Abstract
MicroRNA-122 (miR-122) is abundant in the liver and involved in lipid homeostasis, but its relevance to the long-term risk of developing metabolic disorders is unknown. We therefore measured circulating miR-122 in the prospective population-based Bruneck Study (n = 810; survey year 1995). Circulating miR-122 was associated with prevalent insulin resistance, obesity, metabolic syndrome, type 2 diabetes, and an adverse lipid profile. Among 92 plasma proteins and 135 lipid subspecies quantified with mass spectrometry, it correlated inversely with zinc-α-2-glycoprotein and positively with afamin, complement factor H, VLDL-associated apolipoproteins, and lipid subspecies containing monounsaturated and saturated fatty acids. Proteomics analysis of livers from antagomiR-122–treated mice revealed novel regulators of hepatic lipid metabolism that are responsive to miR-122 inhibition. In the Anglo-Scandinavian Cardiac Outcomes Trial (ASCOT, n = 155), 12-month atorvastatin reduced circulating miR-122. A similar response to atorvastatin was observed in mice and cultured murine hepatocytes. Over up to 15 years of follow-up in the Bruneck Study, multivariable adjusted risk ratios per one-SD higher log miR-122 were 1.60 (95% CI 1.30–1.96; P < 0.001) for metabolic syndrome and 1.37 (1.03–1.82; P = 0.021) for type 2 diabetes. In conclusion, circulating miR-122 is strongly associated with the risk of developing metabolic syndrome and type 2 diabetes in the general population.
Introduction
MicroRNAs (miRNAs) are small noncoding RNA molecules that regulate gene expression (1). The predominant miRNA in the liver, microRNA-122 (miR-122), has been proposed to play a central role in the regulation of lipid and glucose metabolism (2,3). Inhibition of miR-122 in mice (4,5) and nonhuman primates (6,7) induces fatty acid oxidation, reduces lipid synthesis, and thereby leads to lower levels of total cholesterol.
In humans, it has been suggested that miR-122 may have adverse metabolic effects and may be associated with metabolic diseases. However, as highlighted by our recent review (3), evidence from existing epidemiological studies is sparse and has important limitations. Published studies have focused on correlations with major lipids (8), whereas a breakdown into lipid subspecies would add resolution and help improve our understanding of the regulation of lipid homeostasis by miR-122. Importantly, previous studies had cross-sectional or case-control designs (3) and hence were unable to inform about long-term associations of circulating miR-122 with the development of new-onset disease outcomes over time.
To address this gap in the current literature, we conducted a series of analyses in the prospective Bruneck Study, the randomized controlled Anglo-Scandinavian Cardiac Outcomes Trial (ASCOT), and experiments in mice and cell culture, combining lipidomics, proteomics, and miR-122 data. Our aims were fourfold: first, to assess cross-sectional correlates of circulating miR-122, including lipidomics and proteomics profiles; second, to provide mechanistic insight into the putative regulatory function of miR-122 in lipid metabolism with animal studies of antagomiR-122 interventions and statin treatment; third, to study the effect of statin allocation on serum miR-122 in participants of ASCOT; and fourth, to quantify the, to date unknown, associations of circulating miR-122 with the long-term risk of developing metabolic syndrome and type 2 diabetes (T2D). We compared associations to those with cardiovascular disease (CVD).
Research Design and Methods
The Bruneck Study
The Bruneck Study is a prospective, population-based study (9–13). In 1990, 1,000 individuals aged 40–79 years were recruited as a random sample of Bruneck inhabitants and were reexamined every 5 years since, with participation rates exceeding 90% at all surveys. The current study used the 1995 survey as baseline. Full medical records were available on clinical end points occurring between 1995 and 2010 (1995–2005 for metabolic syndrome) for all individuals, including those who did not participate in later evaluations or died during follow-up (100% follow-up for clinical end points). Metabolic syndrome was diagnosed if three out of the five following characteristics were present: 1) waist circumference in men ≥102 cm and women ≥88 cm; 2) fasting triglycerides ≥150 mg/dL or on drug treatment for elevated triglycerides (fibrates and nicotinic acids); 3) HDL cholesterol in men <40 mg/dL and women <50 mg/dL or on drug treatment for reduced HDL cholesterol (fibrates and nicotinic acids); 4) blood pressure ≥130/≥85 mmHg or antihypertensive drug treatment in a patient with a history of hypertension; and 5) fasting glucose ≥100 mg/dL or on drug treatment for elevated glucose. T2D was diagnosed according to 1997 American Diabetes Association criteria. CVD was defined as myocardial infarction, stroke, or vascular death. Fatal and nonfatal myocardial infarction were deemed confirmed when World Health Organization criteria for definite disease status were met. Ischemic stroke and transient ischemic attacks were classified according to the criteria of the National Survey of Stroke. Self-report of disease was always confirmed by reviewing the participant’s medical records available from their general practitioners and the Bruneck Hospital.
Risk factors were ascertained by validated standard procedures as previously described (9–13). Socioeconomic status was defined on a three-category scale (low, medium, or high) on the basis of information on occupational status and educational level of the person with the highest income in the household. High socioeconomic status was assumed if this person had ≥12 years of education or an occupation with an average monthly income ≥$2,000 (baseline salary before tax). Low socioeconomic status was defined by ≤8 years of education or an average monthly income ≤$1,000. Physical activity was assessed using the validated Baecke score (14). Waist and hip circumferences were assessed with a plastic tape measure at the levels of the umbilicus and the greater trochanters, respectively. Blood samples were taken after an overnight fast. Lipidomics profiling in plasma samples of the Bruneck cohort was performed with mass spectrometry, which allowed quantification of 135 distinct lipid species (9). HbA1c was quantified using high-performance liquid chromatography (Diabetes Control and Complications Trial [DCCT]–aligned assay). The degree of insulin resistance by HOMA-IR was estimated using the following formula: fasting plasma glucose in mmol/L × fasting serum insulin in mU/L divided by 22.5, with higher HOMA-IR values indicating higher insulin resistance (15). miR-122 was measured in serum taken at the 1995 examination (n = 810) as well as in serum and plasma taken at the 2000 examination (n = 695).
ASCOT
ASCOT is a double-blind randomized 2 × 2 factorial study of blood pressure–lowering and lipid-lowering treatment (16–18). A total of 14,412 patients (aged 40–79 years) were randomized between 1998 and 2000 using a computer-generated optimum allocation mechanism blinded for any person involved in the undertaking of the study. Patients randomized to the lipid-lowering arm had low to moderate cholesterol levels (serum total cholesterol ≤6.5 mmol/L) and were allocated atorvastatin (10 mg/day) or placebo. Serum miR-122 levels were measured at baseline and 1 year after randomization (median 13 months [range 12–16]) in participants of the hypertension-associated CVD substudy (HACVD-ASCOT) who were of European ancestry and did not have T2D at study entry.
AntagomiR Treatment in Mice
Mice were injected intraperitoneally with antagomiR-122 and control antagomiRs (65 mg/kg; n = 5 per group) on three consecutive days as previously described (19). AntagomiRs were purchased from Fidelity Systems with the following sequences: antagomiR-122, C*A*AACACCAUUGUCACACU*C*C*A*Chol*-T; controls, A*A*GGCAAGCUGACCUGAA*G*U*U*Chol-T; an asterisk indicates a phosphorothioate backbone modification at the preceding nucleotide. Mice were sacrificed at day 7. Liver and serum samples were harvested for analysis. Blood samples were collected by cardiac puncture. Total and HDL cholesterol were enzymatically measured using the T-Cholesterol and HDL-C Assay Kits (Wako Diagnostics). We measured hepatic miRNA-122 expression using Northern blot, and miR-122 and other miRNAs in liver and serum using quantitative reverse transcription real-time PCR (qRT-PCR). Selected genes were also quantified by qRT-PCR (for details, see Supplementary Data).
Statin Treatment in Mice and Primary Murine Hepatocytes
Six-week-old female C57Bl/6 mice were injected once a day with 20 mg/kg atorvastatin intraperitoneally (Sigma-Aldrich, Taufkirchen, Germany) for 5 days and were sacrificed on day 5. Serum was collected and the liver was perfused with ice-cold PBS, and tissue specimens from the left lower lobe were either snap frozen or placed in RNAlater (Qiagen, Hilden, Germany) until further processing. Details are provided in the Supplementary Data.
Proteomics Analyses
Targeted proteomics profiling in plasma samples of the Bruneck Study (year 2000 evaluation) was performed using multiple reaction monitoring (PlasmaDive kits; Biognosys AG), which allowed quantification of 92 proteins (for details, see Supplementary Data). During continuous operation over 2 weeks, the interday relative SD was <20% and <5% without and with adjustment for the peak area of the authentic standard peptides, respectively.
Proteomics analysis of livers from antagomiR-treated mice was performed after an in-solution digest by liquid chromatography tandem mass spectrometry. Differential protein expression was assessed by two methods (for details, see Supplementary Data): by spectral counting using a high-mass accuracy instrument (Q-Exactive HF; Thermo Fisher Scientific) and by labeling with Tandem Mass Tags (TMT) (Thermo Fisher Scientific) using the triple-stage mass spectrometry (MS3) capability on an Orbitrap Fusion Tribrid MS (Thermo Fisher Scientific). MS3 can overcome the inherent interference of more commonly used two-stage (MS2) workflows when isobaric labeling strategies are used with complex samples (20).
miR-122 Measurement Using qRT-PCR
We measured miRNA-122 in samples of the Bruneck Study, ASCOT, and statin experiments using qRT-PCR, as previously described (11,21). In brief, total RNA was extracted using the miRNeasy kit (Qiagen, Hilden, Germany). For plasma, serum or cell culture supernatants, a fixed volume of 3 μL of the 25-μL RNA eluate was used as input for reverse transcription (RT) reactions. For RNA from cells or tissue, 100 ng input material was used for RT. miRNAs were reverse transcribed using Megaplex Primer Pools (Human Pool A version 2.1 or Rodent Pool A; Life Technologies, Darmstadt, Germany), and products were further amplified, if needed, using Megaplex PreAmp Primers (Primers A version 2.1). Both RT and PreAmp products were stored at −20°C. Taqman miRNA assays were used to assess the expression of individual miRNAs. Diluted preamplification product (0.5 μL) or RT product (corresponding to 0.45 ng input) were combined with 0.25 μL Taqman microRNA assay (20×) (Life Technologies) and 2.5 μL Taqman Universal PCR Master Mix No AmpErase UNG (2×) to a final volume of 5 μL. qRT-PCR was performed on an Applied Biosystems 7900HT thermocycler at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. All samples were run in duplicate. Laboratory technicians were blinded to the participants’ disease status. Relative quantification was performed using the software SDS2.2 (Life Technologies). U6 and exogenous Caenorhabditis elegans spike-in control (Cel-miR-39) were used for normalization purposes in cell and tissue experiments. For conditioned media, normalization was achieved by cultivating the same cell number in the same volume of medium with the spike-in control being used to adjust for any experimental variability in the isolation procedure.
Statistical Analysis
The statistical analysis was conducted according to a prespecified analysis plan. miR-122 values were log transformed for analysis. Cross-sectional associations of miR-122 levels with other participant characteristics were quantified using Spearman correlation coefficients and linear regression models adjusted for age and sex. In the survival analysis, the principal outcomes were metabolic syndrome and T2D, and a secondary outcome was CVD. We used Cox proportional hazard regression with updated covariates for CVD and T2D and pooled logistic regression (22) for metabolic syndrome. Both techniques make full use of the repeat measurements of miR-122 available at the 1995 and 2000 examination. Hazard ratios and odds ratios were assumed to represent the same measure of relative risk and are collectively described as risk ratios (RRs). Participants with prevalent disease were excluded from the respective analyses. Models were adjusted for age and sex, plus socioeconomic status (low, medium, or high), smoking (yes or no), physical activity, and alcohol consumption (“multivariable model”). A sensitivity analysis further adjusted for the potential mediators/confounders BMI and waist-to-hip ratio. The proportional hazards assumption for CVD and T2D was tested using Schoenfeld residuals and was met. We investigated effect modification with formal tests for interaction across groups defined by age, sex, statin intake, and obesity. Principal analyses used significance levels of two-sided P < 0.05. Exploratory analyses used Bonferroni-corrected P values to limit the risk of false-positive results (i.e., 0.00037 for analyses of lipid subspecies, 0.00054 for proteins, and 0.0042 for interaction tests). Analyses were performed using Stata software, version 12.1.
Study Approval
The Bruneck Study protocol was approved by the local ethic committee of Bolzano (Comitato Etico del Comprensorio Sanitario di Bolzano; approval number 28-2010). The ASCOT protocol was approved by central and regional ethics review boards in the U.K. and by national ethics and statutory bodies in Ireland and the Nordic countries. Animal experiments were approved by the Austrian authorities (licensed to A.R.M., BMWF-66.011/0040-II/10b/2009) and U.K. authorities (licensed to Q.X., PPL70/7266). The participants’ written informed consent was obtained prior to their inclusion in the Bruneck and ASCOT studies.
Results
miR-122 and Major Clinical Characteristics in Participants of the Bruneck Study
We successfully quantified circulating miR-122 levels in 810 out of 826 participants of the Bruneck Study. miR-122 levels in serum and plasma were strongly correlated (r = +0.86 [95% CI 0.84–0.88]). The within-person correlation of repeat serum miR-122 measurements taken 5 years apart was +0.24 (0.17–0.31) (Supplementary Fig. 1), which is comparable to the range previously reported for other plasma miRNAs (23). Table 1 shows baseline characteristics of the Bruneck participants and their correlations with miR-122. The mean age of participants was 63 years (SD 11) and 50% were female. Circulating miR-122 was associated with higher levels of liver enzymes, adiposity, inflammation, and insulin resistance and an adverse lipid profile (higher triglycerides and lower HDL cholesterol) (Table 1). Participants with a diagnosis of metabolic syndrome compared with those without had 160% higher circulating miR-122 levels (P < 0.001); participants with a diagnosis of T2D compared with those without had 214% higher circulating miR-122 levels (P < 0.001). No difference in circulating miR-122 levels was observed in participants with a history of CVD compared with participants without a history of CVD (P = 0.969).
Table 1.
Baseline characteristics and cross-sectional correlates of miR-122 in the Bruneck Study
| Variable | Mean (SD) or n (%) | Age- and sex-adjusted difference in miR-122 per SD or compared with reference (95% CI) | P value |
|---|---|---|---|
| Questionnaire based | |||
| Age, years | 63 (11) | −3% (−19, 17) | 0.753 |
| Female sex | 405 (50%) | +31% (−10, 89) | 0.153 |
| Current smoker | 159 (20%) | −22% (−51, 26) | 0.314 |
| Physical activity, Baecke score | 2.3 (0.9) | +1% (−18, 24) | 0.932 |
| Alcohol consumption, g/day | 24 (31) | −1% (−21, 23) | 0.919 |
| Statin treatment | 26 (3%) | −12% (−69, 151) | 0.808 |
| Socioeconomic status | |||
| Low | 494 (61%) | (Reference) | |
| Middle | 176 (22%) | −30% (−56, 14) | 0.150 |
| High | 140 (17%) | +65% (−2, 177) | 0.058 |
| Liver enzymes | |||
| Alanine transaminase, units/L | 23 (13) | +112% (76, 155) | <0.001 |
| Aspartate aminotransferase, units/L | 24 (9.3) | +81% (51, 118) | <0.001 |
| Adiposity measures | |||
| BMI, kg/m2 | 26 (3.9) | +41% (17, 69) | <0.001 |
| Waist-to-hip ratio | 0.93 (0.072) | +44% (17, 76) | <0.001 |
| Markers of inflammation | |||
| Log hs-CRP, mg/L | −1.7 (1.0) | +42% (17, 71) | <0.001 |
| Markers of dysglycemia | |||
| Fasting plasma glucose, mg/dL | 102 (25) | +23% (2, 48) | 0.030 |
| HbA1c, % (mmol/mol) | 5.6 (1.8) (38 [20]) | +4% (−14, 25) | 0.704 |
| Log HOMA-IR | 1.1 (0.6) | +67% (39, 101) | <0.001 |
| Major lipids | |||
| Total cholesterol, mg/dL | 230 (43) | +19% (−1, 43) | 0.070 |
| LDL cholesterol, mg/dL | 145 (38) | +21% (1, 46) | 0.043 |
| HDL cholesterol, mg/dL | 59 (16) | −34% (−45, −21) | <0.001 |
| Log triglycerides, mg/dL | 4.8 (0.5) | +62% (35, 94) | <0.001 |
P values significant after Bonferroni correction are shown in boldface type.
miR-122 and Lipidomic and Proteomics Profiles in Participants of the Bruneck Study
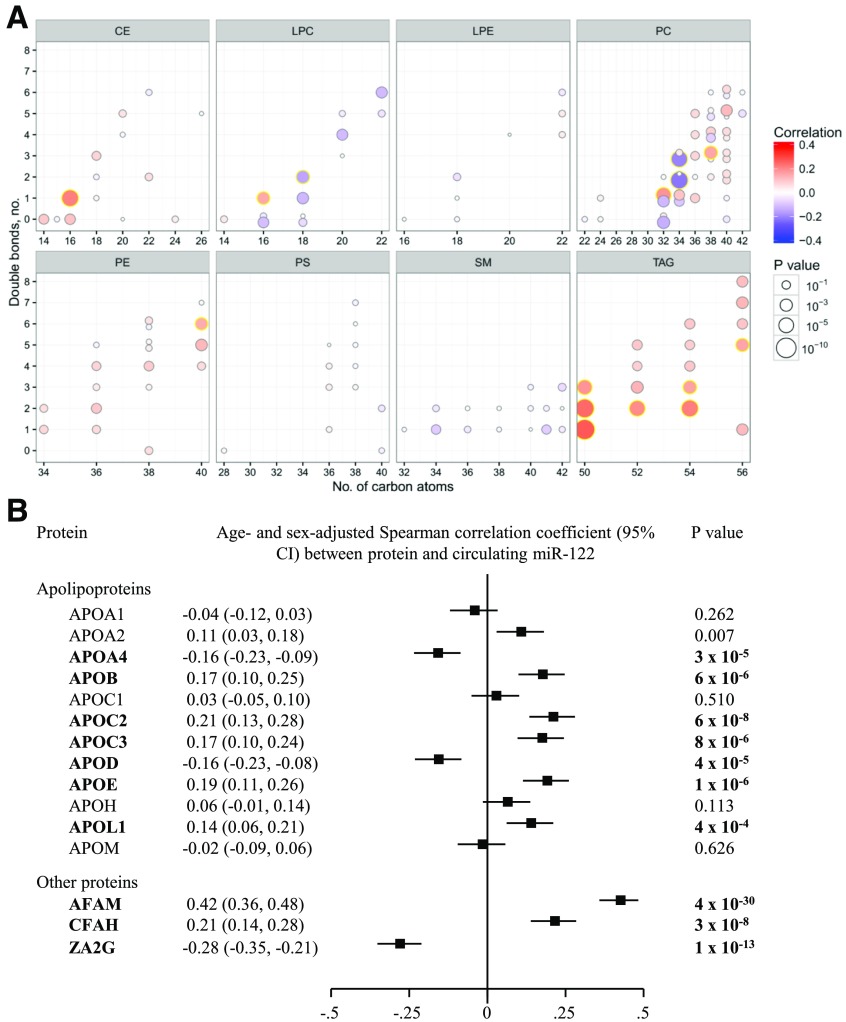
To provide novel insight into the complex correlation patterns of miR-122 beyond those with major clinical characteristics, we quantified cross-sectional correlations of miR-122 with lipidomic and proteomics profiles. Of the 135 distinct lipid subspecies available in the Bruneck Study (9), miR-122 showed a specific correlation with lipid subspecies comprised of monounsaturated and saturated fatty acids within the lipid classes triacylglycerols and cholesterol esters (Fig. 1A).
Figure 1.
Cross-sectional correlation of serum miR-122 levels with lipid subspecies (A) and selected proteins related to lipid metabolism (B) in the Bruneck Study. In A, lipid species are arranged by lipid class in eight panels according to the number of total carbon atoms and number of double bonds. Lipid species highlighted with a yellow halo showed statistically significant correlations after Bonferroni correction. For better visibility, those lipid species with alkyl ether linkage are shifted upwards, whereas their alkyl ether–free counterparts are shifted downward. In B, P values significant after Bonferroni correction are shown in bold. The full panel of proteins is shown in Supplementary Fig. 2. AFAM, afamin; CE, cholesteryl ester; CFAH, complement factor H; LPC, lysophosphatidylcholine; LPE, lysophosphatidylethanolamine; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PS, phosphatidylserine; SM, sphingomyelin; TAG, triacylglycerol; ZA2G, zinc-α-2-glycoprotein.
The proteomics assessment, over four orders of magnitude in abundance by mass spectrometry, covered 92 plasma proteins, including apolipoproteins, complement factors, and coagulation factors (Fig. 1B) (for full results see Supplementary Fig. 2). Circulating miR-122 was most strongly associated with afamin (r = +0.42; P = 4 × 10−30), complement factor H (r = +0.21; P = 3 × 10−8), and zinc-α-2-glycoprotein (r = –0.28; P = 10−13). Focused analyses of apolipoproteins revealed significant positive correlations with APOB, APOC2, APOC3, APOE, and APOL1 and significant inverse correlations with APOA4 and APOD.
Treatment With AntagomiR-122 in Mice and Effect on the Hepatic Proteome
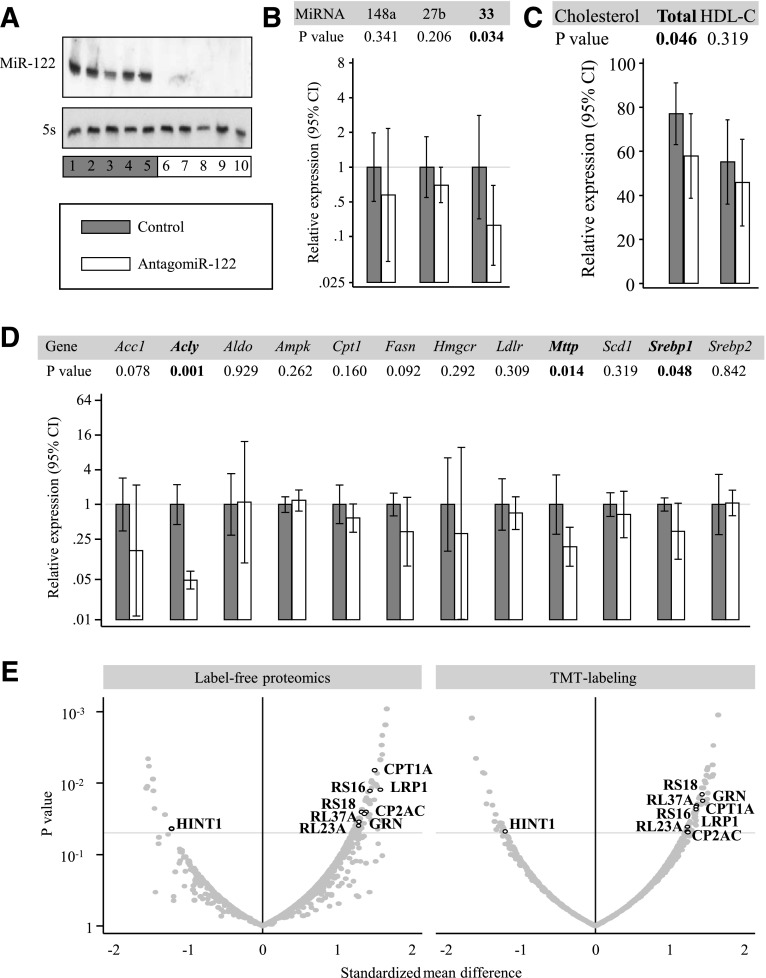
To scrutinize the role of miR-122 in the regulation of hepatic lipid metabolism, we inhibited miR-122 in mice using antagomiRs and studied consequences thereof at the miRNA, lipid, gene expression, and protein level. AntagomiR-122 led to an almost complete inhibition of miR-122 expression (Fig. 2A), with a secondary effect on hepatic miR-33 expression, but no effects on the expression of other miRNAs relevant to the hepatic liver metabolism, including miR-27b and miR-148a (Fig. 2B). The marked inhibition of hepatic miR-122 expression by antagomiR-122 treatment was reflected in a >10-fold reduction in the mean serum levels of circulating miR-122 (P = 0.032; n = 10 mice per group) (data not shown). Consistent with previous reports (3), antagomiR-122 treatment resulted in a reduction of total cholesterol levels (Fig. 2C). Gene expression in the liver was downregulated in the liver for ATP citrate lyase (Acly), microsomal triglyceride transfer protein (Mttp), and sterol regulatory element-binding protein 1 (Srebp1) (Fig. 2D).
Figure 2.
Effects of antagomiR-122 injection in mice. Panels show liver miR-122 expression assessed by Northern blotting (A), expression of other hepatic miRNAs involved in lipoprotein metabolism (B), serum cholesterol (C), gene expression (D), and hepatic proteome profile (E). Two proteomics methods were used for quantitation: a label-free method based on spectral counting and a 10-plex experiment using TMT labeling. Proteins that were returned as differentially expressed by both techniques are highlighted (for details see Supplementary Tables 1 and 2). Acc1, acetyl-CoA carboxylase; Aldo, aldolase; CP2AC, cytochrome P450 2A12; Fasn, fatty acid synthase; GRN, granulins; Hmgcr, HMG-CoA reductase; Ldlr, LDL receptor; RL23A, 60S ribosomal protein L23a; RL37A, 60S ribosomal protein L37a; RS16, 40S ribosomal protein S16; RS18, 40S ribosomal protein S18; Scd1, stearoyl-CoA desaturase-1.
At the protein level, we analyzed consequences of antagomiR-122 treatment using both a label-free and a TMT labeling approach (Fig. 2E and Supplementary Tables 1 and 2). Eleven proteins were returned as differentially expressed in both data sets and included proteins with an apparent connection to lipid metabolism, i.e., carnitine O-palmitoyltransferase 1 (CPT1A), pro-LDL receptor–related protein 1 (LRP1), and histidine triad nucleotide-binding protein 1 (HINT1). Few are predicted miR-122 targets (Supplementary Table 3), and the proteomics changes were not accompanied by corresponding changes in gene expression (Supplementary Fig. 3A). However, we observed a modest but significant downregulation of the GTPase Rab27a (Supplementary Fig. 3B), a key regulator of exosome release (24).
Effect of Statin Therapy on miR-122 Levels
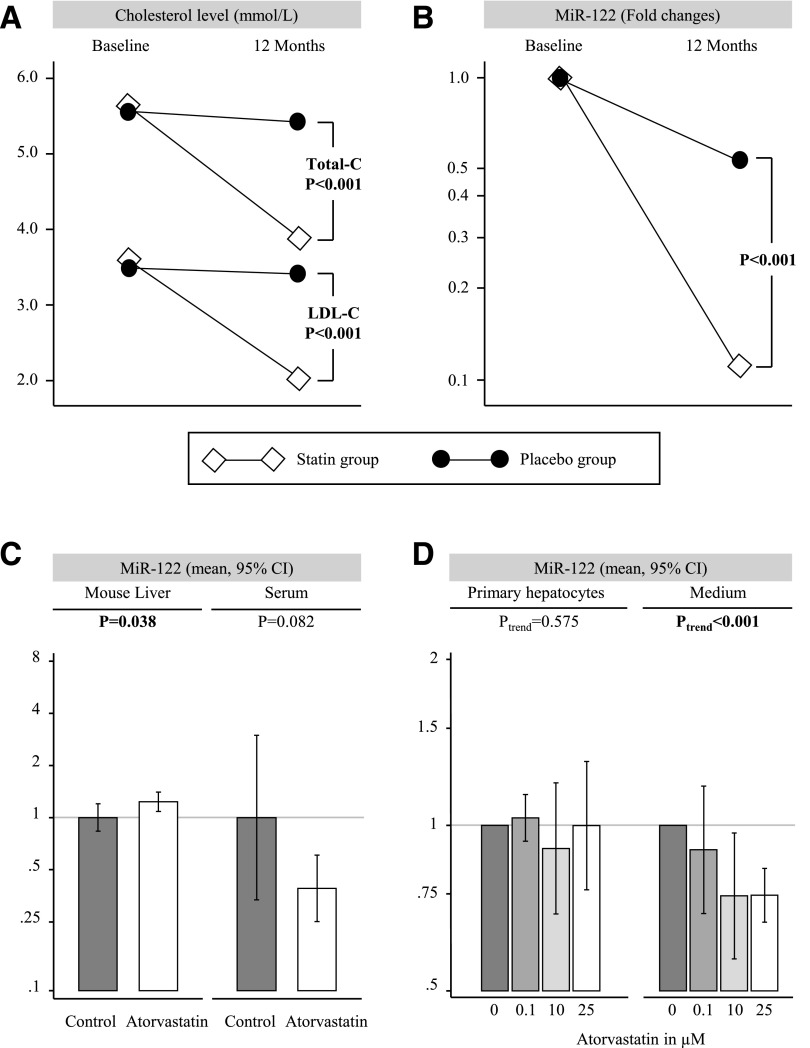
We next assessed the effect of statin treatment on miR-122 levels. In the placebo-controlled clinical HACVD-ASCOT trial, 12-month atorvastatin treatment led not only to the expected reduction in total cholesterol and LDL cholesterol but also to a marked reduction in serum miR-122 levels (all P < 0.001) (Fig. 3A). In contrast, other miRNAs quantified in the same samples remained unchanged (data not shown).
Figure 3.
Effects of atorvastatin treatment on total cholesterol (Total-C), LDL cholesterol (LDL-C) (A), and serum miR-122 in ASCOT participants (B), serum miR-122 in mice (C), and miR-122 secretion from primary hepatocytes (D).
In mice, atorvastatin treatment had only modest effects on hepatic miR-122 expression (+23%; P = 0.038) but reduced serum miR-122 levels (−61%; P = 0.082) (Fig. 3B). Short-term treatment with statins in mice did not result in a reduction of total cholesterol levels (data not shown), which is in agreement with previous reports (25). Similarly, in murine primary hepatocytes, increasing doses of atorvastatin did not affect cellular miR-122 levels (Ptrend = 0.575) but markedly reduced miR-122 in the culture medium (Ptrend < 0.001) (Fig. 3C). Thus, the inhibitory effect of atorvastatin on circulating miR-122 is independent of lipid levels and hepatic miR-122 expression.
Association of miR-122 With Development of New-Onset Metabolic Syndrome and T2D
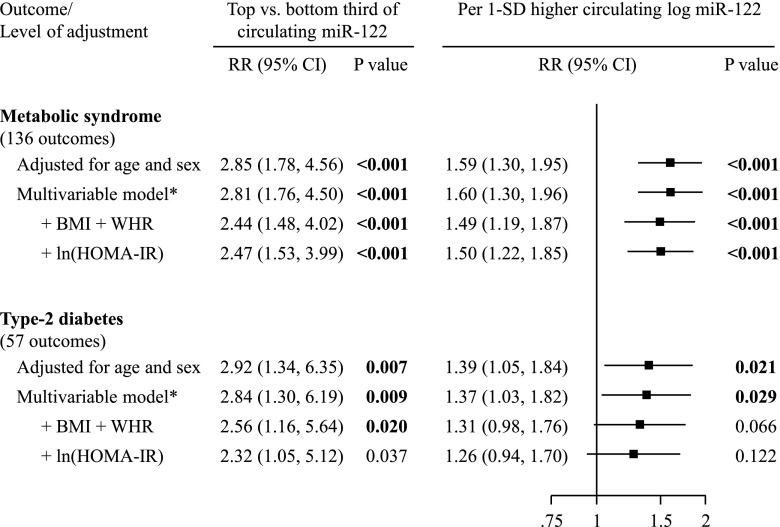
Among participants of the Bruneck free of preexisting disease at baseline, we recorded new onset of 136 events of metabolic syndrome and 57 events of T2D. Age- and sex-adjusted RRs comparing top versus bottom third of miR-122 levels were 2.85 (1.78–4.56; P < 0.001) for metabolic syndrome and 2.92 (1.34–6.35; P = 0.007) for T2D (Fig. 4). Age- and sex-adjusted RRs per one-SD higher log miR-122 were 1.59 (1.30–1.95; P < 0.001) for metabolic syndrome and 1.39 (1.05–1.84; P = 0.021) for T2D. RRs were virtually identical when further adjusted for socioeconomic status, smoking, physical activity, and alcohol consumption: 1.60 (1.30–1.96; P < 0.001) for metabolic syndrome and 1.37 (1.03–1.82; P = 0.029). RRs were somewhat attenuated upon further adjustment for BMI and waist-to-hip ratio or for ln HOMA-IR. Results were broadly similar for women and men, and in subgroups according to statin intake and clinical categories of adiposity (Supplementary Fig. 4). miR-122 was not significantly associated with new-onset CVD events in the overall study (RR 1.10 [0.90–1.33]; P = 0.350), although subgroup analyses indicated a possibly stronger and significant association in participants aged <60 years compared with participants aged ≥60 years (P for interaction = 0.006) (Supplementary Fig. 5).
Figure 4.
Association of miR-122 with new-onset metabolic syndrome and T2D in the Bruneck Study. Multivariable model (*): age, sex, socioeconomic status, smoking, physical activity, and alcohol consumption. WHR, waist-to-hip ratio.
Discussion
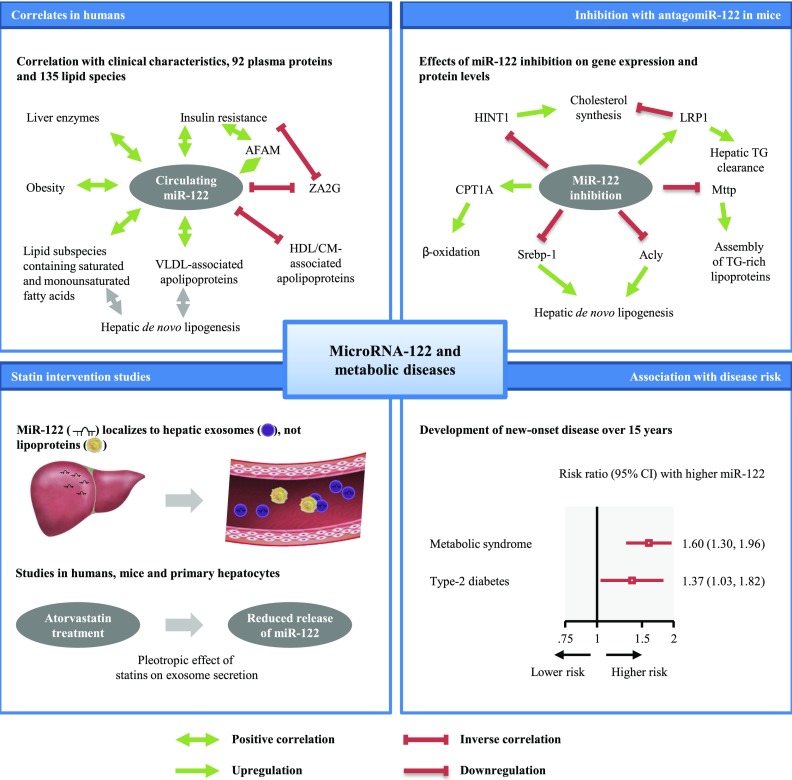
In the current study, we use a multidimensional “omics” approach in a population-based study to identify metabolic signatures associated with miR-122. We report a number of important and entirely novel results based on miRNA measurements in >2,000 human blood samples (from the 1995 and 2000 evaluations in the Bruneck Study plus ASCOT) combined with experimental follow-up to provide a mechanistic context as summarized in Fig. 5.
Figure 5.
Summary of the key findings. AFAM, afamin; CM, chylomicrons; TG, triglycerides; ZA2G, zinc-α-2-glycoprotein.
First, circulating miR-122 levels are elevated in people with prevalent metabolic syndrome or T2D and correlate strongly with lipid subspecies containing saturated and monounsaturated fatty acids. In a prospective setting, elevated serum levels of miR-122 antedate the manifestation of metabolic syndrome and T2D, but not CVD. Second, serum levels of miR-122 positively correlate with major lipids (triglycerides, LDL cholesterol, and HDL cholesterol) in the general community and substantially decline with cholesterol-lowering statin therapy (atorvastatin 10 mg). We further corroborate this observation by in vitro and in vivo experiments demonstrating a reduction of miR-122 in the supernatant of atorvastatin-treated murine hepatocytes and in serum of atorvastatin-treated wild-type mice and confirmed miR-122 effects on enzymes involved in lipid metabolism in the liver. Overall, we provide strong evidence for circulating miR-122 being a marker of hepatic lipid metabolism.
System-Wide Relations of Circulating miR-122
miR-122, which is primarily expressed in the liver, has been suggested to regulate the expression of various genes associated with cholesterol and fatty acid metabolism (2). In mice, inhibition of miR-122 led to markedly lower plasma cholesterol levels, halted hepatic lipid synthesis, and enhanced hepatic fatty acid oxidation (4,5). Two studies in nonhuman primates reported similar reductions in cholesterol (6,7).
In line with these reports, our study showed that inhibition of miR-122 in mice using antagomiR-122 led to a downregulated expression of genes implicated in lipid metabolism (Fig. 2D), such as Acly, Mttp, and Srebp-1 (26). This is further corroborated by the notion that miR-122 knockout mice express less Mttp, an essential enzyme that regulates the assembly of lipoproteins (27,28). Furthermore, our proteomics analysis of liver extracts from antagomiR-122–treated mice (Fig. 2E) revealed increases in CPT1A, a rate-limiting enzyme of fatty acid oxidation, that was not observed at the gene expression level (Fig. 2D, CPT1), and LRP1, which plays key roles in cholesterol homeostasis (29,30) and the removal of atherogenic lipoproteins, including VLDL (31). The proteomics analysis also identified a decrease in HINT1, which, based on data from Hint-1–deficient mice, is expected to contribute to reductions of total and esterified cholesterol (32).
These data provide mechanistic underpinning for our observation in the Bruneck Study that circulating miR-122 levels are positively associated with lipid subspecies that can be produced by hepatic de novo lipogenesis (Fig. 1A). We and others have previously shown that these lipid subspecies, comprised of saturated and monounsaturated fatty acids, are associated with a higher risk of CVD (9) and T2D (33). We also identified strong positive correlations with apolipoproteins found on VLDL (apoB100, apoC2, apoC3, and apoE) and inverse correlations with plasma apoD (present mainly in HDL) and apoA4 (a major component of HDL and chylomicrons) (Fig. 1B). Moreover, our comprehensive assessment of plasma proteins returned a positive correlation with afamin, which was previously linked to prevalent and new-onset metabolic syndrome and all its components (34), a positive correlation with complement factor H, a protein that binds malondialdehyde epitopes and protects from oxidative stress (35), and an inverse correlation with zinc-α-2-glycoprotein, an adipokine that leads to lipid degradation and higher insulin sensitivity in adipocytes (36,37).
Circulating miR-122 as Novel Biomarker
In the current study, we show, for the first time, that baseline levels of miR-122 are associated with the development of metabolic syndrome and with T2D (Fig. 4). Notably, associations did not vary by degree of adiposity, a strong determinant of cardiometabolic risk (38,39). Statin treatment decreased both lipoprotein and miR-122 release from the liver. Since miR-122 is either absent from lipoproteins, including VLDL and HDL (Supplementary Table 4), or only present at very low levels, i.e., in LDL (40), the pronounced effect of statins on circulating miR-122 levels cannot be explained by effects on plasma lipoproteins. Instead, it is probably caused by reduced secretion of liver exosomes (Fig. 5), in which miR-122 has been localized in abundance (41,42). Circulating miR-122 is undetectable in exosome-depleted serum (42). By inhibiting cholesterol synthesis, statins also modulate protein prenylation (43). This posttranslational modification promotes the membrane localization of proteins, in particular of Rab27 proteins that control the different steps of exosome secretion (24,44). Statins may reduce circulating miR-122 levels by inhibiting the prenylation of Rab proteins and hepatic exosome secretion. The latter might constitute a novel part of the beneficial pleiotropic effects of statins. This is further corroborated by our findings in mice, demonstrating a reduction of circulating miR-122 levels after short-term treatment with atorvastatin without concomitant reduction in total cholesterol levels and reduced gene expression of Rab27a in response to antagomiR-122 treatment, the key effector GTPase that drives the exosome release process (Supplementary Fig. 3B).
Strengths and Limitations
The prospective Bruneck cohort is extremely well-characterized with a 100% follow-up and high-quality ascertainment of clinical end points and potential confounders. miR-122 was measured in serum and plasma. We incorporated repeat measurements of miR-122 in our statistical models, which is particularly important given that the within-person variability of miR-122 was high (correlation coefficient over 5 years: 0.24). In contrast to platelet-related miRNAs, which are reduced in patients with diabetes (21,45), miR-122 levels showed positive associations with metabolic syndrome and T2D and were highly correlated in serum and plasma. Expression data in the human liver would be a more direct measure, but, clearly, this is not feasible in population studies. The Bruneck Study was conducted in an entirely Caucasian population. Thus, it remains to be determined whether these findings equally apply to other ethnicities.
Conclusions
High circulating miR-122 levels correlate with complex lipids containing saturated and monounsaturated fatty acids that can be derived from hepatic de novo lipogenesis and an adverse metabolic profile. Inhibition of HMG-CoA reductase by atorvastatin reduces miR-122 release. Circulating miR-122 levels are associated with future development of metabolic syndrome and T2D in the general population.
Supplementary Material
Article Information
Funding. P.W. was supported by an Erwin Schrödinger Fellowship sponsored by the Austrian Science Fund (J-3679-B13). D.K. was supported by a scholarship sponsored by the “Studienstiftung des Deutschen Volkes.” A.Z. and M.M. are Intermediate and Senior Fellows of the British Heart Foundation (FS/13/18/30207 and FS/13/2/29892, respectively). C.M.R. was supported by a grant from the American Heart Association (15SDG23000025). A.D.H. was supported by the National Institute for Health Research (NIHR) University College London Hospitals Biomedical Research Centre. This study was supported by a British Heart Foundation special project grant (SP/12/5/29574), Fondation Leducq (MIRVAD, 13 CVD 02), Diabetes UK (12/0004530), and an excellence initiative (Competence Centers for Excellent Technologies [COMET]) of the Austrian Research Promotion Agency FFG (“Research Center of Excellence in Vascular Ageing – Tyrol, VASCage”; K-Project number 843536). The Bruneck Study was supported by the Pustertaler Verein zur Prävention von Herz- und Hirngefaesserkrankungen, Gesundheitsbezirk Bruneck, and the Assessorat für Gesundheit, Province of Bolzano, Italy. This work was also supported by JDRF and the NIHR Biomedical Research Center based at Guy’s and St Thomas’ National Health Service Foundation Trust and King’s College London in partnership with King’s College Hospital and grants from the National Institutes of Health (R01HL107953 and 1F31AG043318 to L.G. and R01HL106063 to C.F.-H.) and the Foundation Leducq Transatlantic Network of Excellence in Cardiovascular Research (to C.F.-H. and M.M.).
Duality of Interest. The HACVD-ASCOT study was supported by an investigational grant from Pfizer, New York, NY, with additional support provided by Servier Research Group, Paris, France, and Leo Laboratories, Copenhagen, Denmark. The Medical University of Innsbruck and King’s College London filed patent applications on miRNA biomarkers. No other potential conflicts of interest relevant to this article were reported.
Author Contributions. P.W. and S.K. researched, analyzed, and interpreted data and wrote the manuscript. P.Sk. researched and interpreted data and reviewed and edited the manuscript. A.R.M. researched data and reviewed and edited the manuscript. X.Y. researched and analyzed data and reviewed and edited the manuscript. D.K., A.Z., T.B., M.W., C.M.R., L.G., N.R., E.B., A.D.H., P.Sa., C.F.-H., H.T., and J.W. researched data and reviewed and edited the manuscript. M.M. researched and interpreted data and wrote the manuscript. M.M. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db16-0731/-/DC1.
References
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fernández-Hernando C, Ramírez CM, Goedeke L, Suárez Y. MicroRNAs in metabolic disease. Arterioscler Thromb Vasc Biol 2013;33:178–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Willeit P, Skroblin P, Kiechl S, Fernández-Hernando C, Mayr M. Liver microRNAs: potential mediators and biomarkers for metabolic and cardiovascular disease? Eur Heart J 2016;37:3260–3266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esau C, Davis S, Murray SF, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab 2006;3:87–98 [DOI] [PubMed] [Google Scholar]
- 5.Krützfeldt J, Rajewsky N, Braich R, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 2005;438:685–689 [DOI] [PubMed] [Google Scholar]
- 6.Elmén J, Lindow M, Schütz S, et al. LNA-mediated microRNA silencing in non-human primates. Nature 2008;452:896–899 [DOI] [PubMed] [Google Scholar]
- 7.Lanford RE, Hildebrandt-Eriksen ES, Petri A, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science 2010;327:198–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao W, He HW, Wang ZM, et al. Plasma levels of lipometabolism-related miR-122 and miR-370 are increased in patients with hyperlipidemia and associated with coronary artery disease. Lipids Health Dis 2012;11:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stegemann C, Pechlaner R, Willeit P, et al. Lipidomics profiling and risk of cardiovascular disease in the prospective population-based Bruneck study. Circulation 2014;129:1821–1831 [DOI] [PubMed] [Google Scholar]
- 10.Willeit P, Kiechl S, Kronenberg F, et al. Discrimination and net reclassification of cardiovascular risk with lipoprotein(a): prospective 15-year outcomes in the Bruneck Study. J Am Coll Cardiol 2014;64:851–860 [DOI] [PubMed] [Google Scholar]
- 11.Zampetaki A, Willeit P, Tilling L, et al. Prospective study on circulating microRNAs and risk of myocardial infarction. J Am Coll Cardiol 2012;60:290–299 [DOI] [PubMed] [Google Scholar]
- 12.Kiechl S, Wittmann J, Giaccari A, et al. Blockade of receptor activator of nuclear factor-κB (RANKL) signaling improves hepatic insulin resistance and prevents development of diabetes mellitus. Nat Med 2013;19:358–363 [DOI] [PubMed] [Google Scholar]
- 13.Willeit P, Willeit J, Brandstätter A, et al. Cellular aging reflected by leukocyte telomere length predicts advanced atherosclerosis and cardiovascular disease risk. Arterioscler Thromb Vasc Biol 2010;30:1649–1656 [DOI] [PubMed] [Google Scholar]
- 14.Baecke JA, Burema J, Frijters JE. A short questionnaire for the measurement of habitual physical activity in epidemiological studies. Am J Clin Nutr 1982;36:936–942 [DOI] [PubMed] [Google Scholar]
- 15.Bonora E, Kiechl S, Willeit J, et al.; Bruneck Study . Metabolic syndrome: epidemiology and more extensive phenotypic description. Cross-sectional data from the Bruneck Study. Int J Obes Relat Metab Disord 2003;27:1283–1289 [DOI] [PubMed] [Google Scholar]
- 16.Sever PS, Dahlöf B, Poulter NR, et al.; ASCOT investigators . Prevention of coronary and stroke events with atorvastatin in hypertensive patients who have average or lower-than-average cholesterol concentrations, in the Anglo-Scandinavian Cardiac Outcomes Trial--Lipid Lowering Arm (ASCOT-LLA): a multicentre randomised controlled trial. Lancet 2003;361:1149–1158 [DOI] [PubMed] [Google Scholar]
- 17.Poulter NR, Wedel H, Dahlöf B, et al.; ASCOT Investigators . Role of blood pressure and other variables in the differential cardiovascular event rates noted in the Anglo-Scandinavian Cardiac Outcomes Trial-Blood Pressure Lowering Arm (ASCOT-BPLA). Lancet 2005;366:907–913 [DOI] [PubMed] [Google Scholar]
- 18.Stanton A, Fitzgerald D, Hughes A, et al.; Anglo-Scandinavian Cardiac Outcomes Trial . An intensive phenotyping study to enable the future examination of genetic influences on hypertension-associated cardiovascular disease. J Hum Hypertens 2001;15(Suppl. 1):S13–S18 [DOI] [PubMed] [Google Scholar]
- 19.Zampetaki A, Attia R, Mayr U, et al. Role of miR-195 in aortic aneurysmal disease. Circ Res 2014;115:857–866 [DOI] [PubMed] [Google Scholar]
- 20.Werner T, Sweetman G, Savitski MF, Mathieson T, Bantscheff M, Savitski MM. Ion coalescence of neutron encoded TMT 10-plex reporter ions. Anal Chem 2014;86:3594–3601 [DOI] [PubMed] [Google Scholar]
- 21.Willeit P, Zampetaki A, Dudek K, et al. Circulating microRNAs as novel biomarkers for platelet activation. Circ Res 2013;112:595–600 [DOI] [PubMed] [Google Scholar]
- 22.D’Agostino RB, Lee ML, Belanger AJ, Cupples LA, Anderson K, Kannel WB. Relation of pooled logistic regression to time dependent Cox regression analysis: the Framingham Heart Study. Stat Med 1990;9:1501–1515 [DOI] [PubMed] [Google Scholar]
- 23.Bertoia ML, Bertrand KA, Sawyer SJ, Rimm EB, Mukamal KJ. Reproducibility of circulating microRNAs in stored plasma samples. PLoS One 2015;10:e0136665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ostrowski M, Carmo NB, Krumeich S, et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol 2010;12:19–30; sup pp 1–13 [DOI] [PubMed] [Google Scholar]
- 25.Schonewille M, de Boer JF, Mele L, et al. Statins increase hepatic cholesterol synthesis and stimulate fecal cholesterol elimination in mice. J Lipid Res 2016;57:1455–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Menon B, Sinden J, Franzo-Romain M, Botta RB, Menon KMJ. Regulation of LH receptor mRNA binding protein by miR-122 in rat ovaries. Endocrinology 2013;154:4826–4834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wen J, Friedman JR. miR-122 regulates hepatic lipid metabolism and tumor suppression. J Clin Invest 2012;122:2773–2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsai WC, Hsu SD, Hsu CS, et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest 2012;122:2884–2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.El Asmar Z, Terrand J, Jenty M, et al. Convergent signaling pathways controlled by LRP1 (receptor-related protein 1) cytoplasmic and extracellular domains limit cellular cholesterol accumulation. J Biol Chem 2016;291:5116–5127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Basford JE, Wancata L, Hofmann SM, et al. Hepatic deficiency of low density lipoprotein receptor-related protein-1 reduces high density lipoprotein secretion and plasma levels in mice. J Biol Chem 2011;286:13079–13087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Medh JD, Bowen SL, Fry GL, et al. Hepatic triglyceride lipase promotes low density lipoprotein receptor-mediated catabolism of very low density lipoproteins in vitro. J Lipid Res 1999;40:1263–1275 [PubMed] [Google Scholar]
- 32.Beyoğlu D, Krausz KW, Martin J, et al. Disruption of tumor suppressor gene Hint1 leads to remodeling of the lipid metabolic phenotype of mouse liver. J Lipid Res 2014;55:2309–2319 [DOI] [PMC free article] [PubMed]
- 33.Rhee EP, Cheng S, Larson MG, et al. Lipid profiling identifies a triacylglycerol signature of insulin resistance and improves diabetes prediction in humans. J Clin Invest 2011;121:1402–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kronenberg F, Kollerits B, Kiechl S, et al. Plasma concentrations of afamin are associated with the prevalence and development of metabolic syndrome. Circ Cardiovasc Genet 2014;7:822–829 [DOI] [PubMed] [Google Scholar]
- 35.Weismann D, Hartvigsen K, Lauer N, et al. Complement factor H binds malondialdehyde epitopes and protects from oxidative stress. Nature 2011;478:76–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garrido-Sánchez L, García-Fuentes E, Fernández-García D, et al. Zinc-alpha 2-glycoprotein gene expression in adipose tissue is related with insulin resistance and lipolytic genes in morbidly obese patients. PLoS One 2012;7:e33264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang M, Liu R, Li S, et al. Zinc-α2-glycoprotein is associated with insulin resistance in humans and is regulated by hyperglycemia, hyperinsulinemia, or liraglutide administration: cross-sectional and interventional studies in normal subjects, insulin-resistant subjects, and subjects with newly diagnosed diabetes. Diabetes Care 2013;36:1074–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Würtz P, Wang Q, Kangas AJ, et al. Metabolic signatures of adiposity in young adults: Mendelian randomization analysis and effects of weight change. PLoS Med 2014;11:e1001765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fall T, Hägg S, Mägi R, et al.; European Network for Genetic and Genomic Epidemiology (ENGAGE) consortium . The role of adiposity in cardiometabolic traits: a Mendelian randomization analysis. PLoS Med 2013;10:e1001474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol 2011;13:423–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang X, Yuan T, Tschannen M, et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics 2013;14:319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gallo A, Tandon M, Alevizos I, Illei GG. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One 2012;7:e30679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang CY, Liu PY, Liao JK. Pleiotropic effects of statin therapy: molecular mechanisms and clinical results. Trends Mol Med 2008;14:37–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jaé N, McEwan DG, Manavski Y, Boon RA, Dimmeler S. Rab7a and Rab27b control secretion of endothelial microRNA through extracellular vesicles. FEBS Lett 2015;589:3182–3188 [DOI] [PubMed] [Google Scholar]
- 45.Elgheznawy A, Shi L, Hu J, et al. Dicer cleavage by calpain determines platelet microRNA levels and function in diabetes. Circ Res 2015;117:157–165 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.