Abstract
The expression of an antisense RNA revealed that an Mn-catalase was required in Thermus thermophilus for aerobic but not for anaerobic growth. The antisense system is based on the constitutive expression of a “bicistronic” transcript consisting of the kanamycin resistance gene mRNA followed by the antisense RNA against the selected target.
Catalases are detoxifying enzymes (14, 15) that belong to three classes (2, 24). Two of them contain a heme group. The monofunctional heme-catalases are by far the most widely distributed, being present in bacteria, archaea, and eukaryotes, whereas bifunctional heme-catalases, which accept peroxides as well, have not yet been found in higher eukaryotes.
Members of the third group of catalases contain Mn instead of heme in their active sites (23). They are more restricted in distribution but frequently appear among thermophiles and hyperthermophiles. An Mn-catalase was identified in Thermus thermophilus, further purified, and crystallized, and its three-dimensional structure was resolved (1). An apparently monocistronic single gene (AJ551423) seems responsible for this activity in the genome of T. thermophilus HB27 (AE017221) (8). However, the presence in the genome of ∼39% of genes encoding proteins without known homologues does not exclude the putative existence of new enzymes with catalase activity.
Attempts to isolate a cat insertion mutant.
To check the relevance of the Mn-catalase, an insertional mutagenesis approach was assayed (12). After a series of unsuccessful attempts to get knockout mutants in the cat gene, we decided to assay an antisense strategy similar to that described previously for mesophilic bacteria (10, 16, 19).
Antisense expression vector.
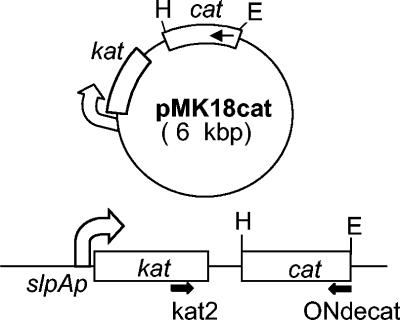
Plasmid pMK18 contains replicative origins for Escherichia coli and T. thermophilus and a marker (kat) for thermostable selection on kanamycin (3). The kat gene is expressed from a promoter (slpAf) which is strongly expressed in T. thermophilus and moderately in E. coli (6) and does not include any transcription terminator (12). In the derived plasmid pMK18r, transcriptions from slpAf pass through its multicloning site (Fig. 1). Therefore, any gene cloned in the appropriate orientation in pMK18r could be cotranscribed in T. thermophilus as a fusion to the kat gene.
FIG. 1.
The pMK18cat plasmid. The 0.9-kb DNA fragment codifying the Mn-catalase from T. thermophilus HB8 was cloned into pMK18r to obtain plasmid pMK18cat. The relative positions of the ONdecat and kat2 primers used for RT-PCR are indicated. H, HindIII; E, EcoRI.
We cloned (18) the coding region of the Mn-catalase gene (cat) into pMK18r (Fig. 1). The construct (pMK18cat) did not show any toxicity for E. coli on Luria-Bertani medium (13) after transformation (7) (data not shown). By contrast, very few colonies were obtained when plasmid pMK18cat was used to transform T. thermophilus HB27 compared to the results obtained with pMK18r. To have reliable data on the toxicity of pMK18cat and its putative relationship with aerobic respiration, we compared the toxicity characteristics of pMK18cat in aerobic and anaerobic conditions in the facultative anaerobes T. thermophilus HB8 and HB27::nar, two strains that can grow with nitrate as an electron acceptor (17).
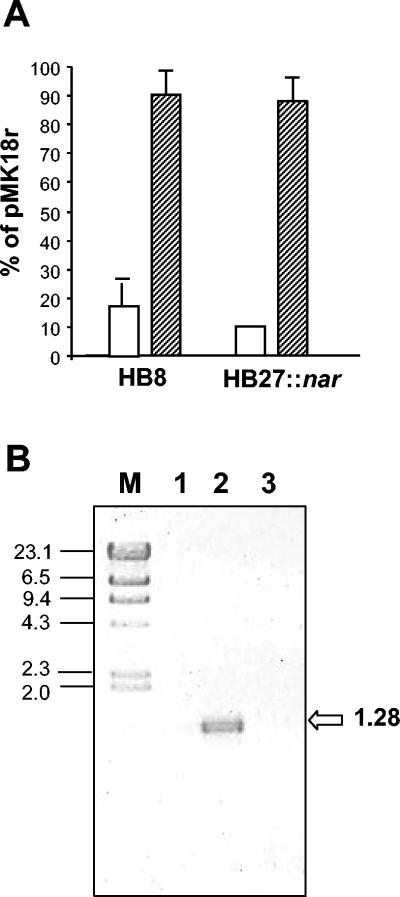
The strains were transformed with either pMK18cat or pMK18r, and the cells were plated on TB medium (5) with KNO3 (40 mM) and kanamycin (30 mg/liter) and further incubated under either aerobic or anaerobic conditions (Anaerocult; Merck). Figure 2A demonstrates that both plasmids rendered similar numbers of transformants in a given strain when the cells were grown anaerobically. By contrast, a significantly lower number of transformants were obtained with pMK18cat when the cells were grown aerobically. Thus, pMK18cat toxicity and aerobic growth were linked.
FIG. 2.
Deleterious effect of pMK18cat. (A) Cell cultures of T. thermophilus HB8 or HB27::nar were transformed with identical amounts of pMK18r and pMK18cat, and the selection plates were grown aerobically (white bars) or anaerobically (stripped bars). The percentages of transformants with pMK18cat with respect to those obtained with pMK18r are represented for each condition and strain. (B) Oligonucleotides ONdecat and kat2 were used for RT-PCR on purified RNA isolated from anaerobically grown cells of T. thermophilus HB8 transformed with plasmid pMK18cat (Lane 2) or pMK18r (Lane 3). Negative controls were carried out with the sample of lane 2 in the absence of the RT step (Lane 1). The sizes (in kilobase pairs) of HindIII-digested lambda phage DNA fragments (M) and of the RT-PCR-amplified fragment are indicated.
Expression of an antisense RNA.
The results described above suggested that an antisense cat-RNA was being synthesized from plasmid pMK18cat, thus limiting the expression of the Mn-catalase and making the cells more sensitive to oxygen. To confirm this hypothesis, total RNA from the anaerobically grown cells was isolated from the plates, and reverse-transcription PCR (RT-PCR) assays were subsequently developed with primers ONdecat (9) for RT and kat2 (5′GAAACTTCTGGAATCGC3′) for the subsequent PCR (Fig. 1). As shown in Fig. 2B, we detected the expected transcript (1,280 bp) on cells with pMK18cat (lane 2) but not on those containing pMK18r (lane 3). Therefore, a “bicistronic” transcript consisting of the sense kat-mRNA fused to an antisense cat-RNA was being expressed from pMK18cat.
Sensitivity to H2O2.
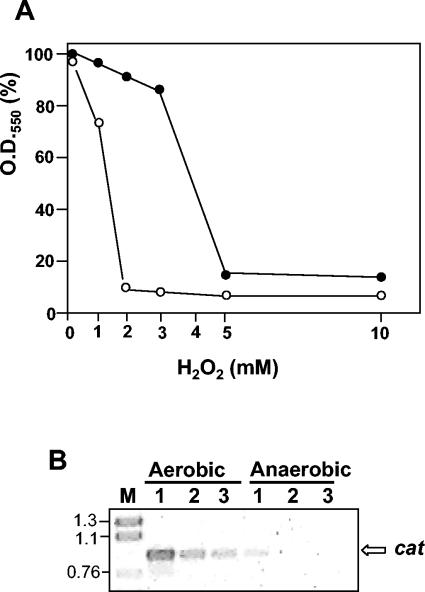
Identical amounts of anaerobically grown cells with either pMK18r or pMK18cat were inoculated into 12-ml culture tubes containing 3 ml of TB medium and were further incubated at 70°C in the presence of different concentrations of H2O2. As shown in Fig. 3, the cells with pMK18cat were more sensitive to H2O2; their growth was inhibited at a concentration (2 mM) at which those with pMK18r were almost unaffected. Therefore, the presence of the cat antisense RNA results in a decrease in the resistance to H2O2.
FIG. 3.
Role of Mn-catalase during aerobic growth. (A) Identical amounts of anaerobically grown cells of T. thermophilus HB27::nar transformed with plasmid pMK18r (black circles) or pMK18cat (white circles) were inoculated in 12-ml tubes containing 3 ml of TB medium in the presence of the indicated concentrations of H2O2. The percentage of the OD550 reached after 10 h of incubation at 70°C (microaerobic growth) with respect to untreated cultures compared to the H2O2 concentration is shown. (B) RT-PCR to detect the cat gene on total RNA from exponential cultures of T. thermophilus HB8 grown aerobically or after 7 h of anaerobic growth by nitrate respiration (anaerobic). The amounts of total RNA used were 4 μg (lane 1), 2 μg (lane 2), and 1 μg (lane 3). Lane M, size markers in kilobase pairs.
Effects on Cat expression.
To check whether the loss of resistance was actually related to the expression of the Cat protein, the catalase activity of untreated cultures of the above experiment was measured (9). Whereas the control strain showed around 0.04 units (in micromoles per minute at 60°C) per ml of culture at an optical density at 550 nm (OD550) of 1, the cells carrying pMK18cat presented less than half of this activity level (∼0.015 units/OD550). Thus, the presence of pMK18cat actually results in a reduction of the catalase activity of the cell.
Repression of cat transcription.
All our results supported the idea that the Mn-catalase was not necessary for anaerobic growth. To check whether its expression was subjected to genetic control by oxygen, total RNA was purified from two parallel cultures of T. thermophilus HB8, one of them grown aerobically in TB medium and the other subjected to a 7-h period of growth in TB medium with nitrate and without shaking. Semiquantitative RT-PCR with primers OHincat and ONdecat (9) revealed that the transcription of the cat gene was repressed under the latter, essentially anaerobic, conditions (Fig. 3B).
Concluding remarks.
The expression of antisense RNA is being used as one of the preferred technologies for the functional analysis of specific genes in eukaryotic systems (4). By contrast, such technology has started to be used in prokaryotes only recently (10, 16, 19) despite natural gene regulation by antisense RNA being well documented (22).
Plasmid pMK18r provides a simple way to check for the physiological relevance of any gene in T. thermophilus. For this, the coding region of the target gene has to be cloned in an inversely oriented position with respect to that of the kat selection marker, leading to the expression of a transcriptional fusion between the mRNA of the latter and the target antisense RNA. By this method we have shown the biological relevance of Mn-catalase during aerobic growth of T. thermophilus but also that it is dispensable for anaerobic growth. The existence of around ∼1/10 of the cells transformed with plasmid pMK18cat that still grew under aerobic conditions (Fig. 2A) is similar to the inhibition figures found for essential genes in other bacteria (11, 20, 21) and could be related to phenotypic expression differences among the transformed bacterial population. Whatever the reason for such leakiness, the absence of a growth bias between cells transformed with plasmids pMK18r and pMK18cat under conditions of anaerobic growth demonstrated a clear-cut relationship between aerobic growth and pMK18cat toxicity. Such a relationship was confirmed by the hypersensitivity to H2O2 shown by cells transformed with pMK18cat and their lower content in catalase activity (Fig. 3A). Moreover, semiquantitative RT-PCR revealed that the transcription of the cat gene was repressed during anaerobic growth by nitrate respiration (Fig. 3B).
In conclusion, our results show that the existence in T. thermophilus of alternative or redundant catalase activities among those open reading frames without homologues in the gene databanks is unlikely and open the field for the physiological analysis of essential genes in Thermus spp. through an antisense expression strategy.
Acknowledgments
The financial support of project BIO2001-1627 to J. Berenguer and projects MATINOES G5RD-CT-2002-00752 from EC and PPQ2002-011231 from Spanish CICYT to J. M. Guisán are acknowledged. A. Hidalgo and R. Moreno are recipients of fellowships from Gobierno Vascco and Biotools B & M, respectively. An Institutional Grant from Fundación Ramón Areces to the CBMSO is also acknowledged.
REFERENCES
- 1.Antonyuk, S. V., V. R. Melik-Adamyan, A. N. Popov, V. S. Lamzin, P. D. Hempstead, P. M. Harrison, P. J. Artymuk, and V. V. Barynin. 2000. Three-dimensional structure of the enzyme dimanganese catalase from Thermus thermophilus at 1 A resolution. Crystallogr. Rep. 45:105-116. [Google Scholar]
- 2.Chelikani, P., I. Fita, and P. C. Loewen. 2004. Diversity of structures and properties among catalases. Cell. Mol. Life Sci. 61:192-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Grado, M., P. Castán, and J. Berenguer. 1999. A high-transformation efficiency cloning vector for Thermus. Plasmid 42:241-245. [DOI] [PubMed] [Google Scholar]
- 4.Dorsett, Y., and T. Tuschl. 2004. siRNAs: applications in functional genomics and potential as therapeutics. Nat. Rev. Drug Discov. 3:18-29. [DOI] [PubMed] [Google Scholar]
- 5.Fernández-Herrero, L. A., M. A. Badet-Denisot, B. Badet, and J. Berenguer. 1995. GlmS of Thermus thermophilus HB8: an essential gene for cell wall synthesis identified immediately upstream of the S-layer gene. Mol. Microbiol. 17:1-12. [DOI] [PubMed] [Google Scholar]
- 6.Fernández-Herrero, L. A., G. Olabarría, and J. Berenguer. 1997. Surface proteins and a novel transcription factor regulate the expression of the S-layer gene in Thermus thermophilus HB8. Mol. Microbiol. 24:61-72. [DOI] [PubMed] [Google Scholar]
- 7.Hanahan, D. 1983. Studies on transformation of E. coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 8.Henne, A., H. Bruggemann, C. Raasch, A. Wiezer, T. Hartsch, H. Liesegang, A. Johann, T. Lienard, O. Gohl, R. Martinez-Arias, C. Jacobi, V. Starkuviene, S. Schlenczeck, S. Dencker, R. Huber, H. P. Klenk, W. Kramer, R. Merkl, G. Gottschalk, and H. J. Fritz. 2004. The genome sequence of the extreme thermophile Thermus thermophilus. Nat. Biotechnol. 22:547-553. [DOI] [PubMed] [Google Scholar]
- 9.Hidalgo, A., L. Betancor, R. Moreno, O. Zafra, F. Cava, R. Fernández-Lafuente, J. M. Guisán, and J. Berenguer. 2004. Thermus thermophilus as a cell factory for the production of a thermophilic Mn-catalase which fails to be synthesized in E. coli. Appl. Environ. Microbiol. 70:3839-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ji, Y., G. Woodnutt, M. Rosenberg, and M. K. Burnham. 2002. Identification of essential genes in Staphylococcus aureus using inducible antisense RNA. Methods Enzymol. 358:123-128. [DOI] [PubMed] [Google Scholar]
- 11.Ji, Y., D. Yin, B. Fox, D. J. Holmes, D. Payne, and M. Rosenberg. 2004. Validation of antibacterial mechanism of action using regulated antisense RNA expression in Staphylococcus aureus. FEMS Microbiol. Lett. 231:177-184. [DOI] [PubMed] [Google Scholar]
- 12.Lasa, I., J. R. Castón, L. A. Fernandez-Herrero, M. A. Pedro, and J. Berenguer. 1992. Insertional mutagenesis in the extreme thermophilic eubacteria Thermus thermophilus. Mol. Microbiol. 6:1555-1564. [DOI] [PubMed] [Google Scholar]
- 13.Lennox, E. X. 1955. Transduction of linked genetic characters of the host by bacteriophage P1. Virology 1:190-206. [DOI] [PubMed] [Google Scholar]
- 14.Li, J., X. Gao, M. Qian, and J. W. Eaton. 2004. Mitochondrial metabolism underlies hyperoxic cell damage. Free Radic. Biol. Med. 36:1460-1470. [DOI] [PubMed] [Google Scholar]
- 15.Loewen, P. C., M. G. Klotz, and D. J. Hassett. 2000. Catalase—an old enzyme that continues to surprise us. ASM News 66:76-82. [Google Scholar]
- 16.Parish, T., and N. G. Stoker. 1997. Development and use of a conditional antisense mutagenesis system in mycobacteria. FEMS Microbiol. Lett. 154:151-157. [DOI] [PubMed] [Google Scholar]
- 17.Ramírez-Arcos, S., L. A. Fernández-Herrero, I. Marín, and J. Berenguer. 1998. Anaerobic growth, a property horizontally transferred by an Hfr-like mechanism among extreme thermophiles. J. Bacteriol. 180:3137-3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook, J., E. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Sturino, J. M., and T. R. Klaenhammer. 2004. Antisense RNA targeting of primase interferes with bacteriophage replication in Streptococcus thermophilus. Appl. Environ. Microbiol. 70:1735-1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vickers, T. A., J. R. Wyatt, and S. M. Freier. 2000. Effects of RNA secondary structure on cellular antisense activity. Nucleic Acids Res. 28:1340-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vickers, T. A., S. Koo, C. F. Bennett, S. T. Crooke, N. M. Dean, and B. F. Baker. 2003. Efficient reduction of target RNAs by small interfering RNA and RNase H-dependent antisense agents. A comparative analysis. J. Biol. Chem. 278:7108-7118. [DOI] [PubMed] [Google Scholar]
- 22.Wagner, E. G., S. Altuvia, and P. Romby. 2002. Antisense RNAs in bacteria and their genetic elements. Adv. Genet. 46:361-398. [DOI] [PubMed] [Google Scholar]
- 23.Yoder, D. W., J. Hwang, and J. E. Penner-Hahn. 2000. Manganese catalases. Met. Ions Biol. Syst. 37:527-557. [PubMed] [Google Scholar]
- 24.Zamocky, M., and F. Koller. 1999. Understanding the structure and function of catalases: clues from molecular evolution and in vitro mutagenesis. Prog. Biophys. Mol. Biol. 72:19-66. [DOI] [PubMed] [Google Scholar]