Abstract
The group A Streptococcus (GAS) is an important pathogen that is responsible for a wide range of human diseases. Fibronectin binding proteins (FBPs) play an important role in promoting GAS adherence and invasion of host cells. The prtF2 gene encodes an FBP and is present in approximately 60% of GAS strains. In the present study we examined 51 prtF2-positive GAS strains isolated from the Northern Territory of Australia, and here we describe two genotypes of prtF2 which are mutually exclusive. The two genotypes have been identified previously as pfbp and fbaB. We show that these genotypes map to the same chromosomal location within the highly recombinatorial fibronectin-collagen-T antigen (FCT) locus, indicating that they arose from a common ancestor, and in this study these genotypes were designated the pfbp type and the fbaB type. Phylogenetic analysis of seven pfbp types, 14 fbaB types, and 11 prtF2-negative GAS strains by pulsed-field gel electrophoresis (PFGE) produced 32 distinct PFGE patterns. Interpretation of evolution based on the PFGE dendrogram by parsimony suggested that the pfbp type had a recent origin compared to the fbaB type. A comparison of multiple DNA sequences of the pfbp and fbaB types revealed a mosaic pattern for the amino-terminal region of the pfbp types. The fbaB type is generally conserved at the amino terminus but varies in the number of fibronectin binding repeats in the carboxy terminus. Our data also suggest that there is a possible association of the pfbp genotype with sof (84.2%), while the fbaB genotype was found in a majority of the GAS strains negative for sof (90.6%), indicating that these two prtF2 subtypes may be under different selective pressures.
One mechanism for adherence of Streptococcus pyogenes (group A streptococcus [GAS]) to host cells and tissues is mediated by the interaction with the host ligand, fibronectin. Strains of GAS encode several proteins that have the capacity to bind fibronectin (9, 10, 16, 18, 19, 31, 32, 36, 37). This in itself strongly suggests that the fibronectin binding protein (FBP)-fibronectin interaction may play an important role in the progression of GAS infection and disease. Whereas many different FBPs in GAS have been described, not all strains are genetically totipotent for each of these FBPs (12, 14, 24, 40). For example, the sfbI, sof, and prtF2 genes encoding FBPs are present in approximately 52, 44, and 60% of GAS strains isolated from the Northern Territory of Australia, respectively (12).
GAS is a human-specific pathogen and can cause a wide range of diseases, from benign mucosal and skin infections to life-threatening diseases and sequelae, such as acute poststreptococcal glomerulonephritis and rheumatic heart disease (11). Diversity in the repertoire of the genes encoding FBPs may have implications for GAS tissue tropism, persistence within the human host, and the spectrum of diseases that the strains can cause. For instance, Neeman et al. (29) have shown that there is an association between sfbI-positive GAS strains and persistence after antibiotic treatment. Likewise, an association between prtF2 and GAS invasive diseases has been observed (12, 37). SfbI, Sof, and PrtF2 are distinct proteins, and while sfbI and prtF2 are located in the same chromosomal location called the fibronectin-collagen-T antigen (FCT) locus (5), the sof gene is situated outside this locus.
PrtF2 was originally described by Jaffe et al. (18). Subsequently, Rocha and Fischetti (32) described another FBP designated PFBP. PrtF2 and PFBP have very high sequence identity and possess similar domains. More recently, Terao et al. (37) identified FbaB, an FBP from the M3 and M18 GAS serotypes. This protein and PFBP also have the same leader sequence and exhibit high sequence similarity in the C-proximal region of the protein, which contains the fibronectin binding domains. These observations raise the important question of the evolutionary relationship between the FBP genes.
In order to address this question and increase our understanding of the evolution of PrtF2, we selected 51 prtF2-positive and 11 prtF2-negative genotypically distinct GAS strains. Here we report characterization of the two distinct genotypes of PrtF2 by PCR and DNA sequence analysis and examination of the strains by pulsed-field gel electrophoresis (PFGE) to determine the evolutionary relationship of the prtf2 genotypes. The epidemiological and evolutionary implications of the data are discussed below.
MATERIALS AND METHODS
Bacterial strains.
Sixty-two GAS isolates belonging to distinct genotypes as judged by Vir typing (17) or emm sequence typing (1) were selected for this study. These strains were isolated from patients in the Northern Territory of Australia and have been described previously (12). In GAS, genomic diversity is predominantly due to recombination (13). Thus, GAS isolates from a defined geographical region, such as the Northern Territory, where the diversity of GAS strains and the disease burden are high, offer an opportunity to discern the lineage of a single locus in relation to the population structure.
Screening for genes encoding fibronectin binding proteins.
All GAS strains were screened for genes encoding FBPs, including prtF2, pfbp, fbaB, sfbI, sof, sfbX, fbp54, and fbaA. The prtF2, sfbI, sof, and fbp54 status of these strains has been described previously (12). However, prtF2 PCR performed with primers situated within the fibronectin binding repeat domains described by Delvecchio et al. (12) does not differentiate between the two genotypes of prtF2 (pfbp and fbaB). Therefore, two sets of PCR primers were designed and utilized in this study. The first amplification with primers VPrtf2-F and VPrtF2-R designed for the signal sequence and cell wall anchor region, respectively, distinguished between the two genotypes of prtF2 and confirmed the mutual exclusiveness of the two prtF2 genotypes, and the second PCR amplification with primers PFBP-F and ManR4 designed for the flanking region of the prtF2 open reading frame also distinguished between the two genotypes and confirmed the location of the genotypes in the chromosome. Primers SfbXF1 and SfbXR1 were used to screen for sfbX in all strains (19), and primers FbaA-F and FbaA-R were used to screen for fbaA in all strains (Table 1). PCRs were carried out in 50-μl (total volume) mixtures containing 2 μl of DNA template extracted with a QIAGEN DNeasy tissue kit, 10 mM Tris-HCl (pH 8.3), 10 mM KCl, 2 mM MgCl2, 50 pmol of each primer, each deoxynucleoside triphosphate at a concentration of 200 μM, and 1 U of Taq DNA polymerase.
TABLE 1.
Primers used for gene detection
| Primer | Sequence (5′ to 3′) | Gene detected | PCR conditions | Size of amplicons (bp) |
|---|---|---|---|---|
| VPrtF2-F | ATAGGATTGTCCGGAGTATCA | pfbp type | 30 cycles of 95°C for 60 s, 50°C for 60 s, and 72°C for 60 s | 3,332-3,322a |
| VPrtF2-R | TTATGTTGCTTCTCACCAG | fbaB type | 2,144-1,922a | |
| PFBP-F | GTACGTTAAGCGCTTGAAAAAG | pfbp type | 30 cycles of 95°C for 60 s, 66°C for 60 s, and 72°C for 60 s | 3,997-4,009a |
| ManR4 | CAACCCATTAGCAGCATCATTCCC | fbaB type | 2,825-2,603a | |
| SfbXF1b | GCAGTGATTCTAGGCTTAGCAAGCATA | sfbX | 30 cycles of 95°C for 60 s, 62°C for 60 s, and 72°C for 60 s | 1,941 |
| SfbXR1b | GTTTTGTCGGTGTTGCGACGTTTTT | |||
| FbaA-F | AGTCTACTACTCAGCCAGTTG | fbaA | 30 cycles of 95°C for 60 s, 50°C for 60 s, and 72°C for 60 s | 541 |
| FbaA-R | CTGTCTTGACAATGAGCGAT |
The amplicon sizes differ depending on the variation in the N-terminal and/or fibronectin binding domains.
The primer sequences used for amplification of sfbX were obtained from reference 19.
DNA sequence analysis.
Complete nucleotide sequences of seven pfbp-type and 11 fbaB-type genes were determined. PCR products obtained with primers PFBP-F and ManR4 (Table 1) were used as templates after the amplicons were purified with a QIAquick PCR purification kit (QIAGEN, Melbourne, Australia). The nucleotide sequences of both strands of DNA were determined by primer walking. DNA sequencing reactions were performed with an ABI Prism BigDye cycle sequencing kit (Applied-Biosystems, Melbourne, Australia), and the products were electrophoresed by using an ABI prism 377 DNA sequencer (Perkin-Elmer, Foster City, Calif.). Compilation and analysis of DNA sequence data were performed by using the Auto Assembler software (Perkin-Elmer). An amino acid analysis was performed by using programs accessed via the Australian National Genomic Information Services (www.angis.org.au). ClustalW (38) was used to produce multiple-sequence alignments of the pfbp and fbaB sequences determined in this study and previously determined sequences for these genes, including those of the M3 GAS strain SSI-1 (accession number AB084272) (37), the M5 GAS strain Manfredo (http://www.sanger.ac.uk), the M12 GAS strain A735 (accession number AF071083) (32), the M18 GAS strain MGAS8382 (accession number AE009964) (33), the M49 GAS strain 100076 (accession number U31980) (18), and the M49 GAS strain B737 (accession number AY049089) (5).
PFGE and phylogenetic analysis.
PFGE was carried out by using the following modification of the method described by Chatellier et al. (7). Briefly, single colonies of each GAS strain were used to inoculate 2 ml of Todd-Hewitt broth (Difco) supplemented with 1% yeast extract and grown overnight at 37°C. Cells were harvested by centrifugation, washed twice with TSE buffer (10 mM Tris-Cl, 1.0 M NaCl, 50 mM EDTA [pH 8.0]), and resuspended in 200 μl of Tris-EDTA buffer (pH 7.5). An equal volume of prewarmed 1.5% low-melting-point preparative-grade agarose (Bio-Rad, Richmond, Calif.) was mixed with each cell suspension, transferred into gel block molds, and allowed to solidify. The blocks were treated for 4 h at 37°C with 400 μl of lysis buffer (6 mM Tris-Cl [pH 7.6], 100 mM EDTA [pH 7.5], 1 M NaCl, 0.5% Brij 58, 0.2% deoxycholate, 0.5% sodium lauroyl sarcosine) containing freshly added lysozyme (1 mg/ml), mutanolysin (100 U/ml), and RNase (20 μg/ml). The lysis buffer was replaced with 300 μl of deproteination solution (1 μg of proteinase K per ml, 1% sodium lauroyl sarcosine, 500 mM EDTA [pH 8.5]), and the blocks were incubated overnight at 50°C. The blocks were prepared for restriction enzyme digestion by washing them three times in Tris-EDTA buffer (pH 7.5) for 30 min each time; the first wash solution contained 0.5 mM phenylmethylsulfonyl fluoride. The blocks were stored in 1 M EDTA (pH 8.5) at 4°C until they were used.
Prior to digestion with restriction enzyme, 2- to 3-mm slices were aseptically cut from the blocks, rinsed for 10 min with sterile distilled water, and equilibrated for 30 min in SmaI restriction buffer. The slices were then incubated overnight at 25°C in fresh restriction buffer containing 20 U of SmaI restriction enzyme (Roche). The digested DNA was resolved by PFGE by using a CHEF-DRTM electrophoresis cell (Bio-Rad, Sydney, Australia) and the following parameters: 0.5× Tris-borate-EDTA running buffer, 6 V/cm, and linearly ramped switch times of 2 to 40 s at 10°C for 23 h. λ DNA concatemers (New England Biolabs, Beverly, Mass.) were included as molecular size standards. The gel was then stained with ethidium bromide (1 μg/ml) for 30 min and visualized with UV illumination by using a GelDoc 1000 image analysis station (Bio-Rad, Sydney, Australia).
PFGE restriction fragment patterns were visually assessed by using the criteria of Tenover et al. (35) and were analyzed by using the GelCompar software (version 4.2; Applied Maths, Kortrijk, Belgium). Genetic similarities were compared by clustering methods (unweighted pair group method with arithmetic means) by using the Dice coefficient. A tolerance in the band positions of 0.2% was used for comparisons of fingerprint profiles. MacClade v. 3.08 (27) was used to assign genotypes to ancestral nodes of the PFGE dendrogram.
Nucleotide sequence accession numbers.
The nucleotide sequences of the 18 genes sequenced in this study have been deposited in the GenBank database under the following accession numbers: NS101, AY612216; NS1140, AY612217; NS125, AY612218; NS178, AY612219; NS179, AY612220; NS192, AY612221; NS195, AY612222; NS210, AY612223; NS235, AY612224; NS240, AY612225; NS265, AY612226; NS436, AY612227; NS506, AY612228; NS53, AY612229; NS564, AY612230; NS581, AY612231; NS730, AY612232; NS803, AY612233.
RESULTS
pfbp and fbaB are two distinct mutually exclusive genotypes of prtF2.
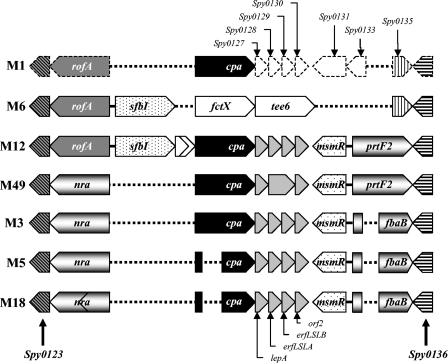
Analysis of GAS chromosomal DNA from 51 prtF2 GAS strains chosen for this study (Table 2) with PCR primers VPrtF2-F and VPrtF2-R revealed amplicons that were in two size classes, an approximately 1.9-kb class and an approximately 3.3-kb class (data not shown). None of these 51 strains yielded both amplicon sizes, suggesting that these classes are mutually exclusive. The prtF2 gene is known to reside within the FCT locus of GAS (5). PCR performed with primers PFBP-F and ManR4 at the known chromosomal position of the prtF2 open reading frame yielded amplicons that were approximately 2.6 and 4.0 kb long (data not shown), indicating that the chromosomal position of prtF2 is within the FCT locus in the 51 prtF2 GAS strains examined, adjacent to the flanking Spy0136 open reading frame (Fig. 1). To confirm that these amplicons are indeed located within the FCT locus, we sequenced a region upstream of the prtF2 open reading frame to confirm the presence of the msmR gene (data not shown) and the junction region between prtF2 and the downstream Spy0136 gene from a representative group of 18 prtF2-positive strains. The nucleotide sequence data generated were aligned with selected FCT junction sequences deposited in the GenBank and other databases. The prtF2-positive nucleotide sequences were highly conserved across this junction sequence (>93%), whereas the prtF2-negative nucleotide sequences (M1 and M6) exhibited significant homology only from nucleotide 207 of the junction sequence (data not shown).
TABLE 2.
Characteristics of GAS strains used in this study
| Isolate | Vir type | emm sequence type | prtF2 type | Other FBP genes
|
||||
|---|---|---|---|---|---|---|---|---|
| sfbI | sof | sfbX | fbp54 | fbaA | ||||
| NS8 | 25 | emm85 | pfbp | + | + | + | + | + |
| NS83 | 37.1 | stns554 | pfbp | + | + | + | + | + |
| NS192 | 3.2 | emm106 | pfbp | + | + | + | + | + |
| NS210 | 34 | emm22 | pfbp | + | + | + | + | + |
| NS226 | 14.1 | emm4.2 | pfbp | + | + | + | + | + |
| NS240 | 116 | st2904 | pfbp | + | + | + | + | + |
| NS730 | 2.2 | emm90 | pfbp | + | + | + | + | + |
| NS878 | 124 | emm76.1 | pfbp | + | + | + | + | + |
| NS1133 | 17.1 | emm101 | pfbp | − | − | − | + | + |
| NS1 | 23 | emm100 | fbaB | − | − | − | + | + |
| NS6 | 12.2 | emm123 | fbaB | − | − | − | + | + |
| NS13 | 24 | emm53 | fbaB | − | − | − | + | + |
| NS50 | 5 | st854 | fbaB | − | − | − | + | + |
| NS80 | 69 | emm70 | fbaB | − | − | − | + | + |
| NS88.2 | 17.4 | emm98.1 | fbaB | − | + | + | + | + |
| NS90 | 52 | stns90 | fbaB | + | − | − | + | − |
| NS101 | 33.1 | emm110 | fbaB | + | + | + | + | + |
| NS125 | 20.1 | emm95 | fbaB | − | − | − | + | + |
| NS178 | 26 | emm54 | fbaB | − | − | − | + | + |
| NS195 | 120 | emm19 | fbaB | + | − | − | + | − |
| NS205 | 20.2 | emm56 | fbaB | − | − | − | + | + |
| NS223 | 4 | emm91 | fbaB | − | − | − | + | + |
| NS225 | 115 | emm99 | fbaB | − | − | − | + | + |
| NS235 | 118 | emm24 | fbaB | − | − | − | + | + |
| NS476 | 39 | emm80 | fbaB | − | − | − | + | + |
| NS501 | 61 | emm14 | fbaB | + | − | − | + | + |
| NS1140 | 101 | emm57 | fbaB | − | − | − | + | − |
| NS20 | 3.4 | emm75.1 | − | + | + | + | + | + |
| NS25 | 18 | emm55 | − | + | − | − | + | − |
| NS204 | 121 | emm2 | − | + | + | + | + | + |
| NS216 | 122 | stns216 | − | − | − | − | + | − |
| NS344 | 127 | emm1 | − | − | − | − | + | + |
| NS931 | 57 | emm65 | − | + | − | − | + | + |
| NS14 | 96 | emm102 | pfbp | + | + | + | + | + |
| NS32 | 29.2 | emm101 | pfbp | − | − | − | + | + |
| NS179 | 7.2 | emm9.1 | pfbp | + | + | + | + | + |
| NS199 | 55 | emm112 | pfbp | + | + | + | + | + |
| NS236 | 111 | emm77 | pfbp | − | + | + | + | + |
| NS436 | 3.3 | emm11 | pfbp | − | + | + | + | + |
| NS564 | 119 | emm44/61 | pfbp | + | + | + | + | + |
| NS672 | 112 | emm28 | pfbp | + | + | + | + | + |
| NS1099 | 126 | emm101 | pfbp | − | − | − | + | + |
| NS1185 | 135 | emm80 | pfbp | − | + | + | + | + |
| NS50.1 | 12.1 | emm108 | fbaB | − | − | − | + | + |
| NS53 | 29.1 | emm71 | fbaB | − | − | − | + | + |
| NS180 | 110 | emm74 | fbaB | − | − | − | + | + |
| NS182 | 60 | emm109 | fbaB | − | + | + | + | + |
| NS265 | 11 | emm56 | fbaB | − | − | − | + | + |
| NS282 | 17.2 | st6030.1 | fbaB | − | − | − | + | + |
| NS506 | 134 | emm14 | fbaB | + | − | − | + | + |
| NS581 | 42 | emm42 | fbaB | − | − | − | + | + |
| NS803 | 131 | emm97.1 | fbaB | − | − | − | + | + |
| NS804 | 130 | emm97.1 | fbaB | + | − | − | + | + |
| NS836 | 46 | stek249 | fbaB | − | − | − | + | + |
| NS1030 | 77 | stck401 | fbaB | + | − | − | + | + |
| NS1033 | 3.22 | stns1033 | fbaB | + | − | − | + | + |
| NS1353 | 44 | stns90 | fbaB | + | + | + | + | + |
| NS351 | 22 | emm58 | − | + | + | + | + | + |
| NS1045 | 117 | emm60 | − | + | + | + | + | + |
| NS1096 | 32 | NDa | − | − | + | + | + | + |
| NS1122 | 65 | emm65 | − | + | + | + | + | + |
| NS1210 | 125 | emm75 | − | + | + | + | + | + |
ND, not determined.
FIG. 1.
Arrangement of the FCT region. The following strains were examined (accession numbers are indicated in parentheses): M1, strain SF370 (AE006482/3); M6, strain D471 (U01312, L10919, and AY049087); M12, strains A735 and A374 (AF447492, AY049088, and AF071083); M49, strains CS101/B737 (U49397 and AY049089) and 100076 (U31980); M3, strain MGAS315 (AE014138); M5, strain Manfredo (http://www.sanger.ac.uk/Projects/S_pyogenes); and M18, strain MGAS8382 (AE009963/4). The open reading frame designations are indicated above the M1 genome. The region is demarcated by two highly conserved open reading frames, Spy0123 and Spy0136 (striped arrows). With the exception of sfbI, which is used instead of prtF1, all other designations are those reported by Podbielski et al. (30) and Bessen and Kalia (5). The dotted lines represent gaps introduced to aid alignment. Open reading frames with no significant homology are not shaded. The chevron within nra of M18 indicates a stop codon caused by a single point mutation. Similarly, the single open reading frame in M49 spanning eftLSLA and eftLSLB is caused by a single point mutation that results in replacement of a stop codon (TAA) with a Glu (Q) codon (CAA).
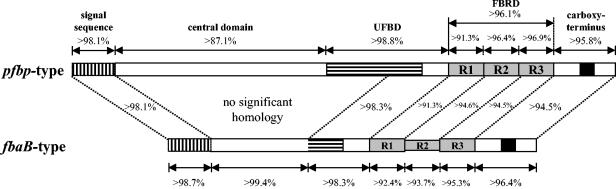
To further characterize these amplicons, we selected seven of the larger amplicons (NS179, NS192, NS210, NS240, NS436, NS564, and NS730) and 11 of the shorter amplicons (NS1140, NS101, NS125, NS178, NS195, NS235, NS265, NS506, NS53, NS581, and NS803) and subjected these amplicons to complete DNA sequence analysis. The prtF2 open reading frame in amplicons that were these two sizes exhibited high levels of nucleotide sequence identity within the N-terminal signal sequence (>98%), part of the upstream fibronectin binding domain (>98%), and the C-terminal anchor region (>94%). A moderate level of identity (>91%) within individual fibronectin binding repeats was also found; there were differences in the number of fibronectin binding repeats within this domain (Fig. 2). The central domain exhibited no significant homology for the two types of amplicons. The larger amplicons displayed a high degree of similarity to the previously published pfbp gene sequence (32), and the smaller amplicons displayed a high degree of similarity to the previously published fbaB gene sequence (37).
FIG. 2.
Comparison of pfbp-type and fbaB-type variants. The percentages indicate the levels of identity observed in pairwise comparisons of DNA sequences both within and between variants. The two mature proteins differ significantly in the central domain but are highly similar in all other domains. The central domain is highly conserved in the fbaB type, but there is considerable variation in the pfbp-type central domain. The pfbp-type variants always possess three nonidentical fibronectin binding repeats in the FBRD, while the fbaB-type variants have variable numbers of repeats due to duplication or deletion of repeat 2 (R2). The overall homology of the FBRD of the fbaB type could not be determined due to the resultant gaps in the alignment; however, the individual repeats are highly homologous both within and between variants.
The central domain of the 11 fbaB-like amplicons sequenced showed a high degree of similarity (>99%) (Fig. 2); however, this domain did not exhibit significant similarity with other sequences deposited in the GenBank database (data not shown). The fibronectin binding repeat domains (FBRD), as defined by Jaffe et al. (18), of the 11 fbaB-like amplicons varied and contained either two (NS265), three (NS53, NS101, NS125, NS178, NS195, NS235, NS506, NS581, and NS803), or four (NS1140) fibronectin binding repeats. By contrast, three intact fibronectin binding repeats were found in all seven pfbp-like amplicons sequenced (Fig. 2). The pfbp-like central domains displayed >87.1% similarity and contained distinct DNA cassettes flanked by highly conserved junction sequences, indicating that horizontal gene transfer may have generated the gene mosaic pattern found within this region (Fig. 3). The nucleotide alignment of the pfbp-like sequences is available at the GenBank website (alignment accession number ALIGN_000736; ftp://ftp.ebi.ac.uk/pub/database/embl/align/ALIGN_000736.dat) (data not shown).
FIG. 3.
Parts of the pfbp-type coding regions of the amino terminus have been horizontally transferred between different pfbp-type genes, producing mosaic alleles. Throughout the sequence highly conserved junction regions were identified (shaded) (>93.1% identity in pairwise comparisons) flanking distinct sections or cassettes, which appear to have been exchanged between alleles, resulting in a mosaic structure. Regions with identity to the pfbp-type gene of M12 strain A735 (prtF2.12) are underlined to illustrate the mobility of individual DNA cassettes.
Given (i) that the two mutually exclusive amplicons from the 51 prtF2-positive strains were amplified by using the same primers sets, (ii) the fact that each of these open reading frames mapped to the same position at the end of the FCT region, and (iii) that the downstream sequences across the junction of the 18 open reading frames analyzed and Spy0136 are virtually identical, we suggest that pfbp and fbaB represent two distinct genotypes of prtF2. These two genotypes were designated the pfbp type and the fbaB type in this study.
The pfbp lineage is more recent than the fbaB lineage.
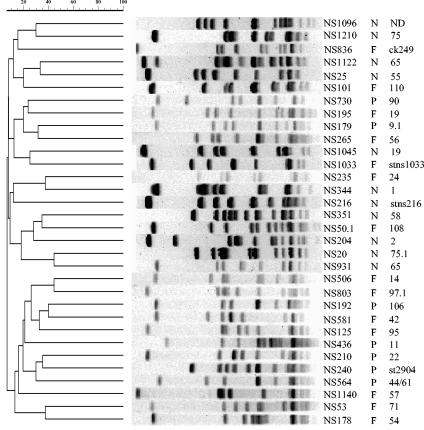
PFGE analysis of 32 strains, including 7 pfbp-positive, 14 fbaB-positive, and 11 prtF2-negative GAS strains, revealed that they produced 32 PFGE patterns, which is consistent with the conclusion that the strains are genetically distinct. Genetic similarity was determined by band-based clustering in the PFGE profiles by using 0.2% tolerance (Fig. 4). The strains were very diverse (<45% similarity) when diversity was measured by using the criteria of Tenover et al. (35). The isolates subjected to PFGE belonged to diverse emm sequence types, with the exception of strains NS1122 and NS931, which were emm sequence type 65. However, despite having the same emm sequence type and a prtF2-negative genotype, NS1122 and NS931 were found to contain distinct Vir types and to produce genetically unrelated PFGE fingerprint profiles, indicating that these strains are genetically distinct.
FIG. 4.
Dendrogram generated by the GelCompar software, showing the genetic relationships among 21 S. pyogenes isolates possessing prtF2 genotypes and 11 isolates not possessing the gene. The dendrogram was constructed by cluster analysis (unweighted pair group method with arithmetic means) of the PFGE patterns obtained after macrorestriction with the SmaI enzyme. A tolerance in the band positions of 0.2% was used for comparison of fingerprint profiles. PFGE fingerprint patterns are shown next to the corresponding branches of the dendrogram. The prtF2 genotypes and emm sequence types are indicated after the strain number. P, pfbp type; F, fbaB type; N, prtF2 not present. The scale indicates percent similarity.
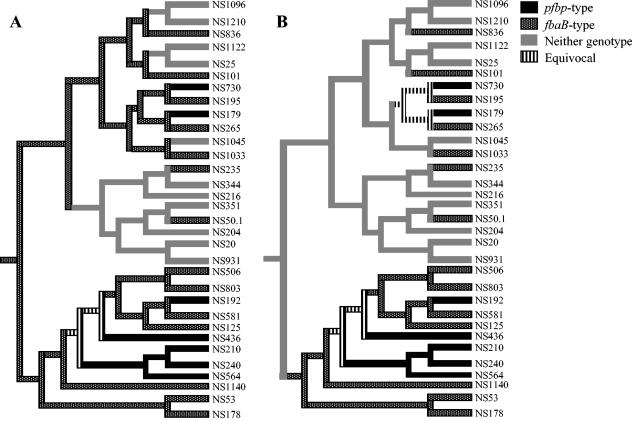
We examined the evolution of different prtF2 genotypes (pfbp type, fbaB type, and prtF2 negative) using MacClade (27) and the PFGE dendrogram (Fig. 4). The MacClade program placed the contemporary genotypes on the tips of the tree, and the evolution of different genotypes was inferred by minimizing the total number of changes over the tree (Fig. 5). When the gain or loss of a genotype was equally weighted, the pattern of evolution shown in Fig. 5A was recovered. This indicated that the fbaB genotype was the older genotype and that the pfbp genotype or the prtF2-negative genotype evolved from an fbaB-positive progenitor. Strains NS50.1 and NS235, which are on a branch occupied predominantly by prtF2-negative strains, may have horizontally acquired the fbaB gene; inter- and intraspecies horizontal acquisition is common in GAS (5, 20, 21, 34). The analysis also indicated that the fbaB type can be lost from the genome, as exemplified by NS1045 and by the ancestral NS1122/NS25 strain and the ancestral NS1096/NS1210 strain. However, in our limited data set, loss of the pfbp type was not observed. The pfbp genotype may have been acquired in an fbaB background, as exemplified by NS179, NS730, NS436, and NS192 and in the ancestor of strains NS564, NS240, and NS210. In this analysis gene gains and losses were equally weighted, and a similar pattern emerged when losses were weighted lower than gains (i.e., when gains were penalized relative to losses).
FIG. 5.
Evolution of prtF2 genotypes, inferred by parsimony by using MacClade v.3.08 (27), based on the PFGE dendrogram (Fig. 4). The trees indicate the evolution patterns obtained when the gains and losses of a genotype were equally weighted (A) and when gains were weighted higher than losses of a genotype (B).
As an alternative evolutionary scenario to examine the effect of positive selection pressure on the pfbp or fbaB genotype, a new MacClade analysis of the PFGE dendrogram was performed with gene gains weighted higher than losses (2:1 or higher). The resulting evolution pattern is shown in Fig. 5B. Here, the prtF2-negative genotype was present originally, the fbaB genotype generally evolved first, and then the pfbp genotype evolved in an fbaB-type genetic background. Potential horizontal gene acquisition of the fbaB type was observed in strains NS50.1, NS235, NS836, NS101, and NS1033, which are parts of branches dominated by prtF2-negative strains. Another difference between the two evolutionary patterns is the equivocal branch giving rise to strains NS265 and NS195 having the fbaB genotype and strains NS179 and NS730 having the pfbp genotype. This could indicate that NS179 and NS730 evolved from an fbaB ancestral genotype or vice versa.
We believe that the analysis shown in Fig. 5A (which is most commonly used) (27) represents the most reasonable working hypothesis for the evolution of different prtF2 genotypes. Nevertheless, as Fig. 5B reveals, a small change in the weight given to gene gains and losses has an impact on the analysis, so that Fig. 5A should be considered only a starting point. Furthermore, these analyses assume that the topology shown in the 0.2% tolerance PFGE dendrogram (Fig. 4) is correct. Although some branches may be misplaced by this analysis, the evolutionary scenario described above represents a reasonable working hypothesis to direct subsequent investigations. Overall, the analysis above suggests that the pfbp type had a more recent origin and arose only in an fbaB-type genetic background.
Distribution of fbaB-type, pfbp-type, and other GAS genes encoding fibronectin binding proteins.
GAS may possess genes encoding several FBPs, including fbaB, pfbp, sfbI, sof, sfbX, fbaA, and fbp54. We investigated the distribution of sfbI, sof, sfbX, fbaA, and fbp54 in the 32 fbaB-type and 19 pfbp-type GAS strains examined in this study. The fbaA and fbp54 genes were found in the majority of GAS strains possessing both the pfbp and fbaB genotypes examined. Thirteen of 19 (68.4%) pfbp-type GAS strains possessed sfbI, sof, and sfbX. In contrast, only 2 of 32 (6.3%) the fbaB-type GAS strains possessed sfbI, sof, and sfbX, while in 21 of 32 (65.6%) fbaB-type GAS strains these genes were not detected (Table 2). GAS strains have been defined as belonging to either class I or class II M types based on epitopes present within the conserved C repeat region of M proteins (2, 3). Class II GAS strains are usually sof positive, while class I GAS strains are generally sof negative (2, 4, 15). Examination of the sof status of the 51 prtF2-positive GAS strains examined in this study revealed that 16 of 19 (84.2%) of the pfbp-positive strains possessed sof. By contrast, only 4 of 32 (13.3%) the fbaB-positive strains contained sof, suggesting that the pfbp type is linked to class II GAS and the fbaB type is linked to class I GAS.
DISCUSSION
Strains causing GAS infections among the aboriginal communities of northern Australia have exhibited high diversity and turnover rates (6, 28). There is also little evidence of the emergence of a dominant clone, which has been a common cause of GAS invasive infections elsewhere in the world (8). Similarly, all strains used in this study were found to be genetically distinct, as indicated by emm typing, Vir typing, and PFGE profiles.
The gene encoding the FBP PrtF2 is situated in the highly recombinatorial FCT region in GAS (5). PrtF2 facilitates the binding of host fibronectin, enabling GAS to adhere to and be internalized by host epithelial cells (18, 25). Epidemiological evidence suggests that the presence of this gene may confer upon the pathogen a greater propensity to cause invasive diseases (12). In the present study we examined 51 prtF2-positive GAS strains isolated from patients in northern Australia, and here we describe two genotypes of prtF2, which are mutually exclusive. Both genotypes map to the same chromosomal location within the FCT locus, between genes encoding the potential gene regulator (msmR) and a hypothetical protein (Spy0136), and the two types are designated the pfbp type and fbaB type.
Other researchers have shown that expression of PrtF2 is influenced by the global negative regulator nra, which is located within the FCT region (30). Recent evidence obtained by Kreikemeyer et al. (25) and the fact nra is not always found in prtF2-positive strains (5) suggest that prtF2 expression is controlled by additional regulatory elements. Immediately upstream of prtF2 is a putative transcriptional regulator gene that has been termed msmR. MsmR belongs to the AraC family of regulators (5). Furthermore, the GAS genes controlled by this putative regulator have not been identified yet. In all strains studied to date, msmR is present upstream of prtF2. This may suggest that msmR affects prtF2 expression directly or regulates other genes that are required for PrtF2 function.
The major difference between the pfbp gene sequence (32) and the original prtF2 gene sequence (18) is the presence of two single-base-pair sequence changes in prtF2. These changes result in a frameshift mutation that results in the loss of the additional 105 amino acids at the N terminus of PFBP, which was not identified in PrtF2. However, the upstream prtF2 gene sequence (GenBank accession number U31980) contains genetic information coding for part of the 105-amino-acid sequence. None of the 18 prtF2 genes sequenced in this study contains a frameshift mutation similar to that reported by Jaffe et al. (18), suggesting that this mutation is infrequent. Clearly, however, the reported sequences of prtF2 (18), pfbp (32), and fbaB (37) are closely related. We therefore suggest that pfbp and fbaB represent two distinct genotypes of prtF2.
Multiple-alignment analysis of the pfbp-type DNA sequences revealed the presence of a mosaic structure within the pfbp-type central domain. This mosaic structure is not present in fbaB-type sequences, suggesting that only the pfbp-type gene sequences are capable of the intergenomic exchange required to produce such arrangements. Several surface-exposed GAS proteins have been shown to display a mosaic gene structure. These include the proteins encoded by emm (41), ska (22), and sfbI (39). It is believed that this type of genetic recombination plays a fundamental role in providing a mechanism for evading host immune responses. This is highlighted by the serotype-specific protection displayed by anti-M-protein antibodies (26). Such horizontal gene transfer may also produce mosaic gene structures that generate functional diversity of surface-exposed proteins. For instance, various M or M-like proteins have been found to bind fibrinogen, Fc domains of various human immunoglobulins, plasminogen, and/or proteins involved in the complement cascade (23).
In this study, we used software that utilized both the PFGE dendrogram and the prtF2 genotype status to infer the evolution of the different genotypes by minimizing the total number of changes over the tree. The results obtained with this methodology suggested that the smaller, more conserved fbaB genotype had a more ancient origin than the pfbp type. This pattern of inheritance was similar when gains or losses were equally weighted or when gains were weighted higher than losses. As the pfbp type appears only in an fbaB-type background, we hypothesize that the pfbp type arose from an insertional event within the fbaB type and that the genetic rearrangement seen in the central domain of this gene is the result of a subsequent process. This also suggests that acquisition of the pfbp-type gene may have some selectable advantage over acquisition of the fbaB-type gene. One possible advantage may be the result of extra functions gained by the protein through the additional pfbp-type central domain sequence. Additionally or alternatively, the mosaic structure of the pfbp-type central domain may allow immune evasion to occur.
Interestingly all pfbp-type genes sequenced contained three repeat regions in the FBRD, while fbaB-type gene sequences had two to four repeats. Jaffe et al. (18) localized the upstream fibronectin binding domain (UFBD) for PrtF2 between amino acids 679 and 783. Residues in the PrtF2 UFBD critical for fibronectin binding were mapped to positions 679 to 717. Homology of the FbaB UFBD region only begins at amino acid 740 of PrtF2. It is not known if the truncated UFBD of FbaB alters fibronectin binding. Perhaps three fibronectin binding repeats are a functional constraint for an intact UFBD in PFBP. Variations in the number of repeats in the FBRD of FbaB may be due loss of this constraint when the UFBD is truncated.
Thirteen of 19 (68.4%) pfbp-type GAS strains investigated in this study possessed sfbI, sof, and sfbX. In contrast, only 2 of 32 (6.3%) fbaB-type GAS strains possessed sfbI, sof, and sfbX. Interestingly, 21 of 32 (65.6%) fbaB-type GAS strains lacked these three genes. The biological implications of the finding that the pfbp type is generally associated with other FBP genes (sfbI, sof, and sfbX) and the biological implications of the finding that the fbaB type is generally associated with fewer FBP genes are not known. A previous study showed that there is linkage association between the sfbI and sof genes (14). In this study we found that there is a linkage association between pfbp and sof. Sixteen of 19 (84.2%) pfbp-type strains possessed sof. By contrast, only 4 of 32 (13.3%) fbaB-type strains contained sof. Class I GAS strains are generally sof negative and have been associated with acute rheumatic fever, unlike class II GAS strains, which are generally sof positive and are usually associated with skin-tropic infections (2, 4, 15). Therefore, our data suggest that the pfbp type may be linked to class II GAS strains and the fbaB type may be linked to class I strains.
GAS is a highly specific yet extremely versatile pathogen of humans. An increasing number of studies are revealing the extent of the genetic diversity displayed by this species and are implicating horizontal gene transfer and recombination as major mechanisms influencing the generation of this diversity. Additional work that leads to a better understanding of the evolution of GAS and, in particular, the emergence of highly virulent strains is warranted and may provide new strategies for predicting GAS disease trends and pathogenic mechanisms.
Acknowledgments
This work was supported by the National Health and Medical Research Council (NHMRC) of Australia.
We thank Kent Wu for his assistance with the analysis of the pulsed-field gel electrophoresis data with the GelCompar software.
REFERENCES
- 1.Beall, B., R. Facklam, and T. Thompson. 1996. Sequencing emm-specific PCR products for routine and accurate typing of group A streptococci. J. Clin. Microbiol. 34:953-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bessen, D. E., K. F. Jones, and V. A. Fischetti. 1989. Evidence for two distinct classes of streptococcal M protein and their relationship to rheumatic fever. J. Exp. Med. 169:269-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bessen, D. E., and V. A. Fischetti. 1990. Differentiation between two biologically distinct classes of group A streptococci by limited substitutions of amino acids within the shared region of M protein-like molecules. J. Exp. Med. 172:1757-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bessen, D. E., L. G. Veasy, H. R. Hill, N. H. Augustine, and V. A. Fischetti. 1995. Serologic evidence for a class I group A streptococcal infection among rheumatic fever patients. J. Infect. Dis. 172:1608-1611. [DOI] [PubMed] [Google Scholar]
- 5.Bessen, D. E., and A. Kalia. 2002. Genomic localization of a T serotype locus to a recombinatorial zone encoding extracellular matrix-binding proteins in Streptococcus pyogenes. Infect. Immun. 70:1159-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carapetis, J. R., A. M. Walker, M. Hibble, K. S. Sriprakash, and B. J. Currie. 1999. Clinical and epidemiological features of group A streptococcal bacteraemia in a region with hyperendemic superficial streptococcal infection. Epidemiol. Infect. 122:59-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chatellier, S., N. Ihendyane, R. G. Kansal, F. Khambaty, H. Basma, A. Norrby-Teglund, D. E. Low, A. McGeer, and M. Kotb. 2000. Genetic relatedness and superantigen expression in group A streptococcus serotype M1 isolates from patients with severe and nonsevere invasive diseases. Infect. Immun. 68:3523-3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cleary, P. P., E. L. Kaplan, J. P. Handley, A. Wlazlo, M. H. Kim, A. R. Hauser, and P. M. Schlievert. 1992. Clonal basis for resurgence of serious Streptococcus pyogenes disease in the 1980s. Lancet 339:518-521. [DOI] [PubMed] [Google Scholar]
- 9.Courtney, H. S., J. B. Dale, and D. I. Hasty. 1996. Differential effects of the streptococcal fibronectin-binding protein, FBP54, on adhesion of group A streptococci to human buccal cells and HEp-2 tissue culture cells. Infect. Immun. 64:2415-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Courtney, H. S., D. L. Hasty, Y. Li, H. C. Chiang, J. L. Thacker, and J. B. Dale. 1999. Serum opacity factor is a major fibronectin-binding protein and a virulence determinant of M type 2 Streptococcus pyogenes. Mol. Microbiol. 32:89-98. [DOI] [PubMed] [Google Scholar]
- 11.Cunningham, M. W. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delvecchio, A., B. J. Currie, J. D. McArthur, M. J. Walker, and K. S. Sriprakash. 2002. Streptococcus pyogenes prtFII, but not sfbI, sfbII or fbp54, is represented more frequently among invasive-disease isolates of tropical Australia. Epidemiol. Infect. 128:391-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feil, E. J., E. C. Holmes, D. E. Bessen, M. S. Chan, N. P. Day, M. C. Enright, R. Goldstein, D. W. Hood, A. Kalia, C. E. Moore, J. Zhou, and B. G. Spratt. 2001. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc. Natl. Acad. Sci. USA 98:182-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodfellow, A. M., M. Hibble, S. R. Talay, B. Kreikemeyer, B. J. Currie, K. S. Sriprakash, and G. S. Chhatwal. 2000. Distribution and antigenicity of fibronectin binding proteins (SfbI and SfbII) of Streptococcus pyogenes clinical isolates from the northern territory, Australia. J. Clin. Microbiol. 38:389-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haanes, E. J., D. G. Heath, and P. P. Cleary. 1992. Architecture of the vir regulons of group A streptococci parallels opacity factor phenotype and M protein class. J. Bacteriol. 174:4967-4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanski, E., and M. Caparon. 1992. Protein F, a fibronectin-binding protein, is an adhesin of the group A streptococcus Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 89:6172-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartas, J., M. Hibble, and K. S. Sriprakash. 1998. Simplification of a locus-specific DNA typing method (Vir typing) for Streptococcus pyogenes. J. Clin. Microbiol. 36:1428-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaffe, J., S. Natanson-Yaron, M. G. Caparon, and E. Hanski. 1996. Protein F2, a novel fibronectin-binding protein from Streptococcus pyogenes, possesses two binding domains. Mol. Microbiol. 21:373-384. [DOI] [PubMed] [Google Scholar]
- 19.Jeng, A., V. Sakota, Z. Li, V. Datta, B. Beall, and V. Nizet. 2003. Molecular genetic analysis of a group A Streptococcus operon encoding serum opacity factor and a novel fibronectin-binding protein, SfbX. J. Bacteriol. 185:1208-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalia, A., M. C. Enright, B. G. Spratt, and D. E. Bessen. 2001. Directional gene movement from human-pathogenic to commensal-like streptococci. Infect. Immun. 69:4858-4869. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Kalia, A., and D. E. Bessen. 2004. Natural selection and evolution of streptococcal virulence genes involved in tissue-specific adaptations. J. Bacteriol. 186:110-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kapur, V., S. Kanjilal, M. R. Hamrick, L. L. Li, T. S. Whittam, S. A. Sawyer, and J. M. Musser. 1995. Molecular population genetic analysis of the streptokinase gene of Streptococcus pyogenes: mosaic alleles generated by recombination. Mol. Microbiol. 16:509-519. [DOI] [PubMed] [Google Scholar]
- 23.Kehoe, M. A., V. Kapur, A. M. Whatmore, and J. M. Musser. 1996. Horizontal gene transfer among group A streptococci: implications for pathogenesis and epidemiology. Trends Microbiol. 4:436-443. [DOI] [PubMed] [Google Scholar]
- 24.Kreikemeyer, B., S. Beckert, A. Braun-Kiewnick, and A. Podbielski. 2002. Group A streptococcal RofA-type global regulators exhibit a strain-specific genomic presence and regulation pattern. Microbiology 148:1501-1511. [DOI] [PubMed] [Google Scholar]
- 25.Kreikemeyer, B., S. Oehmcke, M. Nakata, R. Hoffrogge, and A. Podbielski. 2004. Streptococcus pyogenes fibronectin-binding protein F2: expression profile, binding characteristics, and impact on eukaryotic cell interactions. J. Biol. Chem. 279:15850-15859. [DOI] [PubMed] [Google Scholar]
- 26.Lancefield, R. C. 1962. Current knowledge of type-specific M antigens of group A streptococci. J. Immunol. 89:307-313. [PubMed] [Google Scholar]
- 27.Maddison, W. P., and D. R. Maddison. 1992. MacClade: analysis of phylogeny and character evolution (v. 3.08). Sinauer, Sunderland, Mass. [DOI] [PubMed]
- 28.McDonald, M., B. J. Currie, and J. R. Carapetis. 2004. Acute rheumatic fever: a chink in the chain that links the heart to the throat? Lancet Infect. Dis. 4:240-245. [DOI] [PubMed] [Google Scholar]
- 29.Neeman, R., N. Keller, A. Barzilai, Z. Korenman, and S. Sela. 1998. Prevalence of internalisation-associated gene, prtF1, among persisting group-A streptococcus strains isolated from asymptomatic carriers. Lancet 352:1974-1977. [DOI] [PubMed] [Google Scholar]
- 30.Podbielski, A., M. Woischnik, B. A. Leonard, and K. H. Schmidt. 1999. Characterization of nra, a global negative regulator gene in group A streptococci. Mol. Microbiol. 31:1051-1064. [DOI] [PubMed] [Google Scholar]
- 31.Rakonjac, J. V., J. C. Robbins, and V. A. Fischetti. 1995. DNA sequence of the serum opacity factor of group A streptococci: identification of a fibronectin-binding repeat domain. Infect. Immun. 63:622-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rocha, C. L., and V. A. Fischetti. 1999. Identification and characterization of a novel fibronectin-binding protein on the surface of group A streptococci. Infect. Immun. 67:2720-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smoot, J. C., K. D. Barbian, J. J. Van Gompel, L. M. Smoot, M. S. Chaussee, G. L. Sylva, D. E. Sturdevant, S. M. Ricklefs, S. F. Porcella, L. D. Parkins, S. B. Beres, D. S. Campbell, T. M. Smith, Q. Zhang, V. Kapur, J. A. Daly, L. G. Veasy, and J. M. Musser. 2002. Genome sequence and comparative microarray analysis of serotype M18 group A Streptococcus strains associated with acute rheumatic fever outbreaks. Proc. Natl. Acad. Sci. USA 99:4668-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sriprakash, K. S., and J. Hartas. 1996. Lateral genetic transfers between group A and G streptococci for M-like genes are ongoing. Microb. Pathog. 20:275-285. [DOI] [PubMed] [Google Scholar]
- 35.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Terao, Y., S. Kawabata, E. Kunitomo, J. Murakami, I. Nakagawa, and S. Hamada. 2001. Fba, a novel fibronectin-binding protein from Streptococcus pyogenes, promotes bacterial entry into epithelial cells, and the fba gene is positively transcribed under the Mga regulator. Mol. Microbiol. 42:75-86. [DOI] [PubMed] [Google Scholar]
- 37.Terao, Y., S. Kawabata, M. Nakata, I. Nakagawa, and S. Hamada. 2002. Molecular characterization of a novel fibronectin-binding protein of Streptococcus pyogenes strains isolated from toxic shock-like syndrome patients. J. Biol. Chem. 277:47428-47435. [DOI] [PubMed] [Google Scholar]
- 38.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Towers, R. J., P. K. Fagan, S. R. Talay, B. J. Currie, K. S. Sriprakash, M. J. Walker, and G. S. Chhatwal. 2003. Evolution of sfbI encoding streptococcal fibronectin-binding protein I: horizontal genetic transfer and gene mosaic structure. J. Clin. Microbiol. 41:5398-5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vlaminckx, B. J., E. M. Mascini, J. Schellekens, L. M. Schouls, A. Paauw, A. C. Fluit, R. Novak, J. Verhoef, and F. J. Schmitz. 2003. Site-specific manifestations of invasive group A streptococcal disease: type distribution and corresponding patterns of virulence determinants. J. Clin. Microbiol. 41:4941-4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whatmore, A. M., and M. A. Kehoe. 1994. Horizontal gene transfer in the evolution of group A streptococcal emm-like genes: gene mosaics and variation in Vir regulons. Mol. Microbiol. 11:363-374. [DOI] [PubMed] [Google Scholar]