Abstract.
Functional near-infrared spectroscopy (fNIRS) is a noninvasive neuroimaging technique, which uses light to measure changes in cerebral blood oxygenation through sensors placed on the surface of the scalp. We recorded concurrent fNIRS with magnetoencephalography (MEG) and functional magnetic resonance imaging (fMRI) in order to investigate the group-level correspondence of these measures with source-localized fNIRS estimates. Healthy participants took part in both a concurrent fNIRS-MEG and fNIRS-fMRI neuroimaging session during two somatosensory stimulation tasks, a blocked design median nerve localizer and parametric pulsed-pair median nerve stimulation using interpulse intervals from 100 to 500 ms. We found the spatial correlation for estimated activation patterns from the somatosensory task was , 0.57, and and the amplitude correlation was , 0.52, and for fMRI-MEG, fMRI-fNIRS oxy-hemoglobin, and fMRI-fNIRS deoxy-hemoglobin signals, respectively. Taken together, these results show good correspondence among the fMRI, fNIRS, and MEG with the great majority of the difference across modalities being driven by lower sensitivity for deeper brain sources in MEG and fNIRS. These results provide an important validation of source-localized fNIRS in the context of concurrent multimodal imaging for future studies of the relationship between physiological effects in the human brain.
Keywords: functional near-infrared spectroscopy, multimodal, functional imaging, brain, functional magnetic resonance imaging, magnetoencephalography
1. Introduction
Functional near-infrared spectroscopy (fNIRS) is a noninvasive brain imaging technique that uses visible red to near-infrared light (650 to 900 nm) to measure the spectroscopic optical properties of tissue. In particular, for fNIRS brain imaging, light is used to monitor changes in blood oxygenation in the brain through sensors placed on the surface of the scalp. Hemoglobin changes are estimated based on the differential optical absorption profiles of oxygenated and deoxygenated blood (oxy- and deoxy-hemoglobin, , and Hb, respectively). Since the introduction of this method by Jobsis,1 diffuse optical techniques have been applied to a large variety of different brain-related topics in both clinical and research applications. Several recent reviews have detailed the use of fNIRS brain imaging in the fields of child2,3 and infant4,5 research, clinical monitoring,6,7 and adult brain imaging studies.8,9
Functional NIRS can play an important role in multimodal imaging because its portability readily allows for simultaneous acquisition with larger, immobile neuroimaging methods such as functional magnetic resonance imaging (fMRI) or magnetoencephalography (MEG). In general, multimodal studies can pair modalities based on the compliment of their particular strengths and weaknesses. MEG acquires high temporal resolution images of the brain’s electrophysiological response, but is not sensitive to hemodynamic changes and is restrictive with regards to subject movement. In comparison, fNIRS allows a more portable means to measure cerebral hemodynamic changes and provides additional information about both oxygenated and deoxygenated forms of hemoglobin, but is limited its temporal resolution compared to neural measures. Multimodal brain imaging methods using fNIRS in combination with fMRI, MEG, or electroencephalography (EEG) can lead to a better understanding the physiological basis of human brain activity through investigation of the neural, metabolic, and vascular relationships in the brain. Functional NIRS uses fiber optic cables, which allow remote imaging of a participant inside either fMRI or MEG scanners. Thus, fNIRS can provide a common measurement between sessions to bridge multimodal information. Not only can these methods provide insight into both neural and vascular functionality of the brain, but also can provide independent measurements of the signal allowing for technique cross validation in future studies.
While to date, there have been numerous comparisons of concurrent fMRI and fNIRS (reviewed by Steinbrink et al.10), most of these studies have focused on the temporal correlation of these modalities. In general, a consensus of strong linear correlations between the fNIRS and particular deoxy-hemoglobin component and the fMRI-BOLD signal has been observed. However, there has been considerably less work to examine the spatial agreement of these methods beyond qualitative comparisons. In particular, the reconstruction of images of brain activity in fNIRS is an ill-poised and under-determined problem that requires advanced mathematical techniques to simulate the sensitivity (forward) measurement models of the diffusion of light in the head and preform inversion of the model to construct images. In recent years, our group has developed a number of image reconstruction methods based on cortical surface topology for improving image reconstruction of fNIRS data.11–15 The purpose of this study was to evaluate the performance of these recent image reconstruction methods applied to low-density fNIRS measurements and compare these results to fMRI and MEG source localization. Healthy volunteer participants completed two neuroimaging sessions involving concurrent fNIRS/MEG and concurrent fNIRS/fMRI during a somatosensory stimulation task. Two experimental conditions were examined. First, a 10-s median-nerve localizer task was used to examine the spatial agreement among these three modalities. The spatial location and overlap of the estimate of brain activity from all three modalities was examined in the localizer task. Second, a parametric pulsed-pair study was conducted to look at the relationships of the evoked hemodynamic and electrophysiological responses particularly with regards to nonlinarites in the neural response for interstimulus intervals between 50 to 500 ms. Specifically, the magnitude of the evoked neural and hemodynamic signals in a region-of-interest analysis for the parametric pulsed-pair response was compared.
2. Methods
2.1. Subject Population
In this study, a total of 22 subjects (15 males, 7 females; age range 21 to 45 years; all right handed) were recruited for functional imaging studies. All subjects provided written informed consent prior to study procedures. This study was approved by the University of Pittsburgh Institutional Review Board.
The study consisted of two separate scanning sessions for the MEG-fNIRS and MRI-fNIRS. The MEG-fNIRS scan was done first in all cases due to the potential loss of subjects whose head size or body dimensions precluded them from MEG scanning. A total of four subjects was excluded prior to scanning because their head would not fit inside the MEG scanner while wearing the fNIRS sensors. A further two subjects failed to yield adequate fNIRS signals after initial setup and were excluded. The remaining 16 subjects were scanned with concurrent fNIRS-MEG. Of the 16 remaining subjects, 11 were additionally scanned inside the MRI with concurrent fNIRS. Five subjects were lost to follow-up or withdrew from the study after the first visit. Of the 11 subjects that completed both the MEG and fMRI sessions, a further 4 were rejected due to excessive eye-blink or head motion artifacts in the MEG analysis. Thus, a final total of 7 subjects (4 males, 3 females) were used in the final analysis of the study having completed all aspects of the protocol.
2.2. Experimental Protocol
Unilateral median nerve stimulation was conducted using a computer controlled GRASS S88X stimulator (Natus Neurology Inc., Warwick, Rhode Island). A stimulating bar electrode was positioned on the right median nerve and the voltages were adjusted to match the motor threshold as judged by twitching of the thumb on the stimulated hand. The stimulating voltage was then set at the determined motor-threshold for the study. The median nerve stimulation had duration of with a monophasic pulse. Two experimental tasks were performed. First, a functional localizer task consisting of repeated 10-s blocks of stimulation at 4 Hz with a 10-s interblock rest period (5 min total time; 15 blocks) was administered. The purpose of this task was to provide spatial localization information to define regions-of-interest to the pulsed-pair stimulation task. Second, several 5-min scans of event-related pulsed-pair stimulation were then given. The pulsed-pair condition consisted of a pair of two median nerve pulses separated by an interstimulus interval of 50, 100, 150, 200, 300, 400, or 500 ms. The stimulus conditions were presented in randomized order with a minimum spacing of 8-s between events to avoid potential nonlinearity in the hemodynamic response based on the work by Cannestra et al.,16 which suggested a minimum of 4-s interval was required. A total of 32 pairs of stimulus events was delivered in each 5-min scan. This scan was repeated 4 to 6 times per subject depending on subject tolerance and time constraints. The experimental procedure for both the MEG-NIRS and MRI-NIRS portions of the study were identical. An MEG-compatible bar electrode obtained from Elekta Neuromag (Helsinki, Finland) was used in this study. An identical duplicate electrode was used for the MRI portion of the study, which was verified to have negligible RF-heating and no ferromagnetic characteristics.
In order to control the stimulus timing, a custom dynamically linked library (DLL) was written to digitally interface to the GRASS stimulator via a universal serial bus cable. This allowed digital control of the frequency and duration of the stimulation with microsecond precision. An experimental paradigm was written in Eprime-2 (Psychology Software Tools; Pittsburgh, Pennsylvania) to control the GRASS device via the custom DLL program to prime the stimulator with the correct pulse interval several seconds prior to stimulation. The start of each stimulus event was triggered by Eprime and the GRASS device then controlled the timing of the second pulse. In this way, the pulsed-pair stimulus was delivered with better than millisecond precision as measured by the MEG auxiliary channel. The stimulus output from the GRASS device was sent to both the fNIRS and MEG systems for recording. A current isolation device was used to limit stimulation of the subject.
2.3. Analysis Methods
A schematic overview of our complete multimodal analysis pipeline is shown in Fig. 1. A key feature of this pipeline is the use of the subject anatomy and cortical surface model generated by the FreeSurfer tools available from the Massachusetts General Hospital.17 The extracted cortical surfaces, which are registered among subjects to allow group analysis, are used in all three functional imaging modalities, which allows direct comparisons of each modality. The individual components of this pipeline are described as follows.
Fig. 1.
A schematic overview of the fNIRS/MEG/fMRI analysis pipeline (described in text). Analysis of all three modalities is performed in the space of the segmented and inflated cortical surface obtained from the FreeSurfer tools.
2.4. MEG Acquisition and Analysis
MEG data were collected using a whole-head 306-channel Elekta Neuromag® Vectorview system (Elekta Neuromag Oy, Helsinki, Finland). Concurrent electrooculogram (EOG) was also acquired. This system is composed of 102 triple-sensors consisting of a magnetometer, longitudinal gradiometer, and a latitudinal gradiometer and is contained inside a magnetically shielded room (MSR). During the acquisition, subjects were inside the MSR ( in dimension) sitting comfortably with their head positioned in a helmet containing the sensors (Fig. 2). MEG scans were sampled at 1000 Hz and subsequently down-sampled offline to 250 Hz. The MEG channels were manually inspected and obviously bad channels were removed from further analysis. The MEG signals were then bandpass filtered using cutoffs of 1 and 50 Hz to remove the 60 Hz line noise and slow drifts. Temporal spatial signal separation (tSSS) was employed to remove external magnetic field contributions in the signal recordings, which can lead to spurious signals that appear to have originated from physiological changes but are in fact artifacts external to the MEG sensors.18 Data were then averaged into trials and trials with excess eye blinks or eye movements, as assessed by the maximal difference in the EOG over the trial, were rejected. Data from each condition were subsequently averaged offline.
Fig. 2.
Simultaneous fNIRS with fMRI (left) and fNIRS with MEG (right) were recorded in two separate sessions. The center panels show the localization of the fiducial markers used to register the fNIRS probe in the MRI (top) and MEG (bottom) images.
For source localization, the cortically constrained minimum norm estimate (MNE) inverse solution was employed using the MNE™ software suite v2.7.19 Briefly, a linear inverse operator is applied to the measured signal to calculate the MNE
where represents the MEG channel data at time and is the corresponding current projected onto the cortical surface. The expression of is calculated using the norm, which yields
where is the free source orientation solution of the forward problem calculated using the boundary element method. and are the noise and source covariance matrices, respectively. The noise covariance matrix was calculated using intertrial time. defines the transformation from orthogonal unit current dipoles to measured magnetic fields and uses the boundary element method.20,21 is a weighting factor that is used to avoid the magnification of errors in the data and . We used a value of 3 for this, as is typically done in MEG analysis (MNE software user’s guide version 2.7, 2009;22). Furthermore, because cortical neurons are known to be preferentially oriented perpendicular to the cortical surface, we used a loose orientation constraint. Specifically, the components transverse to the surface were multiplied by 0.4. To compensate for the bias toward superficial currents of the MNE, a scaling factor (i.e., depth weighting) of 0.8 is applied to .23
For group-level maps, the data from each participant was morphed onto a common brain. Statistical values were assessed using a paired -statistic across the seven participants for the MEG response to the median nerve stimulation versus prestimulation baseline. The peak time was chosen individually for each subject based on the time that showed the greatest change relative to baseline averaged across all MEG sensors (; ). For the ROI analysis, ROIs were drawn at the individual subject level. The ROI corresponding to primary somatosensory cortex was determined from the response in the functional localizer scan relative to prestimulus time and applied to the pulsed-pair condition.
2.5. MRI Acquisition and Analysis
Structural and functional MRI was performed using a 3T Siemens TIM TRIO scanner using a 12-channel head coil. A structural T1-weighted image was acquired (Siemens’ MPRAGE; 1 mm isotropic; ; ). A total of 4 to 6 5 min BOLD EPI scans were performed ( 3.5 mm isotropic; ; ; ). Functional MRI analysis was performed using the AFNI and SUMA toolset from the NIH.24,25 In brief, functional EPI data were coregistered to the structural volume and motion corrected to the third TR. The slices were temporally aligned and volumes were blurred using the default 4-mm FWHM kernel. General linear model analysis based on the timing of the seven pulsed-pair and localizer-conditions was performed using the default gamma-variant canonical basis function (GAM)1 with additional motion regressor terms. Group level ANOVA was performed within AFNI.
The structural T1-weighted anatomical MRI image for each subject was processed using the FreeSurfer pipeline in order to generate the segmented anatomical models and registered cortical surface models used for fNIRS, MEG, and fMRI analysis procedures. Following registration and brain segmentation using Freesurfer, the MNE toolkit22 was used to additionally create boundary element models for the skin and outer/inner skull, which allowed construction of a 5-layered head model (skin, skull, CSF, gray matter, and white matter) for each subject which was used to generate the fNIRS forward model using the optical properties described in Fang and Boas26 for the 690- and 830-nm wavelengths. The Monte Carlo program tMCimg27 was used to simulate the optical forward model. The same boundary element and brain surface models were used to generate the MEG forward model as part of the MNE MEG analysis pipeline developed by Hamalainen et al. at the Massachusetts General Hospital.22 The extracted white matter and pial brain surfaces from FreeSurfer were used in the inverse models for both fNIRS and MEG and for the SUMA-AFNI24,25 surface analysis of the fMRI data. In this way, the estimates of brain activation from all three modalities were derived in the same cortical surface space for direct comparisons of the results.
2.6. fNIRS Acquisition
For both MRI and MEG portions of the study, fNIRS signals were recorded using a commercial TechEn CW6 (Milford, Massachusetts) continuous-wave fNIRS system. This system contains up to 16 laser diodes each at 690 and 830 nm (12 and 8 mW, respectively), which are coupled through fiber optic cables mounted onto a head cap. The head cap was constructed of Velcro and plastic materials and was custom built for this study. Due to the limitations of head-clearance inside the MEG scanner, the fNIRS cap had to be restricted to eight detectors and four source positions containing both wavelengths on only one hemisphere of the head. The same design was used for both parts of the study although dedicated identical sets of fiber optics and head caps were used to avoid magnetic contamination of the MEG. The probe extended from left frontal cortex (near 10 to 20 coordinate “F3”) to posterior partial (near 10 to 20 coordinate “P3”) with a source–detector spacing of 3.2 cm as shown in Fig. 2.
2.7. MEG-fNIRS Setup
The fNIRS cap used in this study had a profile height of 11 mm, which reduced the spacing between the MEG SQUID detectors and the subject’s head. Four head position indicator (HPI) coils were glued to the subject’s scalp using collodion gel. These were used to coregister the fNIRS and MEG sensor positions after positioning the fNIRS cap on the subject’s head. Two electrodes were positioned above and below the left eye and above the corner of the left eye and below the corner of the right eye to monitor eye blinks and saccades. Cardiac activity was recorded on an ECG channel using two electrodes that were attached just below the collarbone of the subjects. Prior to MEG scanning, a Polhemus Isotrack (Polhemus, Inc., Colchester, Vermont) positioning system (integrated with the MEG acquisition software) was used to register the HPI coils and to landmark fiducial points (nasion and preauricular points) on the subject’s head. These were then used to register both the fNIRS and MEG sensor locations to the subject’s anatomical MRI data. Based on previous work, we expect the localization error of this method to be around 3 to 5 mm.28 Additional points (up to 100) were digitized on the subject’s head and face to help facilitate coregistration of MEG data with the structural MRI’s. Immediately prior to MEG acquisitions, the HPI coils were energized and their positions localized through the MEG acquisition interface. This registration information was then extracted from the MEG data files and combined with the location of the fNIRS sensors from the isotrack data using a rigid-body transform to localize the fNIRS sensors on the registered MRI volumes from the second session.
For fNIRS recordings, an MEG-dedicated set of fiber optics (10-m long) was passed through the access port (wave-guide) in the side of the MSR. The fNIRS instrument was located next to the MEG acquisition computers. The stimulus timing from the median nerve device was sent to both the MEG and fNIRS instruments through auxiliary recording channels built into both systems. Because the fNIRS instrument was sampled at a lower rate, a 555-timer pulse-elongating circuit had to be constructed to elongate the stimulating pulse to 500 ms for recording by the fNIRS system.
2.8. MRI-fNIRS Setup
Functional NIRS collection during the MRI portion of the study was similar to the MEG collection with the exception that a MRI-dedicated set of 10-m fiber optics and head cap was used to avoid magnetic contamination to the MEG system. The CW6 instrument was positioned in the control room for the MRI scanner and the fiber optics were run through the MR waveguide into the scanner room. The fNIRS head cap used in the MRI portion of study was identical to the one used in the MEG portion of the study. The head cap contained vitamin E capsules in place of the four MEG HPI coil positions, which were used for registration of the cap from the T1-weighted MRI image for each subject. In previous work, we have estimated the registration error of this approach to be about 5 mm.14 The timing of the stimulus paradigm was triggered from the MRI scanner.
2.9. fNIRS Analysis
Analysis of fNIRS results was performed using custom MATLAB scripts. Raw 20-Hz fNIRS data were converted to optical density (absorption) at both 690 and 830 nm for all measurement pairs. The unfiltered optical density data for all measurement pairs were then analyzed using a statistical general linear model as described in Barker et al.11 This model uses an iterative estimation of an autoregressive (AR) whitening filter and weighted-least-squares regression to reduce the effects of serially correlated noise and motion artifacts in the fNIRS data. In brief, a regression model (design matrix; ) was constructed from convolution of a canonical hemodynamic response function (default AFNI GAM1 function;24) with the stimulus onsets and durations of the median nerve stimulation events. Thus, a linear regression model is constructed of the form , where is the vector of time-points for each optical measurement pair (optical density) and is the estimated weights of the regressors. The values of are first estimated from a weighted least-squares procedure, which reduces the influence of noise outliers (robustfit function in MATLAB29). A th-order AR model is then fit to the residual error of the regression fit where is estimated by an AIC criteria search. The AR coefficients are used to construct a whitening filter (), which is applied to both the left and right hand sides of the regression model (e.g., ). The regression model is again solved using robust regression methods and the procedure is iterated until convergence (usually within 2 to 3 iterations). This is repeated for each optical measurement channel and wavelength. Our previous work in developing this model, described in Barker et al.,11 had shown that this model was robust to both spike and shift types of motion artifacts in the fNIRS data. In particular, the presence of infrequent, large amplitude motion artifacts in the fNIRS data results in a heavy-tailed statistical distribution of noise, which can be addressed with robust regression methods to treat these outliers. In addition, we had shown that correction for serially correlated errors due to systemic physiology and drifts produced much more accurate estimates of model errors and type-I error rates compared to least squares estimation. Thus, this model is fairly robust to false-discoveries due to superficial physiological contamination.11 This model performs best when raw (unfiltered) data are used since removing portions of the frequency spectrum can make the prewhitening procedure ill-posed and the algorithm numerically unstable.
The outputs of the regression models (e.g., estimates of and the estimate of the covariance errors in associated with the optical density changes for each fNIRS measurement channel) were then reconstructed into estimates of brain activity. For image reconstruction, we used a random-effects cortical surface topography model, which had been previously described in Abdelnour et al.13 In this model, this fNIRS forward model (sensitivity model) is computed on the cortical surface of the brain using the extracted surfaces from the Freesurfer model described in the previous section. Of note, this same cortical surface is also used in the MEG and fMRI models via AFNI-SUMA. The registered “spherical” inflated surface for each hemisphere from the Freesurfer model is reparameterized using spherical wavelet basis set described in Abdelnour et al.14 Spherical wavelets are used to describe the image (e.g., brain activity map) on the surface of the cortex at different spatial resolutions similar to more conventional wavelet representations with the distinction that these spherical wavelets are specifically designed to model a two-dimensional topography on a closed three-dimensional surface. The spherical wavelets are derived from a recursive subdivision of an icosahedral mesh, which can easily be extracted from the existing Freesurfer cortical surfaces from the icosahedral levels 1 to 4 (2562 nodes per hemisphere with about 6.2-mm vertex spacing and face areas). Since the registered “spherical” surfaces are used in the FreeSurfer model for each subject, the spherical wavelets map on to approximately the same anatomical regions across subjects. For example, the th parameter in the model represents brain activation at the same cortical gyrus for all subjects even though that gyrus may be in a slightly different Cartesian location in the space of the pial brain surface. Of further note, this surface mapping feature of the Freesurfer models allows activations to be transferred and averaged among subjects and is used in both the AFNI-SUMA and MEG-MNE pipelines for group level models. We note that this is the same convention and mesh used in the MEG MNE reconstruction through the Freesurfer pipeline.
Because the spherical wavelet model parameterizes each individual subject’s fNIRS forward model into the same registered brain space, the models for all subjects can be concatenated into a single large random-effects inverse model and solved simultaneously for all subjects. Our random-effects group level fNIRS model was previously described in Abdelnour et al.13 In brief, this group level analysis model is similar to the group-level image reconstruction of MEG data described in Mattout el al.30 The fNIRS forward model for all individual subjects () can be concatenated into a single-inverse operator such that
In this random-effects model, each individual subject’s image is modeled as a perturbation on the group average (e.g., ). Because the fNIRS inverse model is ill-posed (meaning that there are a large number of equally plausible solutions that model the data), this random-effects approach attempts to find an estimate of the group average that is self-consistent with all the individual subjects. Our previous work13 had shown that this approach resulted in a much smaller point-spread function and higher effect sizes in group-level analysis compared to a more conventional two-step approach of reconstructing each individual subject and then averaging the results. While this approach has also been shown to be effective in MEG analysis,30 this is not yet implemented in the MEG pipeline used by MNE and thus, it is important to point out that our MEG processing still used this two-step approach.
The fNIRS random-effects inverse model was solved using a hierarchical Bayesian model as described by Abdelnour et al.13 Restricted maximum likelihood and expectation–maximization recursion was used to estimate the noise hyper-parameters of the model. The covariance of the regression coefficients from the general linear model () was used to construct the covariance prior on the measurement noise, whereas a scalar identity operator was used to model oxy- and deoxy-hemoglobin priors. Thus, a total of three hyper-parameters () are estimated in this model. A description of the algorithm is found in Abdelnour et al.13 and follows the description of the hierarchal inverse model used in the MEG and EEG image analysis in SPM.30,31
3. Results
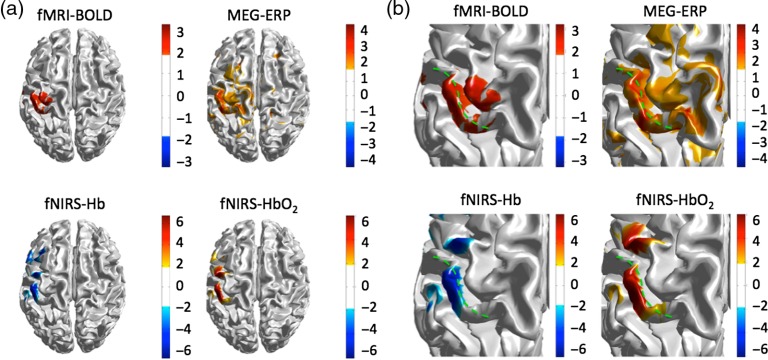
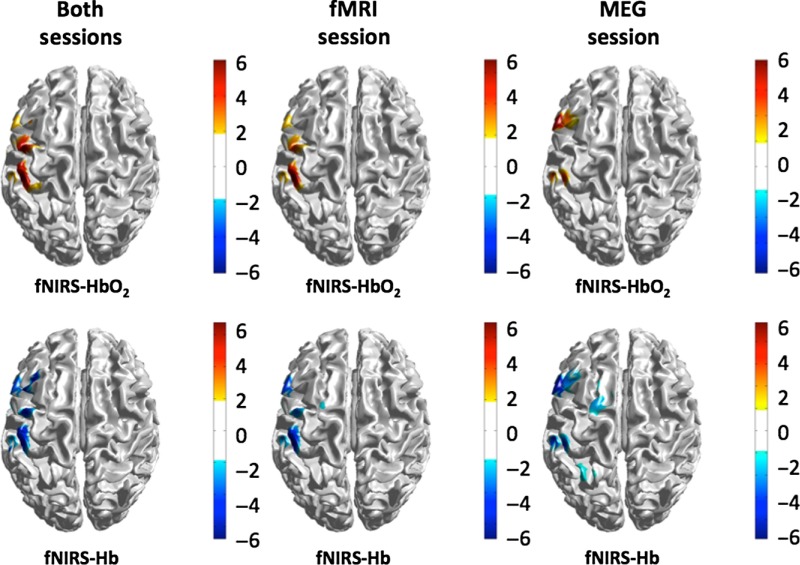
The fMRI, fNIRS, and MEG modalities all showed significant changes in brain activity during the median nerve localizer task. Figure 3 shows the group level results for the three modalities. In all modalities, the activation was located in the left somatosensory cortex as shown in Fig. 3(a). The fNIRS results show the group-level results from across both imaging sessions. A closer view of the somatosensory cortex is shown in Fig. 3(b), which shows the location of all three modalities along the primary somatosensory gyrus. Figure 4 shows the results of the fNIRS image reconstructions for the fMRI-fNIRS and MEG-fNIRS sessions independently to examine the intersession differences in the fNIRS signals. The fMRI-fNIRS and MEG-fNIRS sessions show similar regions of the left somatosensory region particularly for the oxy-hemoglobin images. The MEG-fNIRS session data showed a bit more noise for the deoxy-hemoglobin image compared to the fMRI-fNIRS session or combined data likely related to lower data quality due to compression of the fNIRS cap within the MEG sensor helmet.
Fig. 3.
Reconstructed images of brain activation to the median nerve localizer task for the fMRI, MEG, and fNIRS modalities. The -score for the localizer median nerve task is shown for each modality and thresh-held at . (a) The reconstructed activation maps for the fMRI-BOLD, MEG, and fNIRS signals (average of both the fMRI and MEG sessions) are shown. Activation was observed on the left somatosensory (S1) region of the brain. (b) The same data are shown with a zoomed view of the somatosensory (postcentral) gyrus, which is outlined in green dashed line for each image.
Fig. 4.
The reconstructed fNIRS images of oxy- and deoxy-hemoglobin are shown for the data collected in the concurrent fMRI and MEG sessions and for the combined analysis of both data sessions. The -score for the localizer median nerve task is shown.
Based on the image reconstructions for the three modalities, the center-of-mass of the activation patterns for all images are given in Table 1. The location of the center-of-mass of each modality is shown in Fig. 5(a). The displacement between the centers (Table 2) of the fMRI-BOLD signal was 26, 20, and 19 mm from the MEG, fNIRS oxy-hemoglobin, and fNIRS deoxy-hemoglobin cluster centers, respectively, when examined over the entire brain. This result was consistent with the recent fMRI-MEG study by Stevenson et al. using similar median nerve tasks that showed a displacement between the dipole MEG model and the BOLD signal of .32 The N20 component was used to localize the MEG response from the localizer median nerve task. The center of the MEG was 27 and 29 mm from the oxy- and deoxy-hemoglobin centers. Finally, the center of the oxy-hemoglobin cluster was 3 mm from the center of the deoxy-hemoglobin cluster. Due to both MEG and fNIRS having greater sensitivity to shallower sources, we hypothesized that much of the relative displacement across modalities was due to discrepancies for deeper sources. To test this, we examined the center of mass displacement as a factor of whether or not deeper sources were included in the map [Fig. 5(b)]. This analysis demonstrated that there is strong correspondence across the modalities at shallower depths, peaking at 9 mm (), 16 mm (Hb), 10 mm (MEG) from the fMRI-BOLD center of mass when including the image up to 20 mm from the scalp. As deeper sources were included in the analysis, the displacement of the centers of mass converges to what is shown in Table 2.
Table 1.
Center-of-mass of estimated localizer task responses. The coordinates, spread, and area of the estimated brain responses are provided for the three imaging modalities. FNIRS results from the fMRI-session, MEG-session, and combined results are presented. Coordinates are provided in Talairach space based on the FreeSurfer cortical surface registration.
| Center of activation (cm) | Depth from skin (cm) | Extent of activation (cm) | Cluster size () | Cluster size () | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| (half max) | () | |||||||||
| fMRI-BOLD | 19.5 | 3.47 | 7.6 | 6.8 | 22.5 | 82.9 | 62.8 | |||
| MEG | 36.2 | 2.81 | 13.4 | 11.4 | 10.9 | 38.5 | 195.6 | |||
| fNIRS [oxy-hemoglobin] | Both sessions | 0.9 | 37.1 | 1.97 | 3.6 | 12.5 | 5.7 | 17.2 | 30.2 | |
| fNIRS [deoxy-hemoglobin] | Both sessions | 3.3 | 35.3 | 1.99 | 4.5 | 17.7 | 6.3 | 17.6 | 29.9 | |
| fNIRS [oxy-hemoglobin] | MRI session | 39.9 | 1.96 | 3.3 | 4.2 | 4.6 | 11.5 | 25.6 | ||
| fNIRS [deoxy-hemoglobin] | MRI session | 35.0 | 1.97 | 4.1 | 14.1 | 10.5 | 12.0 | 25.4 | ||
| fNIRS [oxy-hemoglobin] | MEG session | 20.6 | 14.9 | 1.90 | 4.3 | 23.1 | 10.2 | 11.6 | 34.8 | |
| fNIRS [deoxy-hemoglobin] | MEG session | 27.0 | 15.3 | 2.00 | 4.7 | 21.4 | 9.2 | 8.5 | 17.4 | |
Fig. 5.
The center-of-mass of the localizer activation patterns was calculated for the four reconstructed image types and two NIRS sessions. There was a slight displacement of the fNIRS and MEG signals toward the cortical surface compared to the center of BOLD activation. In addition, the fNIRS results from the fNIRS-MEG session were slightly more frontal compared to the other data types. The data are additionally summarized in Tables 1 and 2.
Table 2.
Center-of-mass activation displacements. The displacement in millimeters between the center-of-mass of the estimated localizer task responses for each modality is shown. The upper-right half of the table (italics) shows the minimum displacement between each modality along the depth from the scalp surface [e.g., also see Fig. 5(b)]. The bottom-left half (not italics) shows the center-of-mass displacement over the entire brain, including the subcortical regions. Distances cited in millimeters.
| fMRI-BOLD | MEG | (both sessions) | fNIRS-Hb (both sessions) | (MRI session) | fNIRS-Hb (MRI session) | (MEG session) | fNIRS-Hb (MEG session) | ||
|---|---|---|---|---|---|---|---|---|---|
| fMRI-BOLD | — | 7.1 | 9.0 | 15.1 | 7.2 | 13.5 | 17.7 | 20.9 | |
| MEG | 25.8 | — | 15.1 | 20.3 | 13.6 | 19.0 | 23.6 | 25.4 | |
| Both sessions | 19.3 | 26.8 | — | 5.7 | 1.4 | 3.8 | 9.5 | 13.2 | |
| fNIRS-Hb | Both sessions | 18.7 | 28.7 | 3.1 | — | 7.4 | 1.0 | 3.5 | 6.2 |
| MRI session | 20.8 | 21.4 | 9.9 | 12.8 | — | 5.2 | 10.9 | 14.5 | |
| fNIRS-Hb | MRI session | 16.5 | 25.2 | 5.3 | 7.2 | 7.2 | — | 5.8 | 7.3 |
| MEG session | 29.3 | 51.6 | 30.7 | 27.8 | 39.2 | 32.0 | — | 2.3 | |
| fNIRS-Hb | MEG session | 34.7 | 55.8 | 34.5 | 31.5 | 43.7 | 36.7 | 6.6 | — |
In Fig. 5(c), we show the depth of the activation from the surface of the scalp for each imaging modality. The mass-weighted average depth of the activation from the nearest point on the surface of the scalp as detected by fMRI-BOLD was 35 mm. For MEG, this depth was about 7 mm more shallow at 28 mm and for fNIRS this was almost 15 mm shallower at 19 mm from the scalp. Fig. 5(c) shows the histogram plots of the depth location of the detected activation areas () for each modality. For fNIRS, to 35% of the reconstructed signal was between 18 to 22 mm from the surface of the scalp. There were no signals reconstructed beyond about 28 mm. In contrast, for the fMRI-BOLD, roughly 50% of the signal was located at a depth of 28 mm or greater, which means that the fNIRS method missed about half of the signal detected by BOLD because of the limited fNIRS depth penetration. The depth of the MEG reconstructed signal was deeper than the fNIRS, but still had a fall off around 30 mm compared to the BOLD signal.
To further quantify the relationship among the modalities, we examined the spatial correlation between each modality pair as a factor of whether or not deeper sources were included in the correlation. In Fig. 5(d), we show the correlation of the fNIRS and MEG signals as a function of the inclusion distance from the surface of the scalp. The correlation for both MEG and fNIRS with the BOLD signal was strongest only if the superficial cortical points were included. As the inclusion depth increases, the correlations decrease to approach the whole-brain asymptotic limit, which includes comparisons in deep subcortical areas where neither MEG nor fNIRS have strong sensitivity. Both the whole-brain and peak correlation at depth are provided in Table 3. The whole-brain limit of correlation between fMRI-BOLD and the MEG was . Likewise, the correlation between BOLD and oxy- and deoxy-hemoglobin was and , respectively. The correlation between the MEG and oxy- and deoxy-hemoglobin was and , respectively. However, by restricting the correlation analysis to the superficial cortex only, the maximum correlation between the MEG and fMRI-BOLD was found to be and was and between the BOLD and fNIRS oxy- and deoxy-hemoglobin signals, respectively (Fig. 5). Likewise, the maximum correlation between the MEG and fNIRS reconstructions was 0.46 and 0.37 for oxy- and deoxy-hemoglobin. The maximum correlation between fNIRS oxy- and deoxy-hemoglobin was . The spatial correlation results between all other combinations are provided in Table 3. All correlations (including when deep sources are included in the analysis) were statistically significant ().
Table 3.
Spatial correlation. The correlation of the activation patterns between the estimated localizer responses. The upper-right half of the table (italics) shows the maximum correlation values between each modality along the depth from the scalp surface (e.g., also see Fig. 5). The bottom-left half (italics) shows the correlation over the entire brain, including the subcortical regions that showed activity in fMRI but not fNIRS nor MEG.
| fMRI-BOLD | MEG | (both sessions) | fNIRS-Hb (both sessions) | (MRI session) | fNIRS-Hb (MRI session) | (MEG session) | fNIRS-Hb (MEG session) | ||
|---|---|---|---|---|---|---|---|---|---|
| fMRI-BOLD | — | 0.54 | 0.57 | 0.62 | 0.3 | ||||
| MEG | 0.17 | — | 0.46 | 0.46 | 0.17 | ||||
| Both sessions | 0.28 | 0.16 | — | 0.98 | 0.88 | ||||
| fNIRS-Hb | Both sessions | — | 0.99 | 0.97 | |||||
| MRI session | 0.3 | 0.16 | 0.97 | −0.79 | — | 0.89 | |||
| fNIRS-Hb | MRI session | 0.91 | — | 0.98 | |||||
| MEG session | 0.13 | 0.03 | 0.46 | 0.54 | — | ||||
| fNIRS-Hb | MEG session | 0.63 | 0.69 | — |
3.1. Pulsed Pair Results
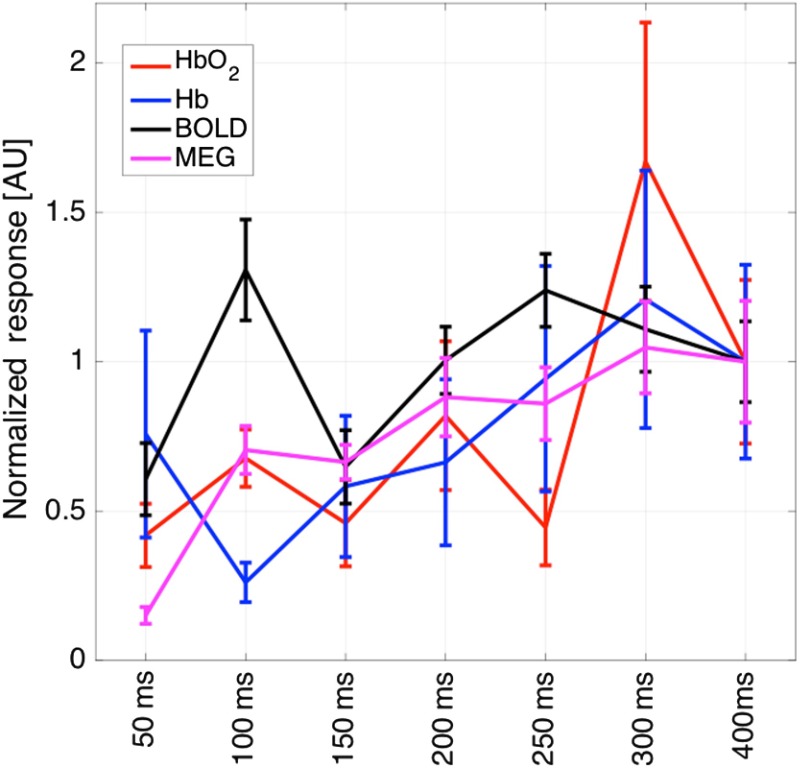
In the second half of the study, we performed an event-related pulsed-pair median nerve study. The pulsed-pair task uses two stimuli placed 50 to 500 ms apart. A slow event-related design with a minimum 8-s ISI was used to avoid additional nonlinearity in the hemodynamic response unrelated to the neural-refractory effect.16 As shown in Fig. 6, when the second pulse was after the first, the MEG ERP response was significantly diminished by the neural refractory period. At 300 ms and later, the second ERP response was again visible. A similar task had previously been used by Ogawa et al.33 showing a similar effect in rodent forepaw stimulation and was more recently examined for multimodal fMRI-MEG by Stevenson et al.34 Using the region-of-interest defined as the union of activations from all of the modalities observed in the localizer task the magnitude of brain activity for all seven interpair intervals was calculated from each modality.
Fig. 6.
MEG ERP responses. The estimated MEG evoked responses for the seven pulsed-pair conditions are shown. For short inter-pulse intervals (), the second pulse falls in the neural-refractory period and does not provide a strong ERP response. After 400 ms, the second ERP response is visible in the MEG data. The plots show the grand average across all subjects.
In Fig. 7, grand average of the normalized magnitude of the MEG [area-under-curve ], fMRI, and fNIRS signals for the pulsed-pair conditions is presented. The signals are shown normalized to the mean of the seven conditions. We found significant correlations of , 0.52, and between the magnitude of the fMRI and fNIRS [oxy-/deoxy-hemoglobin] responses compared with the AUC of the MEG response using colocalized regions-of-interest. The BOLD and fNIRS magnitudes were also significantly correlated for oxy- and deoxy-hemoglobin responses ( and , respectively).
Fig. 7.
Pulsed pair responses. The magnitude of the evoked response to the pulsed-pair stimulation conditions was estimated from a region-of-interest defined as the intersection of the MEG, fNIRS, and fMRI responses. The mean normalized MEG area-under-the-curve and estimated hemodynamic responses are shown for the seven interpulse intervals. The plots show the grand average across all subjects.
In addition, the amplitude of the evoked pulsed-pair response was also estimated using modality-specific regions-of-interest. For the fNIRS data, the region-of-interest was defined in fNIRS channel (source–detector) space as the average value over all statistically significant channels () on the probe. Using the modality-specific regions-of-interest, we still found statistically significant () correlations between the MEG signal (AUC) and the fNIRS channel oxy- and deoxy-hemoglobin data from the MEG-fNIRS session ( and , respectively). The fNIRS channel-based correlations to the MRI-BOLD signal went for down to and for oxy- and deoxy-hemoglobin. These lower correlations observed in fNIRS channel space compared to the reconstructed images may be due to errors introduced from intersubject registration and anatomy, which are accounted for in the image reconstruction process but not in the raw channel space data.
4. Discussion
In this work, we examined the spatial localization of reconstructed fNIRS signals and compared this to both fMRI and MEG measurements. Although previous work has established the general correspondence of these measurements from a physiological and biophysical basis (reviewed in Ref. 10), it is unclear how well spatial agreements can be achieved in practice given the inherent difficulties and ill-posed nature of the reconstruction of low-density fNIRS measurements into spatial images. Previously, we have described and numerically validated several advanced methods for fNIRS image reconstruction that incorporate anatomical information from structural MRI,14 group-level random effects models,12 and hierarchical regularization models.13 In addition, we have recently detailed several methods for improved time-series analysis and generalizations of the linear model to deal with fNIRS specific noise structures.11,35,36 In this work, we have presented an analysis pipeline combining these recent advancements that allows all three modalities (MEG, fMRI, and fNIRS) to be presented and compared in the same cortical brain space.
5. Proposed Analysis Pipeline
As detailed in Fig. 1, our analysis of all three modalities was centered upon the use of the inflated cortical surface from FreeSurfer17 as the registration and reconstruction basis for each modality. In the case of both fNIRS and MEG, this cortical surface was used to restrict the image reconstruction problem to a two-dimensional (manifold) estimation problem directly on the folded three-dimensional cortical surface. Similarly, this FreeSurfer cortical representation is used in the MNE22 package for MEG analysis and as part of the AFNI/SUMA analysis package.24,25 The use a common basis upon the cortical surface for analysis in all three modalities allowed direct comparisons of the estimated brain activation patterns across modalities. However, we also propose that this approach can be useful in future work to better utilize the mutual information among multiple modalities. For example, in our previous work in Adbelnour et al.,13 we described how maximum likelihood methods could be used to introduce multiple regularization priors for fNIRS image reconstruction based on prior fMRI (or MEG) data. In this approach, multimodal data are used to constrain the reconstruction of the fNIRS data as a statistical prior. Since all three modalities are defined in the same spatial basis support, this pipeline will allow future use of such methods.
6. Comparision of fNIRS, fMRI, and MEG Signals
The spatial reconstruction of fNIRS signals requires a solution to the optical inverse problem (e.g., estimation of brain signals from a limited number of surface measurements). Therefore, spatial comparisons of fNIRS and other modalities are sensitive to the means of solving this inverse problem. To get around this issue, previous work by Sassaroli et al.37 and Huppert et al.38 both looked at the consistency of the measured sensor-space fNIRS data with the fMRI image by using the fNIRS sensitivity model to forward-project the fMRI into the measurement space of the fNIRS signals for comparison and thereby avoiding the mathematical image reconstruction process. These works showed that the fNIRS channel-data were spatially consistent with the fMRI data, but did not address the question of how well fNIRS on its own can localize signals within the brain given the current state-of-the-art in data collection and image reconstruction methods for fNIRS imaging. Using high-density optical tomography, Eggebrecht et al.39 showed a spatial correlation between NIRS and fMRI of between (verb generation task) and (reading task) at similar 6-mm fMRI smoothing as used in our current study. However, in contrast to our study, Eggebrecht et al. used a high-density NIRS system with a total of up to 644 NIRS source–detector pairs in comparison to the standard-density probe with 28 pairs used in this current study. Given the lower cost, ubiquity, and increased flexibility of standard density acquisition as was used in this study, it is notable that the correlations reported here compare favorably with those reported in the high-density fNIRS study by Eggebrecht et al. mentioned above. This is particularly relevant to multimodal imaging, as high-density NIRS systems are not compatible with concurrent data collection. More recently, the work by Cui et al.40 looked at both sensor-space and image-based comparisons of fNIRS and fMRI in a variety of cognitive and sensory-motor tasks and found spatial correlations of based on a simple spline interpretation to display the fNIRS data into a standardized brain template.
The relationship between fNIRS and electrophysiology recordings has also been examined. The review by Shibasaki41 discusses the topic of correlation between the hemodynamic and electrophysiological signals, including a discussion of fNIRS. The localization of functional tasks in concurrent MEG-fNIRS and EEG-fNIRS has also been examined by a few groups. Sander et al.42 looked at direct current (DC) MEG and fNIRS signals during a motor task study. A similar study was also done by Mackert et al.43 that also showed qualitative agreement between DC-MEG and fNIRS. Similarly, Seki et al.44 looked at auditory function using a custom built unshielded MEG and fNIRS system. A more comprehensive study using conventional MEG was done by Ou et al.45 Ou et al. used a median nerve electrical stimulation task to look at the relationships of the fNIRS and MEG signals under a parametric stimulation task. They examined the relationship of the amplitude of the optically measured oxy- and deoxy-hemoglobin signals with respect to various physiological peaks in the MEG signal. Specifically, they found that the amplitude of the N20 and P60 peaks best predicted the change in the hemodynamic signal for changes in the duration of the stimulation task. This study also looked at the spatial localization of the fNIRS and MEG signals and found spatial agreement with MEG source localization using a dipole model and the nearest optical channels (e.g., source localization was not used in fNIRS).
Using our proposed pipeline, we found close agreement with all three modalities as presented in Fig. 3. The activations for all three modalities were localized to the primary somatosensory (postcentral) gyrus and primary motor (precentral) gyrus. Although the fNIRS and MEG spatial resolution is generally coarser than this, the additional information added by the cortical surface used in the image reconstruction process adds spatial information to constrain the images. Thus, with the addition of this structural information, we found that the overall spatial resolution of the fNIRS images was comparable to the BOLD fMRI. We found that for shallower regions of the brain, there was only a 9- and 16-mm displacement between the BOLD and fNIRS activations [center-of-mass; and Hb, respectively)] and and correlation between the two maps at a depth of 20 mm. This error was similar to that observed between the MEG and BOLD signal (10.4 mm) with a similar correlation (). To our knowledge, this is the first study to use the pulsed-pair task with fNIRS. However, our results are similar to the work by Stevenson et al.34 that used a similar pulsed-pair median nerve task during independent sessions of fMRI and MEG in seven subjects. They reported good spatial agreement between the two modalities. In their study, the displacement of the average BOLD and MEG (dipole) locations was found to be , which is similar to the 25-mm displacement found in this study for the whole head analysis, but greater than the 7.1-mm displacement found when only the superficial layers of the cortex were considered in the analysis. The relatively large inaccuracy of the signals in MEG and fNIRS for depths beyond 30 mm suggests that it is prohibitively difficult to image deep brain sources, such as subcortical regions, with these methods. Previously, this falloff of signal with depth had been modeled using simulated data,46–48 but the current work provides an empirical examination of this issue. One caveat of the current study is that it is done in the context of median nerve stimulation, which provides a relative context for comparing localization.
7. Test–Retest Reliability
Since the fNIRS data were recorded in two separate sessions (once with fMRI and once with MEG), we also examined the test-retest reliability of the fNIRS signals and image reconstruction. As was presented in Fig. 4, we found close agreement across the sessions for the oxy-hemoglobin estimates of the response to the localizer task. The deoxy-hemoglobin images were slightly noisier, particularly in the case of the MEG-NIRS session data, but were still strongly correlated with the fMRI-NIRS session data (Table 3). This indicates that the fNIRS results were spatially consistent between the two sessions. There was a slight frontal bias of the estimated activation in the MEG-NIRS session in which more activation was observed in the frontal lateral region of the probe (see Figs. 4 and 5).
8. Conclusions
The results of this study show close agreement in spatial localization of fNIRS with MEG and fMRI results. In addition, the magnitude of estimate brain responses for these three modalities was linearly correlated during a parametric task. The main source of difference across the modalities was the decreased sensitivity with respect to depth seen in fNIRS and MEG. These results validate the concurrent recording approach for future studies of neurovascular coupling and multimodal fusion with fNIRS building a bridge between fMRI and MEG.
Acknowledgments
This work was funded by startup funding from the University of Pittsburgh Department of Radiology and the National Institutes of Health (Nos. P30AG024827, R21NS064457, and R01AG029546).
Biographies
Theodore Huppert received his PhD in biophysics from Harvard University. He is an associate professor of radiology and bioengineering at the University of Pittsburgh. He works on signal processing and advanced methods for multimodal imaging including fNIRS, MEG, EEG, and fMRI.
Avniel Ghuman received his PhD in biophysics from Harvard University and is an assistant professor of neurosurgery at the University of Pittsburgh. He is the director of MEG research and works on applications of MEG to psychology and clinical medicine.
Biographies for the other authors are not available.
Disclosures
None of the authors has any conflicts of interest to disclose.
References
- 1.Jobsis F. F., “Noninvasive, infrared monitoring of cerebral and myocardial oxygen sufficiency and circulatory parameters,” Science 198(4323), 1264–1267 (1977). 10.1126/science.929199 [DOI] [PubMed] [Google Scholar]
- 2.Gervain J., et al. , “Near-infrared spectroscopy: a report from the McDonnell infant methodology consortium,” Dev. Cognit. Neurosci. 1(1), 22–46 (2011). 10.1016/j.dcn.2010.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aslin R. N., “Questioning the questions that have been asked about the infant brain using near-infrared spectroscopy,” Cognit. Neuropsychol. (2012). 10.1080/02643294.2012.654773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toet M. C., Lemmers P. M. A., “Brain monitoring in neonates,” Early human development 85(2), 77–84 (2009). 10.1016/j.earlhumdev.2008.11.007 [DOI] [PubMed] [Google Scholar]
- 5.Lloyd-Fox S., Blasi A., Elwell C. E., “Illuminating the developing brain: the past, present and future of functional near infrared spectroscopy,” Neurosci. Biobehav. Rev. 34(3), 269–284 (2010). 10.1016/j.neubiorev.2009.07.008 [DOI] [PubMed] [Google Scholar]
- 6.Scheeren T. W., Schober P., Schwarte L. A., “Monitoring tissue oxygenation by near infrared spectroscopy (NIRS): background and current applications,” J. Clin. Monit. Comput. (2012). 10.1007/s10877-012-9348-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith M., “Shedding light on the adult brain: a review of the clinical applications of near-infrared spectroscopy,” Philos. Trans. Ser. A 369(1955), 4452–4469 (2011). 10.1098/rsta.2011.0242 [DOI] [PubMed] [Google Scholar]
- 8.Ghosh A., Elwell C., Smith M., “Review article: cerebral near-infrared spectroscopy in adults: a work in progress,” Anesth. Analg. 115(6), 1373–1383 (2012). 10.1213/ANE.0b013e31826dd6a6 [DOI] [PubMed] [Google Scholar]
- 9.Calderon-Arnulphi M., Alaraj A., Slavin K. V., “Near infrared technology in neuroscience: past, present and future,” Neurol. Res. 31(6), 605–614 (2009). 10.1179/174313209X383286 [DOI] [PubMed] [Google Scholar]
- 10.Steinbrink J., et al. , “Illuminating the BOLD signal: combined fMRI-fNIRS studies,” Magn. Reson. Imaging 24(4), 495–505 (2006). 10.1016/j.mri.2005.12.034 [DOI] [PubMed] [Google Scholar]
- 11.Barker J. W., Aarabi A., Huppert T. J., “Autoregressive model based algorithm for correcting motion and serially correlated errors in fNIRS,” Biomed. Opt. Express 4(8), 1366–1379 (2013). 10.1364/BOE.4.001366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abdelnour F., Huppert T., “A random-effects model for group-level analysis of diffuse optical brain imaging,” Biomed. Opt. Express 2(1), 1–25 (2010). 10.1364/BOE.2.000001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abdelnour F., Genovese C., Huppert T., “Hierarchical Bayesian regularization of reconstructions for diffuse optical tomography using multiple priors,” Biomed. Opt. Express 1(4), 1084–1103 (2010). 10.1364/BOE.1.001084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abdelnour F., Schmidt B., Huppert T. J., “Topographic localization of brain activation in diffuse optical imaging using spherical wavelets,” Phys. Med. Biol. 54(20), 6383–6413 (2009). 10.1088/0031-9155/54/20/023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huppert T. J., et al. , “HomER: a review of time-series analysis methods for near-infrared spectroscopy of the brain,” Appl. Opt. 48(10), D280–D298 (2009). 10.1364/AO.48.00D280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cannestra A. F., et al. , “Refractory periods observed by intrinsic signal and fluorescent dye imaging,” J. Neurophysiol. 80(3), 1522–1532 (1998). [DOI] [PubMed] [Google Scholar]
- 17.Dale A. M., Fischl B., Sereno M. I., “Cortical surface-based analysis. I. Segmentation and surface reconstruction,” NeuroImage 9(2), 179–194 (1999). 10.1006/nimg.1998.0395 [DOI] [PubMed] [Google Scholar]
- 18.Taulu S., “Spatiotemporal signal space separation method for rejecting nearby interference in MEG measurements,” Phys. Med. Biol. 51, 1759–1768 (2006). 10.1088/0031-9155/51/7/008 [DOI] [PubMed] [Google Scholar]
- 19.Hamalainen M. S., Ilmoniemi R. J., “Interpreting magnetic fields of the brain: minimum norm estimates,” Med. Biol. Eng. Comput. 32(1), 35–42 (1994). 10.1007/BF02512476 [DOI] [PubMed] [Google Scholar]
- 20.Hamalainen M. S., Sarvas J., “Realistic conductivity geometry model of the human head for interpretation of neuromagnetic data,” IEEE Trans. Biomed. Eng. 36(2), 165–171 (1989). 10.1109/10.16463 [DOI] [PubMed] [Google Scholar]
- 21.Oostendorp T. F., Vanoosterom A., “Source parameter-estimation in inhomogeneous volume conductors of arbitrary shape,” IEEE Trans. Biomed. Eng. 36(3), 382–391 (1989). 10.1109/10.19859 [DOI] [PubMed] [Google Scholar]
- 22.Gramfort A., et al. , “MNE software for processing MEG and EEG data,” NeuroImage 86, 446–460 (2014). 10.1016/j.neuroimage.2013.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin F. H., et al. , “Spectral spatiotemporal imaging of cortical oscillations and interactions in the human brain,” Neuroimage 23(2), 582–595 (2004). 10.1016/j.neuroimage.2004.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cox R. W., “AFNI: software for analysis and visualization of functional magnetic resonance neuroimages,” Comput. Biomed. Res. 29(3), 162–173 (1996). 10.1006/cbmr.1996.0014 [DOI] [PubMed] [Google Scholar]
- 25.Saad Z. S., et al. , “Functional imaging analysis contest (FIAC) analysis according to AFNI and SUMA,” Hum. Brain Mapp. 27(5), 417–424 (2006). 10.1002/hbm.20247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fang Q., Boas D. A., “Monte Carlo simulation of photon migration in 3D turbid media accelerated by graphics processing units,” Opt. Express 17(22), 20178–20190 (2009). 10.1364/OE.17.020178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boas D., et al. , “Three dimensional Monte Carlo code for photon migration through complex heterogeneous media including the adult human head,” Opt. Express 10(3), 159–170 (2002). 10.1364/OE.10.000159 [DOI] [PubMed] [Google Scholar]
- 28.Hansen P. C., Kringelbach M. L., Salmelin R., MEG: An Introduction to Methods, Oxford University Press, New York, p. 436 (2010). [Google Scholar]
- 29.Holland P. W., Welsch R. E., “Robust regression using iteratively reweighted least-squares,” Commun. Stat. Theory Meth. 6(9), 813–827 (1977). 10.1080/03610927708827533 [DOI] [Google Scholar]
- 30.Mattout J., et al. , “MEG source localization under multiple constraints: an extended Bayesian framework,” Neuroimage 30(3), 753–767 (2006). 10.1016/j.neuroimage.2005.10.037 [DOI] [PubMed] [Google Scholar]
- 31.Friston K. J., Statistical Parametric Mapping: The Analysis of Funtional Brain Images, 1st ed., Elsevier/Academic Press, Amsterdam, Boston, p. 647 (2007). [Google Scholar]
- 32.Trefethen L., Bau D., Numerical Linear Algebra, Society for Industrial and Applied Mathematics, Philadelphia: (1997). [Google Scholar]
- 33.Ogawa S., et al. , “An approach to probe some neural systems interaction by functional MRI at neural time scale down to milliseconds,” Proc. Natl. Acad. Sci. USA 97(20), 11026–11031 (2000). 10.1073/pnas.97.20.11026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stevenson C. M., et al. , “Paired pulse depression in the somatosensory cortex: associations between MEG and BOLD fMRI,” Neuroimage 59(3), 2722–2732 (2012). 10.1016/j.neuroimage.2011.10.037 [DOI] [PubMed] [Google Scholar]
- 35.Huppert T. J., “Commentary on the statistical properties of noise and its implication on general linear models in functional near-infrared spectroscopy,” Neurow. 3(1), 010401 (2016). 10.1117/1.NPh.3.1.010401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barker J. W., et al. , “Correction of motion artifacts and serial correlations for real-time functional near-infrared spectroscopy,” Neurophotonics 3(3), 031410 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sassaroli A., et al. , “Spatially weighted BOLD signal for comparison of functional magnetic resonance imaging and near-infrared imaging of the brain,” NeuroImage 33(2), 505–514 (2006). 10.1016/j.neuroimage.2006.07.006 [DOI] [PubMed] [Google Scholar]
- 38.Huppert T. J., et al. , “Quantitative spatial comparison of diffuse optical imaging with blood oxygen level-dependent and arterial spin labeling-based functional magnetic resonance imaging,” J. Biomed. Opt. 11(6), 064018 (2006). 10.1117/1.2400910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eggebrecht A. T., et al. , “A quantitative spatial comparison of high-density diffuse optical tomography and fMRI cortical mapping,” Neuroimage 61(4), 1120–1128 (2012). 10.1016/j.neuroimage.2012.01.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cui X., et al. , “A quantitative comparison of NIRS and fMRI across multiple cognitive tasks,” Neuroimage 54(4), 2808–2821 (2011). 10.1016/j.neuroimage.2010.10.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shibasaki H., “Human brain mapping: hemodynamic response and electrophysiology,” Clin. Neurophysiol. 119(4), 731–743 (2008). 10.1016/j.clinph.2007.10.026 [DOI] [PubMed] [Google Scholar]
- 42.Sander T. H., et al. , “Cross-correlation analysis of the correspondence between magnetoencephalographic and near-infrared cortical signals,” Methods Inf. Med. 46(2), 164–168 (2007). [PubMed] [Google Scholar]
- 43.Mackert B. M., et al. , “Neurovascular coupling analyzed non-invasively in the human brain,” NeuroReport 15(1), 63–66 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Seki Y., et al. , “Simultaneous measurement of neuronal activity and cortical hemodynamics by unshielded magnetoencephalography and near-infrared spectroscopy,” J. Biomed. Opt. 17(10), 107001 (2012). 10.1117/1.JBO.17.10.107001 [DOI] [PubMed] [Google Scholar]
- 45.Ou W., et al. , “Study of neurovascular coupling in humans via simultaneous magnetoencephalography and diffuse optical imaging acquisition,” Neuroimage 46(3), 624–632 (2009). 10.1016/j.neuroimage.2009.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Belardinelli P., et al. , “Source reconstruction accuracy of MEG and EEG Bayesian inversion approaches,” PLoS One 7(12), e51985 (2012). 10.1371/journal.pone.0051985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin F. H., et al. , “Assessing and improving the spatial accuracy in MEG source localization by depth-weighted minimum-norm estimates,” Neuroimage 31(1), 160–171 (2006). 10.1016/j.neuroimage.2005.11.054 [DOI] [PubMed] [Google Scholar]
- 48.Goldenholz D. M., et al. , “Mapping the signal-to-noise-ratios of cortical sources in magnetoencephalography and electroencephalography,” Hum. Brain Mapp. 30(4), 1077–1086 (2009). 10.1002/hbm.20571 [DOI] [PMC free article] [PubMed] [Google Scholar]