. Martinez, M.D., M.P.H.; Perry G. Pernicano, M.D. Baylor College of Medicine, Houston, TX: Nicola Hanania, M.D., M.S.; Philip Alapat, M.D.; Venkata Bandi, M.D.; Mustafa Atik, M.D.; Aladin Boriek, Ph.D.; Kalpatha Guntupalli, M.D.; Elizabeth Guy, M.D.; Amit Parulekar, M.D.; Arun Nachiappan, M.D. Brigham and Women’s Hospital, Boston, MA: Dawn DeMeo, M.D., M.P.H.; Craig Hersh, M.D., M.P.H.; George Washko, M.D.; Francine Jacobson, M.D., M.P.H. Columbia University, New York, NY: R. Graham Barr, M.D., Dr.P.H.; Byron Thomashow, M.D.; John Austin, M.D.; Belinda D’Souza, M.D.; Gregory D. N. Pearson, M.D.; Anna Rozenshtein, M.D., M.P.H., FACR. Duke University Medical Center, Durham, NC: Neil MacIntyre, Jr., M.D.; Lacey Washington, M.D.; H. Page McAdams, M.D.
Health Partners Research Foundation, Minneapolis, MN: Charlene McEvoy, M.D., M.P.H.; Joseph Tashjian, M.D. Johns Hopkins University, Baltimore, MD: Robert Wise, M.D.; Nadia Hansel, M.D., M.P.H.; Robert Brown, M.D.; Karen Horton, M.D.; Nirupama Putcha, M.D., M.H.S. Los Angeles Biomedical Research Institute at Harbor UCLA Medical Center, Torrance, CA: Richard Casaburi, Ph.D., M.D.; Alessandra Adami, Ph.D.; Janos Porszasz, M.D., Ph.D.; Hans Fischer, M.D., Ph.D.; Matthew Budoff, M.D.; Harry Rossiter, Ph.D. Michael E. DeBakey, VAMC, Houston, TX: Amir Sharafkhaneh, M.D., Ph.D.; Charlie Lan, D.O. Minneapolis VA: Christine Wendt, M.D.; Brian Bell, M.D. Morehouse School of Medicine, Atlanta, GA: Marilyn Foreman, M.D., M.S.; Gloria Westney, M.D.; M.S.; Eugene Berkowitz, M.D., Ph.D. National Jewish Health, Denver, CO: Russell Bowler, M.D., Ph.D.; David Lynch, M.D. Reliant Medical Group, Worcester, MA: Richard Rosiello, M.D.; David Pace, M.D. Temple University, Philadelphia, PA: Gerard Criner, M.D.; David Ciccolella, M.D.; Francis Cordova, M.D.; Chandra Dass, M.D.; Gilbert D’Alonzo, D.O.; Parag Desai, M.D.; Michael Jacobs, Pharm.D.; Steven Kelsen, M.D., Ph.D.; Victor Kim, M.D.; A. James Mamary, M.D.; Nathaniel Marchetti, D.O.; Aditi Satti, M.D.; Kartik Shenoy, M.D.; Robert M. Steiner, M.D.; Alex Swift, M.D.; Irene Swift, M.D.; Maria Elena Vega-Sanchez, M.D. University of Alabama, Birmingham, AL: Mark Dransfield, M.D.; William Bailey, M.D.; J. Michael Wells, M.D.; Surya Bhatt, M.D.; Hrudaya Nath, M.D. University of California, San Diego, CA: Joe Ramsdell, M.D.; Paul Friedman, M.D.; Xavier Soler, M.D., Ph.D.; Andrew Yen, M.D. University of Iowa, Iowa City, IA: Alejandro Cornellas, M.D.; John Newell, Jr., M.D.; Brad Thompson, M.D. University of Michigan, Ann Arbor, MI: MeiLan Han, M.D.; Ella Kazerooni, M.D.; Carlos Martinez, M.D. University of Minnesota, Minneapolis, MN: Joanne Billings, M.D.; Tadashi Allen, M.D. University of Pittsburgh, Pittsburgh, PA: Frank Sciurba, M.D.; Divay Chandra, M.D., M.Sc.; Joel Weissfeld, M.D., M.P.H.; Carl Fuhrman, M.D.; Jessica Bon, M.D. University of Texas Health Science Center at San Antonio, San Antonio, TX: Antonio Anzueto, M.D.; Sandra Adams, M.D.; Diego Maselli-Caceres, M.D.; Mario E. Ruiz, M.D.
Footnotes
This work was supported by National Institutes of Health grants T32 HL007427 (E.K.S.), R01HL094635 (C.P.H.), R01NR013377 (C.P.H.), P01HL105339 (E.K.S.), R01HL089897 (J.D.C.), and R01HL089856 (E.K.S.). The COPDGene project is also supported by the COPD Foundation through contributions made to an Industry Advisory Board composed of AstraZeneca, Boehringer Ingelheim, Novartis, Pfizer, Siemens, Sunovion, and GlaxoSmithKline.
Neither the National Institutes of Health nor the Industry Advisory Board had a role in the study design, data collection, data analysis, interpretation of the data, writing of the report, or decision to submit the article for publication. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Heart, Lung, and Blood Institute or the National Institutes of Health.
Author Contributions: Conception and design: L.P.H., E.K.S., C.P.H.; acquisition, analysis, or interpretation of data: L.P.H., M.H.C., M.-L.N.M., J.D.C., T.H.B., E.K.S., and C.P.H.; drafting the manuscript or revising it critically for important intellectual content: L.P.H., M.H.C., M.-L.N.M., J.D.C., T.H.B., E.K.S., and C.P.H.; final approval of the version to be published: L.P.H., M.H.C., M.-L.N.M., J.D.C., T.H.B., E.K.S., and C.P.H.; accountability for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved: L.P.H., M.H.C., M.-L.N.M., J.D.C., T.H.B., E.K.S., and C.P.H.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2016-0101OC on August 10, 2016
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Edmond K, Scott S, Korczak V, Ward C, Sanderson C, Theodoratou E, Clark A, Griffiths U, Rudan I, Campbell H. Long term sequelae from childhood pneumonia; systematic review and meta-analysis. PLoS One. 2012;7:e31239. doi: 10.1371/journal.pone.0031239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kronman MP, Hersh AL, Feng R, Huang YS, Lee GE, Shah SS. Ambulatory visit rates and antibiotic prescribing for children with pneumonia, 1994-2007. Pediatrics. 2011;127:411–418. doi: 10.1542/peds.2010-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marrie TJ, Huang JQ. Epidemiology of community-acquired pneumonia in Edmonton, Alberta: an emergency department-based study. Can Respir J. 2005;12:139–142. doi: 10.1155/2005/672501. [DOI] [PubMed] [Google Scholar]
- 4.Almirall J, González CA, Balanzó X, Bolíbar I. Proportion of community-acquired pneumonia cases attributable to tobacco smoking. Chest. 1999;116:375–379. doi: 10.1378/chest.116.2.375. [DOI] [PubMed] [Google Scholar]
- 5.U.S. Department of Health and Human Services The health consequences of smoking-50 years of progress: a report of the surgeon general. Atlanta, GA: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, National Center for Chronic Disease Prevention and Health Promotion, Office on Smoking and Health2014 [Google Scholar]
- 6.Hayden LP, Hobbs BD, Cohen RT, Wise RA, Checkley W, Crapo JD, Hersh CP COPDGene Investigators. Childhood pneumonia increases risk for chronic obstructive pulmonary disease: the COPDGene study. Respir Res. 2015;16:115. doi: 10.1186/s12931-015-0273-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.NHGRI-EBIGWAS catalog. Available from: https://www.ebi.ac.uk/gwas/search?query=pneumonia
- 8.Hayden LP, Cho MH, McDonald MN, Crapo JD, Beaty TH, Silverman EK, Hersh CP COPDGene Investigators. Susceptibility to childhood pneumonia: A genome wide analysis [abstract] Am J Respir Crit Care Med. 2015;191:A3374. doi: 10.1165/rcmb.2016-0101OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Regan EA, Hokanson JE, Murphy JR, Make B, Lynch DA, Beaty TH, Curran-Everett D, Silverman EK, Crapo JD. Genetic epidemiology of COPD (COPDGene) study design. COPD. 2010;7:32–43. doi: 10.3109/15412550903499522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.COPDGenePhase I study documents. Available from: http://www.copdgene.org/phase-1-study-documents
- 11.Ferris BG. Epidemiology standardization project (American Thoracic Society) Am Rev Respir Dis. 1978;118:1–120. [PubMed] [Google Scholar]
- 12.Cho MH, Boutaoui N, Klanderman BJ, Sylvia JS, Ziniti JP, Hersh CP, DeMeo DL, Hunninghake GM, Litonjua AA, Sparrow D, et al. Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat Genet. 2010;42:200–202. doi: 10.1038/ng.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho MH, McDonald ML, Zhou X, Mattheisen M, Castaldi PJ, Hersh CP, Demeo DL, Sylvia JS, Ziniti J, Laird NM, et al. NETT Genetics, ICGN, ECLIPSE and COPDGene Investigators. Risk loci for chronic obstructive pulmonary disease: a genome-wide association study and meta-analysis. Lancet Respir Med. 2014;2:214–225. doi: 10.1016/S2213-2600(14)70002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y, Byrnes AE, Li M. To identify associations with rare variants, just WHaIT: Weighted haplotype and imputation-based tests. Am J Hum Genet. 2010;87:728–735. doi: 10.1016/j.ajhg.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu K, Wang Z, Li Q, Wacholder S, Hunter DJ, Hoover RN, Chanock S, Thomas G. Population substructure and control selection in genome-wide association studies. PLoS One. 2008;3:e2551. doi: 10.1371/journal.pone.0002551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoggart CJ, Clark TG, De Iorio M, Whittaker JC, Balding DJ. Genome-wide significance for dense SNP and resequencing data. Genet Epidemiol. 2008;32:179–185. doi: 10.1002/gepi.20292. [DOI] [PubMed] [Google Scholar]
- 20.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 21.Evangelou E, Ioannidis JP. Meta-analysis methods for genome-wide association studies and beyond. Nat Rev Genet. 2013;14:379–389. doi: 10.1038/nrg3472. [DOI] [PubMed] [Google Scholar]
- 22.Morris AP. Transethnic meta-analysis of genomewide association studies. Genet Epidemiol. 2011;35:809–822. doi: 10.1002/gepi.20630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.NCBI Resource Coordinators. Database resources of the National Center For Biotechnology Information. Nucleic Acids Res. 2015;41:D8–D20. doi: 10.1093/nar/gks1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jenuth JP. The NCBI. Publicly available tools and resources on the Web. Methods Mol Biol. 2000;132:301–312. doi: 10.1385/1-59259-192-2:301. [DOI] [PubMed] [Google Scholar]
- 27.Mishra A, Macgregor S. Vegas2: software for more flexible gene-based testing. Twin Res Hum Genet. 2015;18:86–91. doi: 10.1017/thg.2014.79. [DOI] [PubMed] [Google Scholar]
- 28.Liu JZ, McRae AF, Nyholt DR, Medland SE, Wray NR, Brown KM, Hayward NK, Montgomery GW, Visscher PM, Martin NG, et al. AMFS Investigators. A versatile gene-based test for genome-wide association studies. Am J Hum Genet. 2010;87:139–145. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petersen A, Alvarez C, DeClaire S, Tintle NL. Assessing methods for assigning SNPs to genes in gene-based tests of association using common variants. PLoS One. 2013;8:e62161. doi: 10.1371/journal.pone.0062161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye Z, Vasco DA, Carter TC, Brilliant MH, Schrodi SJ, Shukla SK. Genome wide association study of SNP-, gene-, and pathway-based approaches to identify genes influencing susceptibility to Staphylococcus aureus infections. Front Genet. 2014;5:125. doi: 10.3389/fgene.2014.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nikolsky Y, Ekins S, Nikolskaya T, Bugrim A. A novel method for generation of signature networks as biomarkers from complex high throughput data. Toxicol Lett. 2005;158:20–29. doi: 10.1016/j.toxlet.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 32.Mooney MA, Nigg JT, McWeeney SK, Wilmot B. Functional and genomic context in pathway analysis of GWAS data. Trends Genet. 2014;30:390–400. doi: 10.1016/j.tig.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao SY, Lin X, Christiani DC. Genome-wide association and network analysis of lung function in the Framingham Heart Study. Genet Epidemiol. 2014;38:572–578. doi: 10.1002/gepi.21841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dammann CE, Nielsen HC, Carraway KL., III Role of neuregulin-1β in the developing lung. Am J Respir Crit Care Med. 2003;167:1711–1716. doi: 10.1164/rccm.200205-468OC. [DOI] [PubMed] [Google Scholar]
- 35.Boucherat O, Benachi A, Chailley-Heu B, Franco-Montoya ML, Elie C, Martinovic J, Bourbon JR. Surfactant maturation is not delayed in human fetuses with diaphragmatic hernia. PLoS Med. 2007;4:e237. doi: 10.1371/journal.pmed.0040237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Boer WI, Hau CM, van Schadewijk A, Stolk J, van Krieken JH, Hiemstra PS. Expression of epidermal growth factors and their receptors in the bronchial epithelium of subjects with chronic obstructive pulmonary disease. Am J Clin Pathol. 2006;125:184–192. doi: 10.1309/W1AX-KGT7-UA37-X257. [DOI] [PubMed] [Google Scholar]
- 37.Kettle R, Simmons J, Schindler F, Jones P, Dicker T, Dubois G, Giddings J, Van Heeke G, Jones CE. Regulation of neuregulin 1β1-induced MUC5AC and MUC5B expression in human airway epithelium. Am J Respir Cell Mol Biol. 2010;42:472–481. doi: 10.1165/rcmb.2009-0018OC. [DOI] [PubMed] [Google Scholar]
- 38.Finigan JH, Faress JA, Wilkinson E, Mishra RS, Nethery DE, Wyler D, Shatat M, Ware LB, Matthay MA, Mason R, et al. Neuregulin-1-human epidermal receptor-2 signaling is a central regulator of pulmonary epithelial permeability and acute lung injury. J Biol Chem. 2011;286:10660–10670. doi: 10.1074/jbc.M110.208041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klatt AR, Paulsson M, Wagener R. Expression of matrilins during maturation of mouse skeletal tissues. Matrix Biol. 2002;21:289–296. doi: 10.1016/s0945-053x(02)00006-9. [DOI] [PubMed] [Google Scholar]
- 40.Yoo S, Takikawa S, Geraghty P, Argmann C, Campbell J, Lin L, Huang T, Tu Z, Foronjy RF, Spira A, et al. Integrative analysis of DNA methylation and gene expression data identifies EPAS1 as a key regulator of COPD. PLoS Genet. 2015;11:e1004898. doi: 10.1371/journal.pgen.1004898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Samanta K, Bakowski D, Parekh AB. Key role for store-operated Ca2+ channels in activating gene expression in human airway bronchial epithelial cells. PLoS One. 2014;9:e105586. doi: 10.1371/journal.pone.0105586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suganuma N, Ito S, Aso H, Kondo M, Sato M, Sokabe M, Hasegawa Y. STIM1 regulates platelet-derived growth factor-induced migration and Ca2+ influx in human airway smooth muscle cells. PLoS One. 2012;7:e45056. doi: 10.1371/journal.pone.0045056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wylam ME, Sathish V, VanOosten SK, Freeman M, Burkholder D, Thompson MA, Pabelick CM, Prakash YS. Mechanisms of cigarette smoke effects on human airway smooth muscle. PLoS One. 2015;10:e0128778. doi: 10.1371/journal.pone.0128778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yao TC, Du G, Han L, Sun Y, Hu D, Yang JJ, Mathias R, Roth LA, Rafaels N, Thompson EE, et al. Genome-wide association study of lung function phenotypes in a founder population. J Allergy Clin Immunol. 2014;133:248–255.e241–210. doi: 10.1016/j.jaci.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharma S, Tantisira K, Carey V, Murphy AJ, Lasky-Su J, Celedón JC, Lazarus R, Klanderman B, Rogers A, Soto-Quirós M, et al. A role for Wnt signaling genes in the pathogenesis of impaired lung function in asthma. Am J Respir Crit Care Med. 2010;181:328–336. doi: 10.1164/rccm.200907-1009OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baarsma HA, Spanjer AI, Haitsma G, Engelbertink LH, Meurs H, Jonker MR, Timens W, Postma DS, Kerstjens HA, Gosens R. Activation of WNT/β-catenin signaling in pulmonary fibroblasts by TGF-β₁ is increased in chronic obstructive pulmonary disease. PLoS One. 2011;6:e25450. doi: 10.1371/journal.pone.0025450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Satyanarayana A, Gudmundsson KO, Chen X, Coppola V, Tessarollo L, Keller JR, Hou SX. RapGEF2 is essential for embryonic hematopoiesis but dispensable for adult hematopoiesis. Blood. 2010;116:2921–2931. doi: 10.1182/blood-2010-01-262964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hemnes AR, Trammell AW, Archer SL, Rich S, Yu C, Nian H, Penner N, Funke M, Wheeler L, Robbins IM, et al. Peripheral blood signature of vasodilator-responsive pulmonary arterial hypertension. Circulation. 2015;131:401–409. [Discussion p. 409]. doi: 10.1161/CIRCULATIONAHA.114.013317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okoturo-Evans O, Dybowska A, Valsami-Jones E, Cupitt J, Gierula M, Boobis AR, Edwards RJ. Elucidation of toxicity pathways in lung epithelial cells induced by silicon dioxide nanoparticles. PLoS One. 2013;8:e72363. doi: 10.1371/journal.pone.0072363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Debette S, Kamatani Y, Metso TM, Kloss M, Chauhan G, Engelter ST, Pezzini A, Thijs V, Markus HS, Dichgans M, et al. International Stroke Genetics Consortium; CADISP group; CADISP group. Common variation in PHACTR1 is associated with susceptibility to cervical artery dissection. Nat Genet. 2015;47:78–83. doi: 10.1038/ng.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rautanen A, Mills TC, Gordon AC, Hutton P, Steffens M, Nuamah R, Chiche JD, Parks T, Chapman SJ, Davenport EE, et al. ESICM/ECCRN GenOSept Investigators. Genome-wide association study of survival from sepsis due to pneumonia: an observational cohort study. Lancet Respir Med. 2015;3:53–60. doi: 10.1016/S2213-2600(14)70290-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iversen L, Hannaford PC, Godden DJ, Price D. Do people self-reporting information about chronic respiratory disease have corroborative evidence in their general practice medical records? A study of intermethod reliability. Prim Care Respir J. 2007;16:162–168. doi: 10.3132/pcrj.2007.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gauderman WJ. Sample size requirements for association studies of gene-gene interaction. Am J Epidemiol. 2002;155:478–484. doi: 10.1093/aje/155.5.478. [DOI] [PubMed] [Google Scholar]
- 54.Wood AR, Esko T, Yang J, Vedantam S, Pers TH, Gustafsson S, Chu AY, Estrada K, Luan J, Kutalik Z, et al. Electronic Medical Records and Genomics (eMEMERGEGE) Consortium; MIGen Consortium; PAGEGE Consortium; LifeLines Cohort Study. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet. 2014;46:1173–1186. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]